Abstract
Dynamic regulation of cell shape underlies many developmental and immune functions. Cortical remodeling is achieved under the central control of Rho GTPase pathways that modulate an exquisite balance in the dynamic assembly and disassembly of the cytoskeleton and focal adhesions. Macroautophagy (autophagy), associated with bulk cytoplasmic remodeling through lysosomal degradation, has clearly defined roles in cell survival and death. Moreover, it is becoming apparent that proteins, organelles, and pathogens can be targeted for autophagic clearance by selective mechanisms, although the extent and roles of such degradation are unclear. Here we report a conserved role for autophagy specifically in the cortical remodeling of Drosophila blood cells (hemocytes) and mouse macrophages. Continuous autophagy was required for integrin-mediated hemocyte spreading and Rho1-induced cell protrusions. Consequently, hemocytes disrupted for autophagy were impaired in their recruitment to epidermal wounds. Cell spreading required ref(2)P, the Drosophila p62 multiadaptor, implicating selective autophagy as a novel mechanism for modulating cortical dynamics. These results illuminate a specific and conserved role for autophagy as a regulatory mechanism for cortical remodeling, with implications for immune cell function.
Keywords: selective autophagy, cell spreading, Drosophila, hemocyte, macrophage
Blood cells are subject to high demands for environmentally responsive cell shape changes for both developmental and immune functions that involve cell spreading, migration, and phagocytosis. Drosophila blood cells, called hemocytes, are an evolutionary equivalent of mammalian macrophages in terms of ontogeny, gene expression, and immune roles in pathogen clearance. Hemocytes that arise in the embryo persist into larval stages in free circulation with known surveillance functions and have been studied for in vivo requirements for cell morphology and migration (1–3).
A theme in the remodeling of cell shape involves the reorganization of cortical cytoskeleton and membrane. Members of the Rho GTPase family control the organization and stability of filamentous actin (F-actin) in cell protrusion and membrane dynamics (4). Cell protrusion formation and spreading also are associated with integrin–matrix interactions for surface attachment and focal adhesion function (5). Importantly, both of these examples require multipronged, dynamically regulated cellular pathways for proper cortical remodeling; for instance, Rho GTPase pathway components are subject to regulation via mechanisms that change protein activity through membrane trafficking (6), protein complex formation (7), ubiquitination-mediated protein stability (8), or reversible protein phosphorylation (9).
The process of autophagy, or “self-eating” through cytoplasmic turnover in the lysosome, is a mechanism of cellular recycling and remodeling involved in development, homeostasis, and disease. Autophagy is best understood as a nonselective response to starvation in which the recycling of bulk cytoplasm serves as a pro-survival or pro-death mechanism (10). However, recent work indicates that specific protein aggregates, organelles, and pathogens can be selectively targeted for autophagic degradation. The p62 multiadaptor binds both to ubiquitinated proteins and to Atg8/LC3, a protein central to autophagosome formation, and thus is proposed to act as a receptor for selective autophagic clearance (11–13). Consistent with this, a loss of function of ref(2)P, the Drosophila homolog of p62, was found to result in accumulation of ubiquitinated proteins and disruption of neuronal function (14). However, the extent of roles and identity of targets for selective autophagy are largely unknown.
We identified Atg1 in an RNAi screen for kinase functions required for cell shape change in a Drosophila hemocyte-derived cell line. Previous studies independently discovered the Atg1 Ser/Thr kinase with conserved roles in axonal outgrowth (15–17) and as an essential component for autophagosome formation (18, 19). Here we explore the apparent dual role for Atg1 in the regulation of morphogenesis and autophagy, and evaluate the significance of autophagy in hemocyte cortical behavior and in vivo functions. We show that basal autophagy is not essential for larval hemocyte survival, but is required to promote hemocyte cell spreading and extension of Rho1-induced protrusions. Importantly, autophagy is required for blood cell recruitment to larval wound sites and for cell spreading of mouse macrophages, suggesting conserved roles for autophagy in controlling blood cell shape and function. Our work points to p62-selective autophagy as an additional mechanism regulating cortical dynamics.
Results
Atg1 Is Required Cell-Autonomously for Hemocyte Cell Spreading.
Hemocytes dissected out of Drosophila larvae displayed a cell-spreading response visualized by GFP in live cells (Fig. 1 A and E; 93% cell spread). Starting as round cells, hemocytes flattened over 30 min, while extending spiky radial protrusions containing both bundled F-actin and microtubules (Fig. 1B). In contrast, hemocytes from Atg1Δ3D mutant larvae (18) remained round (Fig. 1C and E; 10% cell spread) and appeared to lack F-actin protrusions (Fig. 1D). To further test the requirement for Atg1 function in hemocytes, we used the Pxn-GAL4 (hemocytes) or Cg-GAL4 drivers (hemocytes and fat body). We found that hemocyte-directed expression of a wild-type Atg1 cDNA could rescue the effects of the Atg1Δ3D mutation on cell spreading (Fig. S1A), whereas the defect in cell spreading was similarly observed with hemocyte-targeted Atg1 RNAi (20) (Fig. S1 B and C). Taken together, these findings indicate a specific, cell-autonomous requirement for Atg1 function for hemocyte spreading.
Fig. 1.
Autophagy is required for hemocyte spreading. (A) Control hemocytes spread with radial protrusions; GFP. (B) Protrusions rich in F-actin and microtubules. (C and D) Atg1Δ3D/ Atg1Δ3D hemocytes did not spread or extend protrusions. (A and C) Cg-GAL4 UAS-GFP. (B and D) F-actin (magenta) and microtubules (green). (E) Percentage of round or spread hemocytes; Cg-GAL4 for RNAi. (Scale bar: 5 μm.)
Autophagy Is Required for Initiation and Maintenance of Cell Spreading.
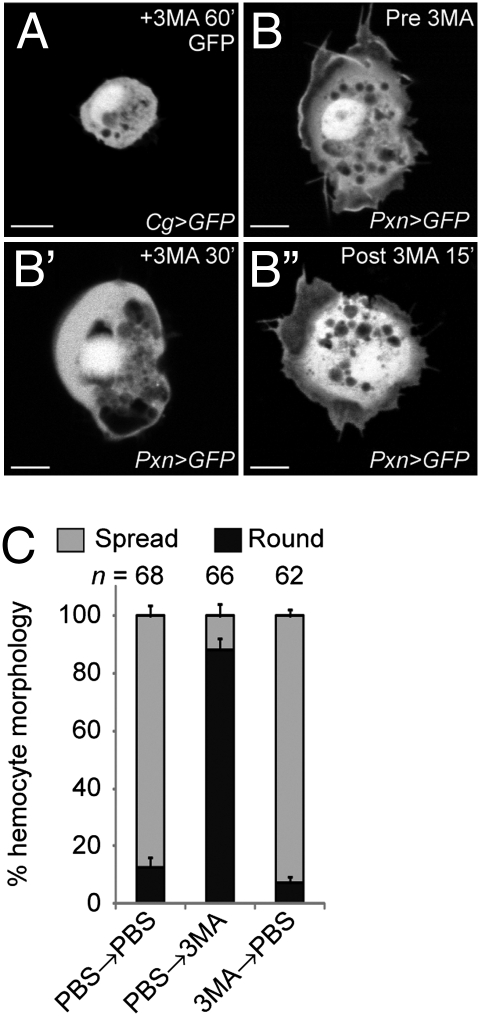
If the role for Atg1 in cell spreading reflects a role for autophagy in cellular morphogenesis, then blocking autophagy in hemocytes by other means also should result in round cells that fail to spread. To test this, we used 3-methyladenine (3MA), an inhibitor of autophagy (21). Wild-type hemocytes treated with 3MA did not spread (2% cell spread), similar to Atg1 mutant hemocytes (Figs. 1E and 2A). In addition, hemocyte-targeted RNAi (22) depletion of other known autophagy-related genes (Atg4, Atg6, Atg7, Atg8a and Atg9) that function directly in autophagosome formation (23) abolished the ability of hemocytes to spread and extend F-actin protrusions (5–8% cell spread; Fig. 1E and Fig. S1 D and E). Thus, the Atg1 function required for hemocyte spreading reflects a general requirement for autophagy in cortical remodeling.
Fig. 2.
Autophagy is required for initiation and maintenance of cell spreading. (A) Hemocytes did not spread when blocked for autophagy with addition of 3MA for 1 h; Cg-GAL4 UAS-GFP. (B–B′′) Example of a single hemocyte imaged over time; Pxn-GAL4 UAS-GFP. Hemocyte spread in PBS (B), retracted protrusions and rounded after 30 min +3MA (B′), and respread 15 min after washout (B′′). (C) All hemocytes were identified as round or spread (mean ± SEM), following 30 min in PBS or +3MA and a 30-min wash with replacement medium, as shown. (Scale bar: 5 μm.)
We used 3MA to temporally distinguish whether autophagy is required for the initiation or maintenance of hemocyte spreading. wild-type hemocytes remained spread when washed with PBS (Fig. 2C), but failed to remain spread once treated with 3MA, instead displaying retracted cell protrusions and a more rounded shape (Fig. 2B′ and C; 12% cells spread). The autophagy dependence was reversible; with hemocytes spreading after 3MA was removed (Fig. 2B′′; 93% cell spread). These findings indicate that continuous autophagy is required for initiation and maintenance of hemocyte spreading.
Autophagy Is Blocked in Atg1Δ3D Hemocytes.
If autophagy is required for hemocyte spreading, then we would expect autophagosomes to be normally present in wild-type but absent in Atg1Δ3D mutant hemocytes. Using transmission electron microscopy (TEM), we identified double-membrane–bound structures typical of autophagosomes in wild-type cells (1.8 per cell; Fig. 3A and Fig. S2 A and B). In contrast, the Atg1Δ3D mutant hemocytes were mostly devoid of double-membrane–bound structures (0.5/cell; Fig. 3A and Fig. S2A). Conversely, blockage of autophagy is predicted to result in an accumulation of proteins normally targeted for autophagic degradation. Atg1Δ3D hemocytes exhibited an increased number of puncta containing Ref(2)P (5.6 vs. 2.4 puncta/cell) (14, 24), the Drosophila homolog of p62 (11–13) (Fig. 3 B and C). The lack of double-membrane structures and the accumulation of Ref(2)P demonstrate that Atg1Δ3D disrupts autophagosome formation and autophagic clearance in hemocytes.
Fig. 3.
Autophagy is blocked in Atg1Δ3D mutant hemocytes. Autophagosome formation and autophagy function are disrupted in Atg1Δ3D mutant hemocytes. (A) Number of double membrane-bound structures per cell in TEM micrographs, 12 hemocytes each condition (bar, mean ± SEM). (B) Number of Ref(2)P puncta per hemocyte (mean ± SEM). (C) Accumulation of Ref(2)P (green, arrowheads), indicating blockage of autophagic clearance in Atg1Δ3D mutant hemocytes. F-actin is shown in magenta. (Scale bar: 5 μm.)
Disruption of Autophagy Has No Effect on Larval Hemocyte Number, Adhesion, or Survival.
Cell morphology is responsive to changes in cell homeostasis, such as cell survival, proliferation, and adhesion. There was no significant difference in the total cell counts, suggesting homeostasis of cell proliferation and survival of Atg1Δ3D mutant hemocytes. Normal cell numbers were present in the two distinct hemocyte populations in the circulating hemolymph, which is easily spilled on dissection (Fig. S2C), and the adherent sessile hemocytes attached to the body wall (Fig. S2D) (25), also indicating that cell adhesion was unaffected. We found no difference in the percentage of dead or dying cells detected by propidium iodide staining (Fig. S2E).
Autophagy provides a means for cells to generate energy when nutrient-deprived, raising the possibility that failure of cell spreading might be due to decreased amino acids or ATP production from blockage of autophagy. We found the same results for control and mutant hemocytes regardless of whether assays were done in buffer or serum-containing medium. The addition of methyl pyruvate, which can be directly incorporated into the Krebs cycle and has been used to overcome the energy-depleting effects of autophagy inhibition (26), did not restore the hemocyte spreading of Atg1 mutants (Fig. S2F). These findings suggest that indirect effects of cell homeostasis or bioenergetics are not responsible for the autophagy-dependent role in cell spreading, further implicating a specific role in cortical remodeling.
Autophagy Is Required for Protrusion Extension but Not Cortical Dynamics.
Cell spreading involves coordinated events in cytoskeletal remodeling for the removal of inhibitory cortical tension and generation of dynamic protrusive forces. A cortical ring of F-actin was present in the autophagy-inhibited hemocytes, similar to unspread wild-type hemocytes immediately after dissection. To address dynamics at the cell surface, we performed time-lapse microscopy imaging of GFP-labeled hemocytes. We found that the hemocyte cortex was dynamic with membrane ruffling under both control and Atg1Δ3D mutant conditions (Fig. 4 and Movie S1), suggesting that autophagy is not essential for total cytoskeletal dynamics or initiation of protrusions. Although the Atg1Δ3D mutant hemocytes could extend and retract short protrusions, the cells did not extend the longer protrusions at the cover glass surface that were characteristic of wild-type hemocytes (Movie S1). The live cell imaging suggests that autophagy might contribute to the cell protrusion attachment or extension that occurs along with cell flattening.
Fig. 4.
Hemocyte cortical dynamics occur but are altered in absence of autophagy. The cell cortex was dynamic in both GFP-labeled wild-type (Upper) and Atg1Δ3D mutant hemocytes (Lower; Movie S1), with stunted protrusions observed in the absence of autophagy. Frames every 40 s from time-lapse microscopy imaging. (Scale bar: 5 μm.)
Autophagy and Integrin Share Functions in Cell Spreading.
Cell protrusion formation and spreading are associated with a focal adhesion function for surface attachment, specifically through integrin–matrix interactions (5). Drosophila βPS1-integrin, encoded by myospheroid (mys), has been shown to be required for the spreading of a Drosophila hemocyte-derived cell line (27). Similar to the autophagy requirement, we found that mys function was required for hemocyte cell spreading (Fig. 5B), but not for association of sessile hemocytes with the body wall (Fig. S3A). Given the shared defect in cell spreading, we investigated βPS1-integrin distribution. In wild-type hemocytes, we found βPS1-integrin along the surface of round cells before spreading and in foci that colocalized with F-actin in spread cells, presumably indicating focal adhesions (Fig. S3B). We found that βPS1-integrin also localized to the plasma membrane in Atg1Δ3D mutant hemocytes and, in rare instances of spread hemocytes with this genotype, in a punctate pattern (Fig. S3C), suggesting that autophagy is not required for βPS1-integrin cell surface localization.
Fig. 5.
Role for Ref(2)P-selective autophagy in integrin-dependent spreading and Rho1-induced cell protrusions. F-actin in hemocytes plated on cover glass (A–D, I and I′) or Con A–coated cover glass (E–H); Pxn-GAL4. Hemocytes with mys βPS1-integrin RNAi failed to spread on cover glass (B), but spread with protrusions on con A (F); in contrast, Atg1Δ3D mutant hemocytes lacked protrusions on con A (H). Cells expressing dominant-negative Rho1N19 remained round on cover glass (Fig. S3D) and on con A (G). (C) Hemocyte expression of constitutively active RhoV14 resulted in longer cell protrusions. (D) Blocking autophagy suppressed RhoV14-induced cell protrusions. (I and I′) Hemocyte spreading is p62-dependent. ref(2)Pod1/+ (I) and mutant hemocytes (I′) on cover glass. (J) Protrusion length (μm, mean ± SEM). (Scale bar: 5 μm; 2.5 μm in zoomed crops of cell edges).
Integrin-independent mechanisms in cell flattening and spreading can be evoked through lectin-mediated surface attachment to Con A (28). Hemocytes disrupted for either mys integrin-mediated adhesion (Fig. 5F) or Atg1-mediated autophagy (Fig. 5H) were able to flatten and spread with an increased cell footprint on Con A–coated cover glass, signifying that an autophagy-dependent function for cell spreading was not a broad essential requirement for cell attachment or flattening. However, unlike wild-type and mys RNAi hemocytes, which extended spiky, filopodial-like protrusions during the first hour after plating on Con A (Fig. 5E and F), the Atg1 RNAi or mutant hemocytes exhibited a discoid shape with lamellipodia rather than the normal protrusions (Fig. 5H). Taken together, these findings indicate that both autophagy and integrin functions in hemocytes are required for integrin-dependent cell spreading, but with a distinct role for autophagy essential to hemocyte protrusion extension. This suggests that autophagy might have two separate roles or, alternatively, that autophagy plays a primary role in cell protrusion that affects integrin-dependent attachment in spreading.
Autophagy Is Required for Extension of Rho1-Induced Cell Protrusions.
The hemocyte-targeted expression of a dominant negative Rho1N19 allele or disruption of the Rho1 effectors dia and rok resulted in hemocytes that maintained a round cell shape (Fig. S3 D–F). Normal Rho1 pathway function was essential for hemocyte spreading on both cover glass and (unlike autophagy) Con A substrates (Fig. 5G). In contrast, hemocytes expressing the constitutively active Rho1V14 allele resulted in enhanced cell spreading, with cells exhibiting longer and more prominent F-actin cell protrusions (Fig. 5 C and J; 2.3 ± 0.1 μm for control vs. 5.5 ± 0.3 μm for Rho1V14). Although hemocytes could spread, the extensive protrusions induced by the Rho1V14 allele were suppressed by simultaneous inhibition of autophagy with the Atg1Δ3D mutation (Fig. 5 D and J; 1.0 ± 0.1 μm). This finding demonstrates that autophagy can modulate Rho1 pathway functions and implicates a specific effect of autophagy at the level of cortical protrusion and remodeling.
Ref(2)P Function Reveals a Role for p62-Selective Autophagy in Hemocyte Spreading.
Autophagosome formation requires common assembly factors and the addition of new membrane, regardless of the encapsulated contents (23), which could indirectly disrupt cortical remodeling through effects on intracellular membrane dynamics. Alternatively, autophagy possibly could play a role in cortical remodeling through the sequestration and/or lysosomal degradation of a specific factor that normally either antagonizes protrusions or promotes a round cell shape. p62 has been proposed to serve as an adaptor for delivery of ubiquitinated cargo to the autophagosome in selective forms of autophagy (11, 29). We found that hemocyte spreading was dependent on ref(2)P encoding Drosophila p62 (Fig. 5I′) (30). This points to selective autophagy of an ubiquitinated substrate as a likely target of an autophagy-dependent mechanism for cortical remodeling.
Autophagy Is Required for Hemocyte Recruitment to Wound Sites.
Wounding of the larval epidermis leads to melanized clot formation and recruitment of hemocytes from circulation (31). At 6 h after puncture wounding, microscopy imaging through the cuticle of live wild-type larvae revealed GFP-positive hemocytes accumulating around and within the clot (50.5 cells; Fig. 6 A, A′, and D). In contrast, fewer hemocytes surrounded the wound site in Atg1Δ3D mutant larvae (22.2 cells) and in larvae with hemocyte-targeted Atg1 RNAi (18.2 cells) (Fig. 6 B–D). This finding demonstrates that autophagy in hemocytes is important for physiologically relevant blood cell behavior within the intact animal.
Fig. 6.
Autophagy is required for hemocyte recruitment to larval wound sites. Whole larva indicates approximate position of wound sites (arrow) and region of low magnification images (white box). Recruitment of GFP-positive hemocytes 6 h. after larval wounding in control (A and A′), Atg1Δ3D mutant (B and B′), and hemocyte-targeted Atg1 RNAi larvae (C and C′), with Pxn-GAL4 GFP expression. White outlines depict the regions (Upper) shown at higher magnification below, and the sites of melanized wounds (Lower). (D) Mean number of hemocytes (± SEM) within two wound diameters per larva. (Scale bar: 100 μm.)
A Role of Autophagy in Regulated Cell Shape Changes in Primary Mouse Macrophages.
To address whether autophagy is broadly required for blood cell remodeling, we tested the effects of disruption of autophagy on cell shape changes in mammalian macrophages. Primary mouse macrophages plated on glass demonstrated cell spreading with dramatic cell elongation (Fig. 7 A and D; 2.8 major:minor axis); however, macrophages disrupted for autophagy with siRNA depletion of ULK1 (Atg1) or Beclin-1 (Atg6) did not undergo extensive cell spreading or elongation and remained predominantly circular in shape (Fig. 7 B and D; major:minor axis, 1.9 for ULK1 siRNA and 2.2 for BECN1 siRNA; P < 0.005). We found similar cell spreading and elongation defects with 3MA disruption of autophagy in two different activated mammalian macrophage cell lines (Fig. S4). These data suggest a broad role for autophagy in regulating hemocyte and macrophage cell shape changes, with significance for conserved immune cell functions.
Fig. 7.
Autophagy plays a role in mouse macrophage spreading and elongation. (A–C) F-actin–stained primary mouse macrophages following siRNA. (A) Control siRNA macrophages elongated on glass. (B and C) RNAi depletion of ULK1 (B and B′) or BECN1 (C and C′) inhibited macrophage spreading and elongation. (D) Cell shape as ratio of major:minor cell axis lengths (1 = round; mean ± SEM) for >94 cells. (Scale bar: 25 μm.)
Discussion
We have identified a continuous role for autophagy in blood cell cortical remodeling, with involvement in the extension of cell protrusions during initiation and maintenance of integrin-mediated cell spreading. The cortical regulation of actin-based membrane protrusions, such as lamellipodia or filopodia, and cell–matrix attachments are linked through shared pathways (5), and both play major roles in cell migration and probing cell contacts relevant to immune cell function. The cell mechanics of cortical remodeling involve cyclic processes for protrusive force and contractions, as well as focal adhesion assembly and disassembly. Responsive molecular mechanisms must dynamically monitor and balance competing activities through macromolecular turnover and rapid re-novation of cellular architecture. Examples of this are seen at multiple levels within Rho GTPase pathways that alter localization, activity, and/or destruction of key regulators that control microfilament network organization and dynamics. We propose protein sequestration in autophagosomes and autophagic clearance as additional mechanisms through which cellular remodeling for dynamic cell states can be achieved.
Previous studies of autophagy have focused on its roles in cell growth, survival, and death as responses to starvation and stress (10). We found that basal autophagy does not appear to affect cell survival, numbers, differentiation, bioenergetics, or adhesion of larval hemocytes. Autophagy is known to encapsulate bulk cytoplasmic contents and to selectively target specific protein aggregates and organelles. The regulation and targets for selective autophagy are not well understood, however. An emerging mechanism appears to involve parallels to the ubiquitin-proteosome system (29), in which p62 is envisioned to serve as a bridge for the delivery of ubiquitinated cargo to autophagosomes. Interestingly, p62 also has been implicated in growth factor–induced neurite outgrowth with possible roles in recruitment of ULK1 to cortical growth receptors (16). This lends further support to the idea that autophagy might play a role in neurite outgrowth (15–17) and synaptic development at the Drosophila neuromuscular junction (32). One known target of selective autophagy is in the cell renovation that follows the morphogenetic event of cytokinesis, whereby an ubiquitinated midbody ring complex is disposed of through p62-mediated autophagic clearance (33). Given the prevalence and possibilities for ubiquitin protein modifications, ubiquitination could play roles in both proteosomal and selective autophagic turnover (29).
The role of autophagy in blood cell spreading could work at the level of facilitating removal of cortical tension or promoting extension of protrusions, which might be coordinated with substrate attachment. We envision that autophagy could serve as a rapid means to sequester or dampen specific signaling activities in response to a changing environment or cues. This is analogous to other posttranslational mechanisms that use ubiquitination or phosphorylation. The shared role for ref(2)P suggests a model involving p62-mediated autophagosome sequestration or autophagic degradation of a potentially ubiquitinated substrate. We identified the Rho1 pathway as a potential point of intersection. Interestingly, a growing list of Rho pathway components with roles relevant to control of cell protrusion and spreading have been found to undergo ubiquitination and changes in protein stability (34–36), and recent connections suggest a potential reciprocal regulatory relationship between the Rho and autophagic pathways (37, 38). Components that are involved in changing levels of focal adhesion activity and that correspond to a rounding-up of cells also have been identified as being under ubiquitin regulation (39, 40). Connections also have been made linking focal adhesion components directly to the process of autophagy (20, 41, 42), as well as to cell spreading. Conceptually, numerous candidate targets of selective autophagy remain to be investigated.
Autophagy has known functions in immune cells in the mounting of antimicrobial responses through its roles in antigen presentation and the targeted destruction of intracellular pathogens (43). Our present findings demonstrate that autophagy also controls immune cell remodeling, which has significance in immune cell surveillance. We found that hemocytes that were disrupted for autophagy and the ability to undergo cell spreading ex vivo also showed decreased recruitment to wound sites in the animals. Consistent with this finding, wild-type hemocytes recruited to larval wound sites exhibited cell spreading, extension of cell processes, and more persistent attachment, in contrast to the sessile hemocyte population with rounded-up morphology (31). We have shown that mouse macrophages subject to blockage of autophagy exhibited defects in induced cell shape changes, with similar morphological phenotypes to those of effects on signaling through integrin and Rho pathways also important for macrophage migration in inflammatory infiltration (44). The requirement for continuous autophagy in hemocyte spreading also might be relevant in other contexts that require a switch to modulate cell morphology, such as rounding-up and respreading during mitosis and events in metastasis.
Materials and Methods
For additional information, see SI Materials and Methods.
Hemocyte Isolation.
Wandering third instar larvae were used for all experiments. To isolate hemocytes, larvae were rinsed briefly in 70% ethanol and then in PBS, and placed in 100 μL of PBS or medium on a cover glass (Fisher Scientific). The cuticle was gently ripped open with fine forceps, and released hemocytes were allowed to settle and spread at 25 °C for 30 min.
Hemocyte Treatments.
Studies were done using 10 μM 3MA (Sigma-Aldrich), 400 μM methyl pyruvate (Sigma-Aldrich), or 1 μg/mL of propidium iodide (Sigma-Aldrich) in PBS. Cell counts were determined using a hemocytometer.
Wounding Procedure and Live in Vivo Imaging.
Wandering third instar larvae were immobilized ventral side down on double-sided tape. Larvae were wounded with a pulled injection needle in the middle of the A5 segment. A drop of water was used to recover wounded larvae, then incubated on grape-agar plates at 25 °C for 6 h. The larvae were remounted on a glass slide with double-sided tape, and GFP-labeled hemocytes around the melanized wound site were visualized by wide-field fluorescence on a Leica DM1600 with 5× (NA 0.15) and 20× (NA 0.5) objectives.
Live Imaging of Hemocyte Dynamics.
Hemocytes expressing eGFP driven by Cg-GAL4 or Pxn-GAL4 were dissected as above into 100-μL complete medium and allowed to settle on the cover glass for 30 min at 25 °C before imaging. For time-lapse recordings, images were captured every 5 s for a total of 180 s on an Olympus FV1000 point scanning microscope.
Primary Macrophage Isolation, siRNA, and Spreading Assay.
Primary macrophages were elicited by i.p. injection of thioglycollate, as described previously (45). For siRNA knockdown, 2.5 × 106 cells plated per 35-mm dish were transfected with 30 nM Dharmacon siGENOME SMARTpool siRNAs (control, #D-001206-13-05; ULK-1, #M040155-00-0005; and Beclin1, #M055895-01-0005). The DeliverX siRNA Transfection Kit (Panomics) was used in accordance with the manufacturer's instructions, with modifications. After incubation with the transfection mix, cells were added to 1.5 mL of OptiMEM Reduced-Serum medium (Invitrogen), incubated at 37 °C for 60 h, and then harvested for cell-spreading assays or protein isolation for Western blot analyses (SI Materials and Methods). For the macrophage-spreading assay, 5 × 105 cells after siRNA transfection were seeded on 12-mm circular cover glass (Fisher Scientific), incubated at 37 °C for 3 h, and then fixed and stained for F-actin.
Microscopy Image Analysis.
To quantitate cell spreading, images of cell fields were manually counted for spread and round cells. To quantitate the sessile hemocytes, CellProfiler was used to automatically identify hemocytes. Lengths of cell protrusions were manually traced and recorded using Volocity (PerkinElmer). The 20× images of wounded larvae were analyzed in Photoshop (Adobe Systems) by manually counting hemocytes within two-wound diameters. All data were exported to Microsoft Excel, and means, SEs, and Student t test values were determined.
Supplementary Material
Acknowledgments
We thank T. Neufeld (University of Minnesota), D. Contamine and S. Gaumer (University Versailles-St. Quentin), I. Andó (Hungary Academy of Sciences), G. Chen (Institute of Biological Chemistry, Taipei), M. Galko (University of Texas, M.D. Anderson Cancer Center), the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and Developmental Studies Hybridoma Bank for reagents; P. Wiesner and Y. Miller (University of California, San Diego) for providing mmLDL and J774A.1 cells; K. Lau and T. Nguyen for technical support; and members of the A.A.K. laboratory for comments. We are grateful to T. Meerloo for technical assistance with electron microscopy preparation and M. Farquhar for access to the CMM Electron Microscope Facility (National Institutes of Health Grant DK17780). Imaging was conducted in part in the University of California San Diego Neuroscience Microscopy Shared Facility (National Institutes of Health Grant P30 NS047101). This work was supported by funds from the Human Frontier Science Program and The David & Lucille Packard Foundation (to A.A.K.), NIH LIPID MAPs consortium Grant GM069338 (to C.K.G.) and Ruth L. Kirschstein National Research Service Award NIH/NCI T32 CA009523 (to J.D.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914168107/-/DCSupplemental.
References
- 1.Paladi M, Tepass U. Function of Rho GTPases in embryonic blood cell migration in Drosophila. J Cell Sci. 2004;117:6313–6326. doi: 10.1242/jcs.01552. [DOI] [PubMed] [Google Scholar]
- 2.Stramer B, et al. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J Cell Biol. 2005;168:567–573. doi: 10.1083/jcb.200405120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams MJ, Habayeb MS, Hultmark D. Reciprocal regulation of Rac1 and Rho1 in Drosophila circulating immune surveillance cells. J Cell Sci. 2007;120:502–511. doi: 10.1242/jcs.03341. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.DeMali KA, Burridge K. Coupling membrane protrusion and cell adhesion. J Cell Sci. 2003;116:2389–2397. doi: 10.1242/jcs.00605. [DOI] [PubMed] [Google Scholar]
- 6.Palamidessi A, et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 7.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 10.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 13.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 14.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura K, et al. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, et al. Unc-51–like kinase 1/2–mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 18.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen GC, et al. Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy. 2007;4:37–45. doi: 10.4161/auto.5141. [DOI] [PubMed] [Google Scholar]
- 21.Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 23.Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 24.Wyers F, Dru P, Simonet B, Contamine D. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol. 1993;67:3208–3216. doi: 10.1128/jvi.67.6.3208-3216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 26.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 27.Kiger AA, et al. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2 doi: 10.1186/1475-4924-2-27. article 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jani K, Schöck F. Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 30.Gay P, Contamine D. Study of the ref(2)P locus of Drosophila melanogaster, II: Genetic studies of the 37DF region. Mol Gen Genet. 1993;239:361–370. doi: 10.1007/BF00276934. [DOI] [PubMed] [Google Scholar]
- 31.Babcock DT, et al. Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proc Natl Acad Sci USA. 2008;105:10017–10022. doi: 10.1073/pnas.0709951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 34.Wang HR, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 35.DeWard AD, Alberts AS. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J Biol Chem. 2009;284:20061–20069. doi: 10.1074/jbc.M109.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo Y, Ho HJ, Wang C, Guan JL. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through the ubiquitin-proteasome pathway. Oncogene. 2010;29:263–272. doi: 10.1038/onc.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baisamy L, Cavin S, Jurisch N, Diviani D. The ubiquitin-like protein LC3 regulates the Rho-GEF activity of AKAP-Lbc. J Biol Chem. 2009;284:28232–28242. doi: 10.1074/jbc.M109.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer PO, et al. Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J Biol Chem. 2009;284:13153–13164. doi: 10.1074/jbc.M809229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dao VT, Dupuy AG, Gavet O, Caron E, de Gunzburg J. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J Cell Sci. 2009;122:2996–3004. doi: 10.1242/jcs.041301. [DOI] [PubMed] [Google Scholar]
- 40.Shao Y, Elly C, Liu YC. Negative regulation of Rap1 activation by the Cbl E3 ubiquitin ligase. EMBO Rep. 2003;4:425–431. doi: 10.1038/sj.embor.embor813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000;149:423–430. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okigaki M, et al. Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc Natl Acad Sci USA. 2003;100:10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascual G, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.