Abstract
Although much has been learned about the design of models of β-sheets during the last decade, modest fold stabilities in water and terminal fraying remain a feature of most β-hairpin peptides. In the case of hairpin capping, nature did not provide guidance for solving the problem. Some observations from prior turn capping designs, with further optimization, have provided a generally applicable, “unnatural” beta cap motif (alkanoyl-Trp at the N terminus and Trp-Thr-Gly at the C terminus) that provides a net contribution of 6 + kJ/mol to β-hairpin stability, surpassing all other interactions that stabilize β-hairpins including the covalent disulfide bond. The motif, made up entirely of natural residues, is specific to the termini of antiparallel β-strands and reduces fraying at the ends of hairpins and other β-sheet models. Utilizing this motif, 10- to 22-residue peptide scaffolds of defined stereochemistry that are greater than 98% folded in water have been prepared. The β-cap can also be used to staple together short antiparallel β-strands connected by a long flexible loop.
Keywords: beta sheets, capping stabilization, peptide hairpins, Trp/Trp interactions
Capping motifs are well-known features of protein and peptide secondary structure; specifically, terminal alpha helix caps (especially N caps) are common both in proteins and designed peptides (1–4). The basis for helix capping is straightforward: countering the helix macrodipole and providing additional H-bonding interactions (1). Caps increase the fold population of isolated α-helical peptides and have been a boon to the design of α-helix models. β-Structures have capping requirements wholly different from those of helices; the ends of canonical β-sheets and hairpins do not have dipole moments or unsatisfied H bonds.
The a priori design of model β-sheet systems (5), focused on hairpins, has lagged behind that of α-helix counterparts. The key discoveries that improved β-hairpin stabilities outside of protein contexts have been sequences with good turn propensities, for example D-Pro-Gly (pG) (6), heterochiral pP (7), and Aib-Gly (8) [or less favorably, Asn-Gly (NG) (5, 9)] and the incorporation of optimized cross-strand pairings [most notably Trp/Trp pairs flanking the turn (10–14)]. However, longer hairpin models are typically still frayed at the termini; to date, fully folded spectroscopic reference values have only been attained via cyclization (15–17). With the exception of cyclization, mutations at terminal sites have yielded only modest changes in hairpin stability; terminal coulombic effects (ΔΔGU = 1.5–2.5 kJ/mol) standing as the only generally observed capping effect (11, 18, 19). Pi-cation interactions have also been shown to provide significant hairpin stabilization (20), but instances in which the interaction appears near the ends of hairpins have provided only marginal stability increases (18, 21–23). An unnatural π-cation interaction has also been shown to stabilize the turn in a tripeptide (24). There has been limited evidence for hairpin fold stabilization by hydrophobic effects involving terminal residues of hairpins (25–27): the stabilizing effects of cross-strand hydrophobic pairs has been reported to decrease with increasing contact order (28).
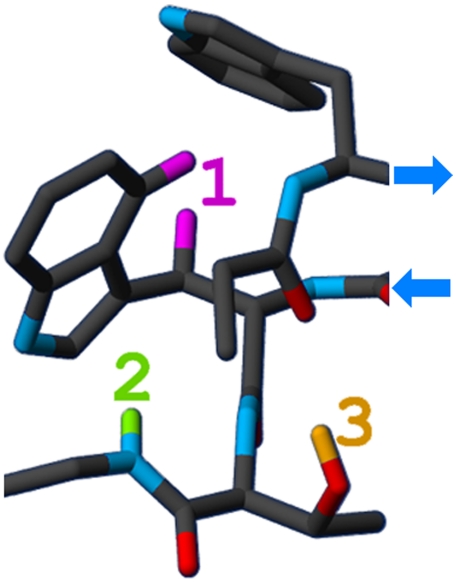
From the perspective of fold design, a β capping strategy should prove useful, serving as a complimentary stabilizing feature remote from optimized turns. To be designated as a “β-cap,” the motif would need to be specific to the termini and provide stabilization sufficient for overcoming the entropic cost of its formation. We recently designed a very short hairpin with a ΔGU in excess of 9 kJ/mol (14). The sequence of this “microprotein” incorporated a β-turn flanking motif (Fig. 1) involving terminal residues that includes an FtE (face-to-edge) Trp/Trp interaction, as well as H-bonding interactions between a threonine hydroxyl and an N-terminal acetyl, and a backbone amide and an indole ring.
Fig. 1.
Interactions within a β capping motif [taken from lowest energy structure of Pr-WIpGIWTGPS (14)]. Protons that provide NMR diagnostics are as follows: 1 (Hε3 and Hβ3 of the edge Trp) and 2 (HN of the i + 2 Gly) experience significant ring current shifts; proton 3 (Hγ1 of the i + 1 Thr) is involved in a bifurcated H bond with the N-terminal C = O and is exchange protected (visible by NMR).
In the present study, we establish that this capping motif qualifies as a β-cap: It has a significant stabilizing effect (> 6 kJ/mol) on a wide variety of β-structures, it is effective in abolishing fraying, and it specifically caps the termini of antiparallel β-strands. This β capping motif can even effect closure of quite long loops with no intrinsic tendency to form direction reversing turns.
Results
The present application of a specific type of flanked Trp/Trp motif as a stabilizing cap for β-sheets builds on our work with short “capped-loop” hairpins such as Ac-WINGKWTG-NH2 (23). All peptides of the form “(acyl)-W-loop-WTG…” (where “loop” is any sequence capable of forming a tight turn; e.g., IpGL, IHGK, ENGR, etc.) exhibited the remarkable stability and diagnostic spectroscopic features expected of stable hairpin folds with cross-strand Trp/Trp pairs at a loop-flanking non-H-bonded position, including a strong exciton couplet visible by circular dichroism (CD) (λmax = 228 nm) (10, 11, 13, 23). However, unlike prior examples (7, 10–13, 22, 23, 29, 30) of turn-flanking Trp/Trp interactions (SI Appendix), in which the upfield chemical shifts observed for the Hε3 and Hβ3 protons for the “edge” tryptophan were observed for the N-terminal Trp, in the acyl-W-loop-WTG species, the C-terminal Trp was the edge species (Fig. 1). The far upfield (shifted as much as 3.8 ppm from its random coil value) glycine amide proton and an NMR-visible threonine hydroxyl proton observed with this motif were unique. The expected chemical shift deviations due to structuring (CSDs) for hairpin turn and strand positions (31) were also observed. All of these spectroscopic features could be used as probes to determine fold population (χF).*
Gly4 to L-Ala and D-Ala mutations (SI Appendix) had negligible effects on fold stability of Ac-WIpGKWTG-NH2 even though these enforce type II′ and I′ turns, respectively (6, 32). This indifference to turn type prompted us to view Ac-W - - WTG as a general capping motif and to examine its efficacy at positions remote from the turn. Because neither NMR or CD melts can effectively distinguish between 90% and 99% folded hairpins (14), we turned to amide H/D exchange protection experiments to determine the fold populations of our more stable constructs. Five constructs bearing our β-capping motif display amide protection factors for cross-strand H-bonded sites in excess of 80 (≥98.7% folded).
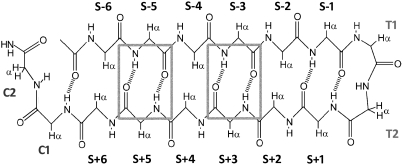
First, we aimed to systematically study the effect of distance of the capping group from the turn. The locations examined are indicated by the distance between the Trp residues and the central turn residues (NG, HG, or pG in the examples that follow); non-H-bonded (NHB) locations are thus S ± even sites (see Fig. 3) (31). One attempt to place the W/W pair at a hydrogen-bonded (HB) site (S ± 5) is also recorded. The peptides employed for this initial survey, together with some analogs and a control, appear in Table 1.
Fig. 3.
A schematic of a 14-residue β-hairpin with an added cap (C1 = Thr and C2 = Gly). Strand sites are designated (S ± #) by their distance from the turn. For the optimized S ± 6 Trp/Trp peptide (Table 1, entry 7), the enboxed cross-strand H-bonded sites display maximal protection of the NHs to H/D exchange. Somewhat reduced protection was observed at other HB sites; further details appear in the SI Appendix.
Table 1.
Cap viability versus tryptophan location
| NMR measures of extent of folding | ||||||||
| Peptides Examined | CSDs | |||||||
| Entry no. | Sequence | W/W site | Turn GΔδHα | CSDs X2HN | C-term W Hβ3 | C-term W Hε3 | C-term G HN | Capped |
| 1 | Ac-WI- - - - - -NG- - - - - -KWTG-NH2 | S ± 2 | 0.404 | 1.008 | −1.216 | −1.556 | −2.705 | Yes |
| 2 | Ac-WVTI- - - -pG- - - -KKIWTG-NH2 | S ± 4 | N/A | 1.492 | −1.307 | −2.023 | −2.944 | Yes |
| 3 | Ac-WVSI- - - -NG- - - -KKIWTG-NH2 | S ± 4 | 0.551 | 1.323 | −1.183 | −1.783 | −2.704 | Yes |
| 4 | WVSI- - - -NG- - - -KKIWTG-NH2 | S ± 4 | 0.152 | 0.062 | −0.074 | −0.029 | −1.038 | No |
| 4b | KKLWVSI- - - -NG- - - -KKIWTGA | S ± 4 | 0.465 | 0.77 | −0.49 | −0.54 | −0.78 | No |
| 5 | Ac-WTVSI- - -NG- - -KKITWTG-NH2 | S ± 5 | 0.201 | 0.256 | −0.202 | −0.09 | −1.155 | No |
| 6 | Ac-WLSVTI- -NG- -KTIKVWTG-NH2 | S ± 6 | 0.569 | 1.46 | −1.125 | −1.973 | −2.796 | Yes |
| 7 | Ac-WITVTI- -HG- -KKIRVWTG-NH2 | S ± 6 | 0.624 | 1.46 | −1.507 | −1.997 | −3.116 | Yes |
| 8a | Ac-WITATI- -HG- -KKARVWTG-NH2 | S ± 6 | 0.545 | 1.07 | −1.08 | −1.44 | −2.5 | Yes |
| 8b | WITVTI- -HG- -KKIRVWTG-NH2 | S ± 6 | 0.466 | 0.475 | −0.032 | −0.412 | −0.95 | No |
| 8c | ATWITVTI- -HG- -KKIRVWTG-NH2 | S ± 6 | 0.601 | 1.28 | −1.163 | −1.363 | −1.206 | No |
| 9 | Ac-WRWVKVWIpGKWIQVPQWTG-NH2 | S ± 8 | N/A | 1.015 | −1.244 | −1.63 | −2.326 | Yes |
| control Ac-WTG-NH2 | 0.01 | 0.07 | −1.264 | No | ||||
| 10 | Ac-WRWQYV- -NG- -KFTPQWTG-NH2 | S ± 6 | 0.488 | 0.8 | −0.952 | −1.5 | −2.19 | Yes |
| 11 | WRWQYV- -NG- -KFTPQWTG-NH2 | S ± 6 | 0.362 | −0.04 | −0.168 | −0.262 | −1 | No |
| 12 | Ac-TRWQYV- -NG- -KFTPQWTG-NH2 | S + 6 | 0.314 | 0.08 | −0.022 | −0.018 | −1.03 | No |
The tryptophan pair location is indicated using the nomenclature appearing in Fig. 3; all data for 280 K. N/A, not applicable.
Capping at S ± 4.
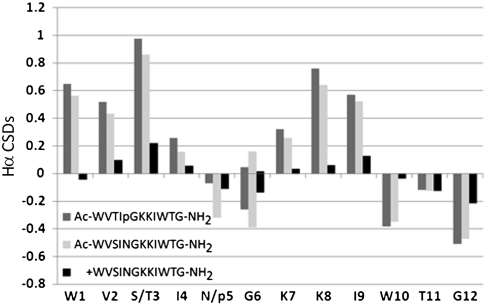
Peptide Ac-WVSINGKKIWTG-NH2 proved to be remarkably stable, tolerated substitutions at the new strand positions (bold), and displayed, in addition to the W/W exciton couplet, all of the CSDs expected for the capping interaction such as the upfield shifts at G12HN and W10 Hε3 and Hβ3, see Table 1 (entry 3). In analogy to our prior capped-turn microprotein motif (14), terminal Ac → Pr and G-NH2 → GPS mutations increased fold stability and the exchange protection factor for the Thr-OH. Removal of the acetyl moiety was drastically destabilizing (ΔΔG: 6.9 kJ/mol), as evidenced by the reduction in diagnostic CSDs for both the strand (Fig. 2) and sites within the cap.
Fig. 2.
Hα CSDs for the acetylated and nonacetylated versions of hairpin WVSINGKKIWTG-NH2 (β-cap motif at the S ± 4 position). Peptide Ac-WVTIpGKKIWTG-NH2 is included as a positive control; it was shown (by amide exchange experiments) to be 99% folded (SI Appendix). CSDs at the varied (pG and NG) turn locus cannot be used to estimate fold populations.
W/W placement at S ± 5.
A single Thr insertion in each strand moves the Trp residues from NHB to H-bonded sites. Ac-WTVSINGKKITWTG-NH2 proved to be ≤ 20% folded at 280 K and exhibited none of the diagnostic structuring CSDs; the CSD of the terminal Gly HN was on par with shifts seen for the “random” coil control, Ac-WTG-NH2 (Table 1). This result suggested that the β-cap requires aryl functions located at a NHB strand site and this placement was used for the remaining examples.
Capping at S ± 6.
Further extension restored hairpin stability: Peptide Ac-WLSVTINGKTIKVWTG-NH2 exhibited improved stability over the shorter analogs; see Table 1 (entry 6). We designed an optimized peptide of this length by maximizing the number of β branched and positively charged residues to increase intrinsic β propensity and solubility. Even though turns with an HG, rather than NG locus, are somewhat less fold stabilizing in other hairpin models, the resulting peptide (Ac-WITVTIHGKKIRVWTG-NH2, Table 1, entry 7) was shown to be > 98.5% folded at 280 K by H/D exchange: the amides of I2, V4, I11, and V13 displayed (94 ± 8)-fold exchange protection (Fig. 3).
Capping at S ± 8.
Designing a longer peptide hairpin which might display folding enhancement by β capping presents a number of challenges: The strand twisting could place the termini too far apart for the essential cross-strand interactions in the cap and aggregation becomes increasingly likely. Starting with a very stable Trp-flanked turn (WIpGKW) (14), and inserting a proline at a non-H-bonded strand position to discourage aggregation (33) (with a rescuing cross-strand Trp), we arrived at Ac-WRWVKVW-IpGK-WIQVPQWTG-NH2. The appearance of the diagnostic CSDs (Table 1, entry 9) indicated that the β-cap was present.
Quantifying Capping Effects on Fraying.
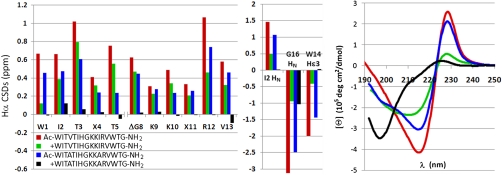
The optimized S ± 6 capped hairpin (Table 1, entry 7) failed to display evidence of thermal fraying on warming 40 °C (SI Appendix). We reasoned that specific stabilization of the termini by the cap should be more evident if the intrinsic tendency toward β-strand formation and association was decreased by central mutations in the strands. This was effected by introducing alanines at a cross-strand HB site: CSDs from the sites in the capping motif (Table 1, entry 8a versus 7), as well as the CD exciton couplet magnitude (Fig. 4, Far Right), indicated a fold population of 0.75 ± 0.03 for the cap in the resulting peptide. The CSDs of this peptide, Ac-WITATIHGKKARVWTG-NH2, are compared to the fully folded control and the corresponding desacetyl species in Fig. 4. The comparison between this peptide (blue bars, intact β-cap) and +WITVTIHGKKIRVWTG-NH2 (green bars, β-cap removed by deacetylation) illustrates the cap’s ability to overcome the inherent propensity for the ends of β-hairpins to partially dissociate. Even though the V/I → A mutations result in CSDs at midstrand positions (T3, T5, and K10) that are smaller than those of +WITVTIHGKKIRVWTG-NH2, the species with an intact β-cap has the larger CSDs at the terminal sites. The stabilizing effect of the β-cap is most pronounced as the elimination of end fraying.
Fig. 4.
Comparisons of CSDs for peptides of the form (Z)WITXTIHGKKXRVWTG-NH2. Peptide Ac-WITVTIHGKKIRVWTG-NH2 (> 98.5% folded) is included as the folded control. The decreased fraying in the Z = Ac, X = A peptide, relative to the otherwise comparably folded des-Ac species, is most readily apparent in the I2HN CSD and those for the Hαs of W1, R12, and V13. The far right panel shows the CD spectra of the same four peptides.
In the peptide with a reduced β propensity in the strands, the capping motif is absolutely required for detectable hairpin formation: Removal of the acetyl group of the β-cap results in a peptide with no NMR or CD detectable hairpin characteristics.
Capping Specificity.
To ascertain whether our motif was a specific end cap or only an accumulation of cross-strand interactions, we examined the effect of polypeptide extension past the N- and C-terminal Trp residues of the motif. C-terminal extension was tolerated (SI Appendix), but N-terminal extensions decreased hairpin fold stability and eliminated (or greatly reduced) the diagnostic CSDs within the capping motif. In the case of the cap with an S ± 4 W/W pair, this is illustrated by peptide KKLWVSINGKKIWTGA (55% folded at 280 K, Table 1, entry 4b). This peptide has a higher fold population than the desacetyl species (Table 1, entry 4) but the diagnostic CSDs for the cap are absent. N-terminal extension also decreased the fold population for an S ± 6 capped system (see Table 1, entry 8c). It is apparent that the W..(loop)..WTG motif is less effective when the N terminus is extended by a polypeptide rather than an alkanoyl group: The least disruptive extension (a Gly-Gly unit) results in a 4 kJ/mol fold destabilization (SI Appendix). To quantitate the effects of N-terminal versus C-terminal extension, we linked two identical capped units with a -Gly-Gly linker. In the resulting peptide (Ac-WIpGKWTGPK-GG-WIpGKWTGPK-NH2), the β-capped motif at the N terminus is 97 ± 2% folded, whereas the fold population of the GG-WIpGKWTGPK-NH2 unit is only 87 ± 5% (ΔΔGU = -4 ± 1 kJ/mol); the complete shift comparisons appear in the SI Appendix.†
To establish that a β-capped unit can be incorporated into more complex multidomain structures, we prepared Ac-WIpGKWTGP-KG-KTWNPATGKWTE with a KG linker between the same β-capped motif (χF ≈ 0.975, in isolation) and a previously examined hairpin stabilized (χF = 0.89 at 280 K) (13) by a turn-flanking W/W interaction. NMR shift comparisons (SI Appendix) indicated that both units folded to the same extent (χF = 0.95 ± 0.025 and 0.86 ± 0.06, respectively, for the β-capped and W/W-flanked-turn segments) when linked.
Quantitating the Folding Contribution of the β-Cap.
Because the Ac-W - - WTG capping motif requires both the acetyl function and the site-specific cross-strand W/W interaction, the ΔΔGF contribution of the cap can be gauged by deacetylation, Trp mutations, or residue swaps that move Trp residues from the terminal to the adjacent non-H-bonded site. The latter has the advantage of retaining identical composition, not changing the net β propensity of the strands. Data presented in Fig. 2 indicated a 7 kJ/mol destabilization by deacetylation. For another 12-residue construct, 5–9 kJ/mol destabilizations were observed (SI Appendix) by replacing one or the other of the two Trp residues by Phe, Leu, or His. In the 16-residue series featured in Fig. 4, 7–9 kJ/mol destabilizations results upon either deacetylation or swapping the W/W cross-strand pair with neighboring nonterminal residues (SI Appendix).
To demonstrate that the β-capping effect we observe is not linked to other features of our hairpin designs, we added the cap to a well-studied hairpin peptide from the literature (16), RWQYVNGKFTVQ-NH2 (χF = 0.28 ± 0.04, SI Appendix). However, to avoid complication due to aggregation at NMR concentrations in the capped peptides, we had to introduce a V12P mutation (33). Diagnostic chemical shifts indicated that the resulting species (Table 1, entry 10), Ac-WRWQYVNGKFTPQWTG-NH2, was 84% folded at 280 K, a 6.1 kJ/mol stability increase relative to RWQYVNGKFTPQ-NH2. Consistent with a β-capping interaction, deacetylation (Table 1, entry 11) reduced the fold stability (χF = 0.42) and resulted in a frayed N terminus; whereas deacetylation of an uncapped control (Table 1, entry 12) increases fold stability.
Nonhairpin β-Sheets.
The capping motif also stabilizes turnless, homodimeric β-sheets of the form (Pr /Ac)-W…C…WTG(PK)-NH2. For example, peptide ((WTTVCIRKWTGPK-NH2)2 was only 72% folded, but N-propanoylation produced the capped version which was shown to be > 98.5% folded by amide exchange studies at 280 K (ΔΔG≥7.5 kJ/mol). Complete chemical shift data, including a monomeric random coil control, appear in the SI Appendix.
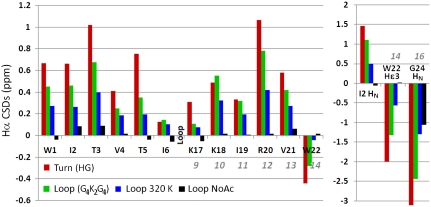
Capping Loops with No Propensity to Form a Reversing Turn.
As a final test of the capping unit, we examined whether the capping functions could bring together segments joined by a long flexible loop. We replaced HG in Ac-WITVTI-HG-KKIRVWTG-NH2 with -GGGGKKGGGG-. Based on the magnitude of the CD exciton couplet and the W22Hε3 and G24HN ring current shifts (Fig. 5), Ac-WITVTI-G4K2G4-KKIRVWTG-NH2 was 66% folded at 280 K. Association of the β-strands was evident in the backbone CSD sequence histogram (Fig. 5), providing a confirming measure of fold stability (χF = 0.70 ± 0.04). The structure melts out cooperatively and deacetylation removes all evidence of hairpin formation for this peptide (ΔΔG > 7.7 kJ/mol).
Fig. 5.
Chemical shift deviation comparisons for (Ac-)WITVTI-XX-KKIRVWTG-NH2, XX = HG (red bars, > 98.5%-folded control) and XX = GGGGKKGGGG (Loop). The complete set of chemical shifts appear in the SI Appendix.
Discussion
A synergistic set of interactions between an alkanoyl-Trp and, optimally, a -WTG- sequence provides 6–9 kJ/mol of stabilization to hairpin, sheet, or loop structures connecting these functions. This stabilization is larger than the effect of a covalent disulfide bond (∼4 kJ/mol) (34). In the present study, stable hairpins of sequences Ac-W-(ZZ)n-loop-(ZZ)n-WTG-X (wherein the loop regions are four or more residues, the majority of the Z residues have some propensity for β-strand conformations, and X can be any amino acid residue or polypeptide extension) have been prepared for n = 0,1,2, and 3. This capping motif appears to stabilize and reduce end fraying for all antiparallel sheet models so long as the Trp residues are at the terminal non-H-bonded strand sites. The components and spectroscopic diagnostics of the AcW/WTG cap are shown in Fig. 1. The likely sources of stabilization are an edge-to-face cross-strand W/W interaction [which provides 2.3–5 kJ/mol of stabilization in other systems (14, 23, 35)], a Gly-HN → indole H bond, and a bifurcated H-bonding pattern at the carbonyl of the acetyl (from both the Thr backbone amide and hydroxyl protons). In accord with this view, W → Y and W → F mutations, at the N-terminal “face” indole result in significant folding destabilization; however, even though the indole in the C-terminal WTG unit also functions as a π base (accepting an H bond from the i + 2 Gly HN), W → Y/F mutations in the WTG unit have smaller effects on the stability of capped hairpins (for Trp → X mutation data, see SI Appendix).
Given that all of the residues of the β-cap occur in proteins and peptides, we looked for cap features in the structural proteome. Acetylation is very common in proteins [estimated at > 50% for eukaryotes (36)], but we could find no example of the capping motif presented herein. Even loop flanking W/W interactions are rare; to our knowledge, there is only one example in a protein (7). Extreme upfield shifts for a Gly-HN have been observed for FXG/YXG/WXG units in proteins (37, 38), but this feature does not appear to have served as a terminating unit for a β-strand.
The β-cap should have wide applicability in the design of folded peptides. We have been able to construct hairpins and other β-sheet models that are 98 + % folded based on amide H/D exchange protection data. With the demonstrated benefits of amide H/D exchange as a method for accurately characterizing well-folded β-hairpins, we expect that it will be widely adopted in future work in the field. Stable scaffolds of defined geometry have potential for stereospecific pharmacophore display. Our demonstration that the β-cap effects closure of a hairpin structure from two β-strands linked by a long flexible loop (Fig. 5), which we designated as a “stapled loop,” is perhaps the most surprising finding in the present study. Thus we expect that the cap will impose β-structure to the nearby portion of longer biorecognition loops with the resulting constructs suitable for target discovery.
The β-cap opens other possibilities for biophysical studies, many of which are currently being examined. The introduction of NMR probes at loop termini may also provide an alternative dynamic NMR measure of loop closure dynamics. This cap may have the potential to staple together proteins with termini close in space, especially if those termini are in β-strand conformations.
The β capping motif we have presented is not just a useful tool for rational design, but also an indication that sequences composed entirely of natural amino acids can be held in stable, specific conformations by unnatural motifs. The existence of other unnatural structuring motifs—not discoverable from bioinformatics but accessible via the analysis of microprotein folds—is likely.
Methods
Peptide Synthesis.
All peptides in the study were synthesized on an ABI 433A peptide synthesizer using fast 9-fluorenylmethoxycarbonyl chemistry, and acetylated (or propionylated) by shaking the resin-bound peptide with 4.3% triethylamine and 3% acetic or propionic anhydride in dimethylformamide. Cleavage from the resin was accomplished by shaking for 1.5 h in TFA with 2.5% water and 2.5% triisopropylsilane. Purity and molecular weight were confirmed using a Bruker Esquire ion trap spectrometer; sequences were confirmed via NOESY connectivities.
NMR.
NMR spectra were recorded on a 500 MHz NMR Bruker DRX spectrometer. A WATERGATE (39) pulse train was used to suppress the H2O solvent signal. Mixing times for NOESYs were 100–125 ms, and total correlation spectroscopy used a 60 ms MLEV-17 spinlock (40). Samples consisted of 1–3 mM peptide diluted in 0.6 mL 50 mM sodium phosphate buffer, with 10% D2O and trace 2,2-dimethyl-2-silapentane-5-sulfonate standard. The pH was set to 2.5 for peptides containing histidine and 6.5 for most other peptides. Sharp line widths consistent with small, fast-folding monomeric peptides were observed (line widths of highly shifted protons of the longer hairpins were broadened slightly, due to exchange broadening associated with the folded/unfolded equilibrium on a microsecond timescale). Unless otherwise indicated, NMR presented herein was recorded at 280 K.
A series of 1D experiments for Ac-WITVTIHGKKIRVWTG-NH2 (pD: 5.89) and Ac-WVTIpGKKIWTG-NH2 (pD: 5.97) were run at 280 K. Protection factors were determined by fitting the log of the peak integrals (normalized to a nonexchanging peak) to a linear equation, and comparing to neighbor-corrected intrinsic rates described by Bai et al. (41).
NMR fold population measures are corroborated by CD melts determined at 30 μM peptide concentrations. This provides additional support for the monomeric state of all peptide folds examined. Fold populations were obtained from CSDs, for each family of structures, the 100%-folded values were established by having a well-folded example that displayed HN protection factors > 50.
Circular Dichroism.
The 30 uM peptidic CD samples were made using 20 mM phosphate buffer; (pH 2.5 for hairpins with HG turns; 6.5 for all others) concentrations were quantified using the combined expected UV absorption coefficients of Trp and Tyr at 280 nm. Units of molar (rather than residue-molar) ellipticity are used, because the capping unit has a single exciton couplet that dominates the spectrum. An updated random coil value of 25,000°/Trp was subtracted from the value at the exciton couplet’s maxima (∼228 nm) when estimating the fold population using CD.
NMR Structure Ensemble Elucidation.
The NOESY spectra for NMR ensemble generation of Ac-WITVTIHGKKIRVWTG-NH2 was acquired on a 750 MHz NMR at 280 K with a mixing time of 120 ms. NOE intensities were converted to distant ranges using an in-house program (di8) which corrects for multiple chemically equivalent protons and sharp aromatic peaks (42, 43). NMR structure generation and acceptance criteria were described previously (13) No structures with NOE constraint violations greater than 0.35 Å were included in the accepted ensembles. Ensemble statistics and the lists of NOE constraints appear in the SI Appendix. MOLMOL (44) was used to calculate rmsd values. The resulting structure ensembles and the full chemical shift assignments appear in the SI Appendix.
Supplementary Material
Acknowledgments.
The studies performed by B.L.K. were funded by a National Institutes of Health (NIH) grant (GM059658) during his period of support on a Molecular Biophysics Training Grant (NIH Grant 5T32-GM008268). Initial lead compounds came from studies funded by an National Science Foundation grant (CHE-0650318).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NMR chemical shifts and restraints can be found in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913534107/-/DCSupplemental.
*Mutational studies (23) indicated that the C-terminal Gly could be replaced with a terminal NH2, and the motif’s i-1 Thr was partly amenable to mutation, but the tryptophan residues and an N-terminal acyl moiety were strictly required; W1T and W6T mutations both resulted in the near-total loss of β-hairpin folding diagnostics.
†In both motifs, the C-terminal Trp displays an upfield Hε3: -2.09 ppm in the first repeat, -1.94 ppm in the second repeat versus -2.01 ppm in Ac-WIpGKWTGPS. In contrast, the extent of upfield shift for the Gly NH in the WTG motifs varies: G20HN -1.92 ppm versus -3.35 ppm for G8HN and -3.17 ppm for G8 of Ac-WIpGKWTGPS. Thus, the cross-strand W/W interaction is intact in both motifs, but the full features of the cap are present only in the N-terminal motif.
References
- 1.Serrano L, Fersht AR. Capping and α-helix stability. Nature. 1989;342:296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 2.Harper ET, Rose GD. Helix stop signals in proteins and peptides: The capping box. Biochemistry. 1993;32:7605–7609. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- 3.Lyu PC, Wemmer DE, Zhou HX, Pinker RJ, Kalenbach NR. Capping interactions in isolated alpha helices: Position-dependent substitution effects and structure of a serine-capped peptide helix. Biochemistry. 1993;32:421–425. doi: 10.1021/bi00053a006. [DOI] [PubMed] [Google Scholar]
- 4.Viguera AR, Serrano L. Experimental analysis of the Schellman motif. J Mol Biol. 1995;251:150–160. doi: 10.1006/jmbi.1995.0422. [DOI] [PubMed] [Google Scholar]
- 5.Ramírez-Alvarado M, Blanco FJ, Serrano L. De novo design and structural analysis of a model β-hairpin peptide system. Nat Struct Biol. 1996;3:604–612. doi: 10.1038/nsb0796-604. [DOI] [PubMed] [Google Scholar]
- 6.Haque TS, Gellman SH. Insights into β-hairpin stability in aqueous solution from peptides with enforced type I′ and type II′ β-turns. J Am Chem Soc. 1997;119:2303–2304. [Google Scholar]
- 7.Favre M, Moehle K, Jiang L, Pfeiffer B, Robinson JA. Structural mimicry of canonical conformations in antibody hypervariable loops using cyclic peptides containing a heterochiral diproline template. J Am Chem Soc. 1999;121:2679–2685. [Google Scholar]
- 8.Masterson LR, et al. Nonstereogenic alpha-aminoisobutyryl-glycyl dipeptidyl unit nucleates type I′ beta-turn in linear peptides in aqueous solution. Biopolymers. 2007;88:746–53. doi: 10.1002/bip.20738. [DOI] [PubMed] [Google Scholar]
- 9.Pastor MT, López de la Paz M, Lacroix E, Serrano L, Pérez-Payá E. Combinatorial approaches: A new tool to search for highly structured β-hairpin peptides. Proc Natl Acad Sci USA. 2002;99:614–619. doi: 10.1073/pnas.012583999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cochran AG, Skelton NJ, Starovasnik MA. Tryptophan zippers: Stable, monomeric β-hairpins. Proc Natl Acad Sci USA. 2001;98:5578–83. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fesinmeyer RM, Hudson FM, Andersen NH. Enhanced hairpin stability through loop design: The case of the protein GB1 domain hairpin. J Am Chem Soc. 2004;126:7238–7243. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- 12.Dhanasekaran M, Prakash O, Gong YX, Baures PW. Expected and unexpected results from combined β-hairpin design elements. Org Biomol Chem. 2004;2:2071–82. doi: 10.1039/b315228f. [DOI] [PubMed] [Google Scholar]
- 13.Andersen NH, et al. Minimization and optimization of designed β-hairpin folds. J Am Chem Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kier BL, Andersen NH. Probing the lower size limit for fold protein-like fold stability: Ten-residue microproteins with specific, rigid structures in water. J Am Chem Soc. 2008;130:14675–14683. doi: 10.1021/ja804656h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syud FA, Espinosa JF, Gellman SH. NMR-based quantification of β-sheet populations in aqueous solution through use of reference peptides for the folded and unfolded states. J Am Chem Soc. 1999;121:11577–11578. [Google Scholar]
- 16.Espinosa JF, Syud FA, Gellman SH. Analysis of the factors that stabilize a designed two-stranded antiparallel β-sheet. Protein Sci. 2002;11:1492–1505. doi: 10.1110/ps.4140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell SJ, Blandl T, Skelton NJ, Cochran AG. Stability of cyclic β-hairpins: Asymmetric contributions from side chains of a hydrogen-bonded cross-strand residue pair. J Am Chem Soc. 2003;125:388–395. doi: 10.1021/ja028075l. [DOI] [PubMed] [Google Scholar]
- 18.Olsen KA, Fesinmeyer RM, Stewart JM, Andersen NH. Hairpin folding rates reflect mutations within and remote from the turn region. Proc Natl Acad Sci USA. 2005;102:15483–15487. doi: 10.1073/pnas.0504392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huyghues-Despointes BMP, Xiaotoa Q, Tsai J, Scholtz JM. Terminal ion pairs stabilize the second β-hairpin of the B1 domain of protein G. Proteins. 2006;63:1005–1010. doi: 10.1002/prot.20916. [DOI] [PubMed] [Google Scholar]
- 20.Riemen AJ, Waters ML. Design of higly stabilized β-hairpin peptides through cation-π interactions of lysine and N-methyllysine with an aromatic pocket. Biochemistry. 2009;48:1525–1531. doi: 10.1021/bi801706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatko CD, Waters ML. The geometry and efficacy of cation-π interactions in a diagonal position of a designed β-hairpin. Protein Sci. 2003;12:2443–2452. doi: 10.1110/ps.03284003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiehna SE, Waters ML. Sequence dependence of β-hairpin structure: Comparison of a salt bridge and an aromatic interaction. Protein Sci. 2003;12:2657–2667. doi: 10.1110/ps.03215403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eidenschink L, Kier BL, Huggins KNL, Andersen NH. Very short peptides with stable folds: Building on the inter-relationship of Trp/Trp, Trp/cation, and Trp/backbone-amide interaction geometries. Proteins. 2009;75:308–322. doi: 10.1002/prot.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbaras D, Gademann K. Stable β turns of tripeptides in water through cation-π interactions. Chem Biochem Eng Q. 2008;9:2398–2401. doi: 10.1002/cbic.200800344. [DOI] [PubMed] [Google Scholar]
- 25.Eidenschink L, Crabbe E, Andersen NH. Terminal sidechain packing of a designed β-hairpin influences conformation and stability. Biopolymers. 2009;91:557–564. doi: 10.1002/bip.21177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riemen AJ, Waters ML. Stabilization of the N-terminal β-hairpin of ubiquitin by a terminal hydrophobic cluster. Biopolymers. 2008;90:394–398. doi: 10.1002/bip.20840. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Z, Miskolzie M, Campbell RE. In vivo screening identifies a highly folded β-hairpin peptide with a structured extension. Chem Biochem Eng Q. 2007;8:880–883. doi: 10.1002/cbic.200600565. [DOI] [PubMed] [Google Scholar]
- 28.Espinosa JF, Munoz V, Gellman SH. Interplay between hydrophobic cluster and loop propensity in β-hairpin formation. J Mol Biol. 2001;306:397–402. doi: 10.1006/jmbi.2000.4349. [DOI] [PubMed] [Google Scholar]
- 29.Honda S, Yamasaki K, Sawada Y, Morii H. Ten residue folded peptide designed by segment statistics. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Honda S, et al. Crystal structure of a ten-amino acid protein. J Am Chem Soc. 2008;130:15327–15331. doi: 10.1021/ja8030533. [DOI] [PubMed] [Google Scholar]
- 31.Fesinmeyer RM, et al. Chemical shifts provide fold populations and register of β hairpins and β sheets. J Biomol NMR. 2005;33:213–231. doi: 10.1007/s10858-005-3731-7. [DOI] [PubMed] [Google Scholar]
- 32.Raghothama SR, Awasthi SK, Balaram P. β-Hairpin nucleation by Pro-Gly β-turns: Comparison of D-Pro-Gly and L-Pro-Gly sequences in an apolar octapeptide. J Chem Soc Perkin Trans 2. 1998;1:137–144. [Google Scholar]
- 33.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santiveri CM, León E, Rico M, Jiménez MA. Context-dependence of the contribution of disulfide bonds to β-hairpin stability. Chem—Eur J. 2008;14:488–499. doi: 10.1002/chem.200700845. [DOI] [PubMed] [Google Scholar]
- 35.Jäger M, Dendle M, Fuller AA, Kelly JW. A cross-strand Trp/Trp pair stabilizes the hPin1 WW domain at the expense of function. Protein Sci. 2007;16:2306–2313. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez A, et al. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- 37.Kemmink J, van Mierlo CPM, Scheek RM, Creighton TE. Local structure due to an aromatic-amide interaction observed by 1H-nuclear magnetic resonance spectroscopy in peptides related to the N-terminus of bovine pancreatic trypsin inhibitor. J Mol Biol. 1993;230:312–322. doi: 10.1006/jmbi.1993.1144. [DOI] [PubMed] [Google Scholar]
- 38.Toth G, Murphy RF, Lovas S. Investigation of aromatic-backbone amide interactions in the model peptide acetyl-Phe-Gly-Gly-N-methyl amide using molecular dynamics simulations and protein database search. J Am Chem Soc. 2001;123:11782–11790. doi: 10.1021/ja011245u. [DOI] [PubMed] [Google Scholar]
- 39.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 40.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 41.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barua B, et al. The Trp-cage: Optimizing the stability of a globular miniprotein. Protein Eng Des Sel. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fesinmeyer RM. Seattle: University of Washington; 2005. Chemical shifts define the structure and folding thermodynamics of polypeptides. PhD thesis. [Google Scholar]
- 44.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.