Abstract
Prions are infectious, self-propagating protein aggregates that have been identified in evolutionarily divergent members of the eukaryotic domain of life. Nevertheless, it is not yet known whether prokaryotes can support the formation of prion aggregates. Here we demonstrate that the yeast prion protein Sup35 can access an infectious conformation in Escherichia coli cells and that formation of this material is greatly stimulated by the presence of a transplanted [PSI+] inducibility factor, a distinct prion that is required for Sup35 to undergo spontaneous conversion to the prion form in yeast. Our results establish that the bacterial cytoplasm can support the formation of infectious prion aggregates, providing a heterologous system in which to study prion biology.
Keywords: Sup35, [PSI+] inducibility factor, amyloid
Prions are infectious, self-propagating protein aggregates that have been implicated in a group of devastating mammalian neurodegenerative diseases (1). The discovery of a prion-like phenomenon in yeast and other fungi has led to profound advances in the understanding of prion biogenesis (2, 3). Prion proteins in mammals as well as fungi typically form highly structured β sheet-rich fibrils, known as amyloids, upon conversion to the infectious, prion form (4, 5, 1–3). However, unlike mammalian prions, yeast prions do not result in cell death, but instead act as heritable, protein-based genetic elements, conferring on the cell new phenotypic traits that are propagated epigenetically (6, 7). Although work over the last 15 years has uncovered a growing number of prions and prospective prion proteins in evolutionarily divergent members of the fungal kingdom (8–14), it is not yet known how pervasive prions are in nature; more specifically, it is not known whether bacteria contain prions or whether the bacterial cytoplasm can support the formation of prions. The study of yeast prions has revealed an essential interplay between prion proteins and cellular chaperone proteins (15, 16); thus, it is of particular interest to learn whether the bacterial chaperone environment is permissive for the formation of prion-like aggregates.
To investigate whether the bacterial cytoplasm can support the formation of infectious amyloid, we sought to determine whether a yeast prion protein could access an infectious conformation in Escherichia coli cells. A particularly well-characterized prion in Saccharomyces cerevisiae, the [PSI+] prion, is formed by the essential translation termination factor Sup35, which assembles into amyloid aggregates when it converts to the prion form (17; see also refs. 3 and 6). Upon conversion, the ability of Sup35 to participate in translation termination is impaired, and as a result, strains containing Sup35 in the prion form (designated [PSI+] strains) manifest a nonsense-suppression phenotype due to significant stop-codon read-through (6). Like other yeast prion proteins, Sup35 has a modular structure, with a distinct prion-determining region (PrD) that is necessary to enable the protein to convert to the prion form (6, 18). In the case of Sup35, an amyloidogenic N-terminal region (N) contains the essential prion-forming determinants including a Q- and N-rich segment and five complete copies of an imperfect oligopeptide repeat (3, 6). The C-terminal domain of Sup35, which is responsible for its essential translation termination activity, does not contribute to prion behavior, whereas a highly charged middle region (M) enhances the solubility of Sup35 in the nonprion form and contributes to the stability of the prion form (i.e., the [PSI+] phenotype) (6). Together, Sup35 N and M (designated NM) can serve as a separable prion-forming module that is transferable to heterologous proteins, conferring prion behavior on the resulting fusion proteins (19).
Here we show that Sup35 NM can access the prion conformation in E. coli cells, and that this process recapitulates certain features of the conversion process as it occurs in the native yeast context. Our findings thus establish the feasibility of using bacteria-based models for studying prion formation.
Results
Aggregation State of NM-GFP Fusion Protein in E. coli Cells.
To investigate whether the prion-forming module of Sup35 (hereafter referred to as NM) can form amyloid aggregates in E. coli cells, we constructed a plasmid vector designed to direct the inducible synthesis of high levels of NM fused to GFP. To facilitate our analysis, we used two well-studied NM variants in addition to the WT NM moiety: one (NMRΔ) that has a greatly reduced ability to undergo spontaneous conversion to the prion form in yeast cells and another (NMR2E2) that has a greatly enhanced ability to convert to the prion form (20). These variants differ from the WT (NMWT) in having either fewer (NMRΔ) or more (NMR2E2) copies of the critical oligopeptide repeat sequence (Fig. 1A).
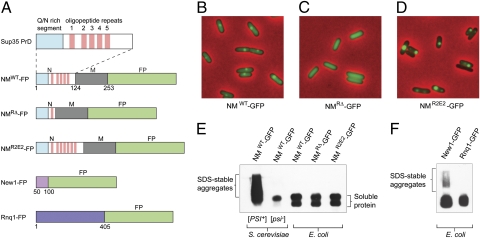
Fig. 1.
Behavior of yeast prion-forming domain fusion proteins in E. coli cells. (A) Fusion constructs. Indicated residues from yeast prion-forming proteins, Sup35 (NM), New1, or Rnq1, were fused to a fluorescent protein (FP). FPs used are GFP, YFP, and CFP. The prion-forming domain of NM is enlarged to highlight its Q/N-rich segment and five complete copies of an oligopeptide repeat sequence; NMRΔ-FP lacks repeats 2–5, and NMR2E2-FP has two additional copies of repeat 2. Constructs containing CFP have three HA-epitope tags fused at the C-terminal end. Fusion genes encoding either the GFP or YFP moiety were expressed under the control of the same arabinose-inducible promoter, whereas genes encoding the CFP moiety were expressed under the control of an isopropyl beta-D-thiogalactopyranoside (IPTG)-inducible promoter. (B–D) Fluorescence images of cells containing the indicated NM-GFP fusion protein. The cells were transformed with plasmids encoding each fusion protein under the control of an inducible promoter. The images show cells examined after the induction of fusion protein synthesis for 5 h. Foci were not observed in cells containing the NMRΔ-GFP fusion protein. Images are representative fields; in each case, several hundred cells were examined. (E) SDD-AGE analysis of cell extracts containing the indicated NM-GFP fusion protein. Blot was probed with an anti-GFP antibody. SDS-stable aggregates migrate as smears above soluble material. Soluble NM-GFP fusion protein (from S. cerevisiae as well as E. coli) migrates as multiple species. Extracts were examined by SDD-AGE analysis in at least three experiments, with similar results. (F) SDD-AGE analysis of cell extracts containing the indicated fusion protein. Blot was probed with an anti-GFP antibody. SDS-stable aggregates migrate as smears above the soluble material. SDS/PAGE and Western blot analysis of cell extracts containing these fusion proteins as well as the NM-GFP fusion protein (A) suggests that observed differences in aggregation behavior are unlikely due to differences in intracellular fusion protein levels (Fig. S1B). Extracts were examined by SDD-AGE analysis in at least three experiments, with similar results.
Cells containing each of the fusion proteins were examined by fluorescence microscopy at various times after the induction of fusion protein synthesis. Over time, cells containing the NMR2E2-GFP fusion protein developed one or two brilliant fluorescent foci (consistent with fusion protein aggregation); cells containing the NMWT-GFP fusion protein developed somewhat less brilliant foci; and cells containing the NMRΔ-GFP fusion protein exhibited diffuse fluorescence only (Fig. 1 B–D). The presence of visible fluorescent foci in cells containing either the NMWT-GFP or the NMR2E2-GFP fusion protein, but not in cells containing the NMRΔ-GFP fusion protein, is consistent with what is observed in [PSI+] yeast cells producing these fusion proteins (20). SDS/PAGE and Western blot analysis of the cell extracts indicated that these differences in aggregation behavior were not due to differences in intracellular fusion protein levels (Fig. S1A).
To determine whether E. coli cells exhibiting fluorescent foci reminiscent of those seen in [PSI+] yeast cells also contained NM-GFP fusion protein in an amyloid-like conformation, we analyzed the bacterial cell extracts using semidenaturing detergent agarose gel electrophoresis (SDD-AGE), which permits the visualization of SDS-stable amyloid polymers (21–23). Consistent with previous observations, we readily detected the higher-molecular-weight smear characteristic of Sup35-derived amyloid when we examined extract derived from [PSI+] yeast cells containing plasmid-encoded NM-GFP; in contrast, we detected only soluble protein when we examined the bacterial extracts (Fig. 1E). Examination of E. coli extracts containing a similar NM-YFP fusion protein also revealed mostly soluble material, although faint higher-molecular-weight smears were detectable in some experiments (Fig. 2). The latter observation suggests that although the bulk of the NM-GFP fusion protein has evidently not accessed a mature amyloid conformation in E. coli despite the appearance of fluorescent foci in cells containing either the NMWT-GFP or the NMR2E2-GFP fusion protein, a small fraction (below the threshold of detection in the experiment of Fig. 1E) may be in the amyloid conformation. Our results below are consistent with this inference.
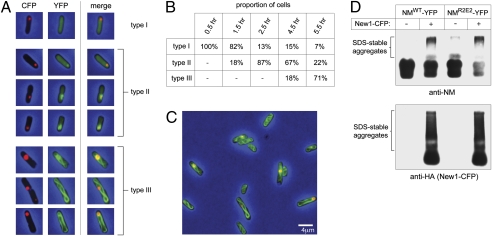
Fig. 2.
Behavior of NM-YFP fusion protein in E. coli cells also containing New1-CFP fusion protein. Cells were transformed with two compatible plasmids, one encoding the NMWT-YFP fusion protein under the control of an arabinose-inducible promoter and the other encoding the New1-CFP fusion protein under the control of a leaky IPTG-inducible promoter. The cells were grown in the absence of IPTG and arabinose was added to induce the synthesis of the NMWT-YFP fusion protein. Sufficient New1-CFP fusion protein was produced under these conditions to form detectable fluorescent foci and SDS-stable material. (A) Fluorescence images of representative cells containing both fusion proteins, 1.5 h after induction of NM-YFP fusion protein synthesis (type I), 2.5 h after induction of NM-YFP fusion protein synthesis (type II), and 5.5 h after induction of NM-YFP fusion protein synthesis (type III). Shown are CFP channel images, YFP channel images, and merged images. (B) Table showing proportion of cells of each type (I, II, or III, as shown in A) at indicated times after induction of NM-YFP fusion protein synthesis. (C) Fluorescence image of a field of cells containing both fusion proteins, 5.5 h after induction of NM-YFP fusion protein synthesis. Similar ribbon-like structures were observed in cells containing New1-CFP together with NMR2E2-YFP (Fig. S3) (D) SDD-AGE analysis of cell extracts containing either NM-YFP or both NM-YFP and New1-CFP, 5.5 h after induction of NM-YFP fusion protein synthesis. SDS-stable NM-YFP aggregates migrate near the top of the gel. Blot was probed with anti-NM antibody to detect NM-YFP (Upper) and anti-HA antibody to detect New1-CFP (which bears three copies of an HA tag at its C terminus) (Lower). Extracts were examined by SDD-AGE in at least two experiments, with similar results.
Yeast-Derived [PSI+] Inducibility Factor Alters Aggregation State of NM Fusion Protein in E. coli Cells.
In yeast, but not in vitro, the spontaneous conversion of Sup35 to the prion form requires the presence of a [PSI+] inducibility (PIN) factor, which is itself a prion (24, 10, 11). Several different yeast proteins, including Rnq1 and New1, can serve as PIN factors in their prion forms (10, 11), and yeast strains containing one or another PIN factor are referred to as [PIN+] strains. The PIN dependence of spontaneous conversion of Sup35 to the prion form in yeast led us to ask whether the introduction of a potential PIN factor into E. coli cells might enhance conversion of Sup35 NM to a prion form in those cells. Accordingly, we sought to examine the aggregation status of NM in E. coli cells also containing either an Rnq1 or a New1 fusion protein. Because Rnq1 and New1 must convert to their respective prion forms to serve as PIN factors in yeast, we first constructed Rnq1-GFP (25) and New1-GFP (26) fusion proteins (Fig. 1A) and examined the behavior of these fusion proteins in E. coli cells. After the induction of fusion protein synthesis, fluorescence microscopy revealed bright foci in cells containing the Rnq1-GFP fusion protein and more irregular aggregate-like structures in cells containing the New1-GFP fusion protein (typically one focus or aggregate per cell in each case) (Fig. S2). When we subjected the cell extracts to SDD-AGE, we readily detected higher-molecular-weight, SDS-stable material in the case of the New1-GFP fusion protein but not in the case of the Rnq1-GFP fusion protein (Fig. 1F).
Proceeding with New1, we investigated its effect on the ability of the Sup35 NM moiety to form amyloid-like aggregates in E. coli cells. To facilitate the analysis by fluorescence microscopy of cells containing both NM and New1, we constructed a New1-CFP fusion protein and an NMWT-YFP fusion protein. Using compatible plasmids directing the synthesis of the two fusion proteins, we followed the cells over time after the induction of NM-YFP synthesis. Initially, NM-YFP fluorescence was diffuse throughout the cells and only foci of New1-CFP were visible (Fig. 2 A, type I, and B); however, by 2.5 h postinduction, most of the cells contained New1-CFP and NM-YFP foci that appeared to colocalize or NM-YFP fluorescence that appeared to be coalescing around the New1-CFP foci (Fig. 2 A, type II, and B). Strikingly, the NM-YFP fusion protein subsequently formed twisted ribbons that traversed the length of the cell (Fig. 2 A, type III, and B and C); although these ribbon-like structures appeared to emanate from New1-CFP foci, they contained detectable YFP, but not CFP, fluorescence, as would be expected if the presence of a PIN factor were stimulating the formation of NM-YFP homopolymers. Control experiments indicated that the NM-YFP fusion protein did not form twisted ribbons in cells lacking the New1 fusion protein (Fig. S3). Furthermore, SDD-AGE analysis indicated that the presence of the New1-CFP fusion protein together with the NM-YFP fusion protein stimulated the conversion of the latter to a higher-molecular-weight, SDS-stable form (Fig. 2D). Thus, in the case of NMWT-YFP, we detected a higher-molecular-weight smear (using an anti-NM antibody) specifically in the presence of New1-CFP; and in the case of NMR2E2-YFP, we detected a faint smear that became more prominent in the presence of New1-CFP. As expected, we also detected SDS-stable New1-CFP aggregates in cells containing both fusion proteins (Fig. 2D).
Yeast-Derived PIN Factor Stimulates Production of Infectious NM Fusion Protein in E. coli Cells.
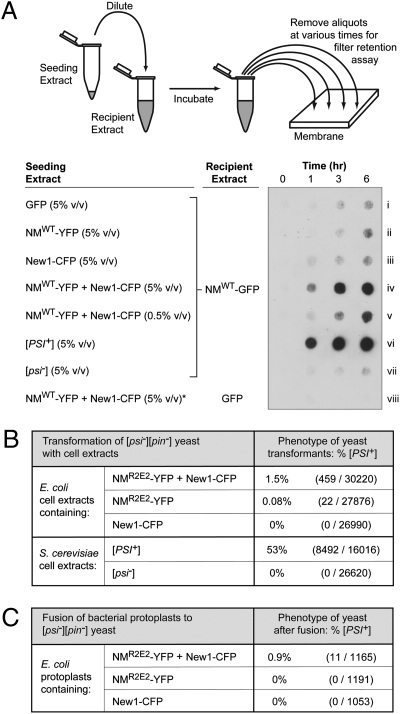
We next sought to determine whether the NM-YFP fusion protein had accessed an infectious prion conformation in E. coli cells containing twisted ribbons and higher-molecular-weight, SDS-stable NM material. As a first test, we investigated whether the cells contained material capable of seeding the conversion of nonamyloid NM fusion protein to the higher-molecular-weight, SDS-stable form. To do this, we diluted extract prepared from cells containing both the New1 and the NM fusion proteins (i.e., cells containing NM fusion protein in the twisted ribbon conformation and in a higher-molecular-weight, SDS-stable form; Fig. 2) into extract prepared from cells containing the NM-GFP fusion protein only (i.e., cells containing predominantly nonamyloid NM fusion protein). Using a filter retention assay (14), we monitored the appearance of higher-molecular-weight, SDS-stable NM material over time (detectable with an anti-NM antibody). For comparison, we tested the seeding activity of extract prepared from cells containing either the New1 fusion protein only (i.e., New1-CFP) or the NM fusion protein only (i.e., NM-YFP); as a negative control, we used extract prepared from cells containing overproduced GFP as mock seed. NM is known to undergo slow, spontaneous conversion to the amyloid form in vitro, a reaction that is accelerated in the presence of preassembled seed particles (17, 27). As expected based on these observations, we detected the accumulation of a relatively small amount of NM-GFP fusion protein in a higher-molecular-weight, SDS-stable form in the negative control reaction (Fig. 3A, row i) and also when either the NM-YFP extract or the New1-CFP extract was used as seed (Fig. 3 A, rows ii and iii). Unlike the extracts containing one or the other fusion protein, the extract prepared from cells containing both the NM-YFP and the New1-CFP fusion proteins dramatically stimulated the conversion of the soluble NM-GFP fusion protein to a higher-molecular-weight, SDS-stable form, and this effect was dose dependent (Fig. 3A, rows iv and v). We conclude that the presence of the New1 fusion protein, together with the NM fusion protein in E. coli cells, facilitates the conversion of the latter to a higher-molecular-weight, SDS-stable form that can seed the conversion of soluble NM-GFP fusion protein to the same form. Additional control reactions confirmed the expected presence and absence of seeding activity in [PSI+] and [psi–] yeast extracts, respectively (Fig. 3A, rows vi and vii), and demonstrated that no detectable higher-molecular-weight, SDS-stable material accumulated when an aliquot of the seeding competent E. coli extract was diluted into extract containing GFP only (Fig. 3A, row viii).
Fig. 3.
Cells with both NM-YFP and New1-CFP aggregates contain seeding-competent, infectious material. (A) E. coli cell extracts containing SDS-soluble NM-GFP were seeded with E. coli cell extract containing overproduced GFP, overproduced NM-YFP, overproduced New1-CFP, or overproduced NM-YFP and New1-CFP; yeast cell extract prepared from either a [PSI+] or a [psi–] strain was also used as seed (strains YJW96 and SG775, respectively). A seed-only control sample (*) consisted of E. coli cell extract containing overproduced NM-YFP and New1-CFP diluted into E. coli extract containing overproduced GFP only. Cartoon depicts experimental protocol. Samples from seeded reactions were removed at various time points, treated with 2% SDS, and filtered through a cellulose acetate (low-binding) membrane. SDS-stable aggregates that were retained were probed with anti-NM antibody. Extracts were examined for seeding activity in three experiments, with similar results. (B) Infection of [pin–][psi–] yeast spheroplasts with extract prepared from E. coli cells containing the indicated fusion proteins, or with extract prepared from either [PSI+] or [psi–] yeast cells (strains YJW96 and SG775, respectively). The New1-CFP fusion protein was encoded on the chromosome under the control of an IPTG-inducible promoter, and NMR2E2-YFP fusion protein was encoded on a plasmid under the control of an arabinose-inducible promoter. Extracts were prepared from cells 6.5 h after induction of fusion protein synthesis with IPTG and arabinose. Yeast spheroplasts were cotransformed with a yeast shuttle vector containing a URA+ selectable marker. [PSI+] transformants exhibited primarily a “strong” phenotype (3). (C) Fusion of [pin–][psi–] yeast spheroplasts with protoplasts prepared from E. coli cells containing the indicated fusion proteins. The New1-CFP fusion protein was encoded on the chromosome under the control of an IPTG-inducible promoter; the NMR2E2-YFP fusion protein was encoded on a plasmid under the control of an arabinose-inducible promoter. Protoplasts were prepared 6.5 h after induction of fusion protein synthesis with IPTG and arabinose. E. coli cells also contained a yeast shuttle vector with a URA+ selectable marker. Analysis of these data by Fisher's exact test suggests that the observed difference in the frequencies of [PSI+] transformants is statistically significant (P = 10−5). All [PSI+] transformants exhibited a “strong” phenotype (3) (Fig. S4).
As a second test for the presence of infectious NM-YFP in cells containing twisted ribbons, we used a previously developed protocol for introducing prion aggregates into yeast cells (28) to investigate whether the bacterially derived NM-YFP fusion protein was capable of infecting [pin–][psi–] yeast cells. Because transient overproduction of Sup35 (or Sup35 NM) in [PIN+] [psi–] strains greatly stimulates the conversion to [PSI+] (24), the use of a [pin–][psi–] recipient strain was critical for this experiment. We prepared extracts from cells containing the New1 and NM fusion proteins in combination, the NM fusion protein only, or the New1 fusion protein only; we then used these extracts (after the addition of plasmid DNA encoding a yeast selectable marker) to transform yeast spheroplasts prepared from a suitable [pin–][psi–] strain. [PSI+] transformants were obtained when we transformed the yeast cells with extract prepared from cells containing both the NM-YFP and the New1-CFP fusion proteins (at a frequency of 1.5%; Fig. 3B) or, at a lower frequency, the NM-YFP protein only (0.08%; Fig. 3B); in contrast, no [PSI+] transformants were obtained when we transformed the yeast cells with extract prepared from cells containing the New1-CFP fusion protein only (Fig. 3B). These results support the idea that the NM-YFP fusion protein can access the infectious prion conformation in E. coli cells; furthermore, comparison of the conversion frequencies (0.08% vs. 1.5%) indicates that the presence of the New1 fusion protein together with the NM fusion protein in E. coli cells facilitates the conversion of the NM fusion protein to the infectious form. As positive and negative controls, we also transformed the yeast spheroplasts with extract prepared from either [PSI+] or [psi–] yeast cells and obtained results that were consistent with the previously published literature (Fig. 3B) (29, 30).
As a third test, we took advantage of a previously developed bacterial protoplast fusion procedure (31) that permits the direct transfer of bacterial cell contents into yeast spheroplasts to ask whether the E. coli cells containing NM-YFP fusion protein in the twisted ribbon conformation harbor material capable of infecting [pin–][psi–] yeast cells. This procedure enabled us to minimize the possibility that any infectious material detected was formed after cell lysis. We prepared bacterial protoplasts from cells containing the New1 and NM fusion proteins in combination, the NM fusion protein only, or the New1 fusion protein only; in addition, these cells contained a shuttle vector encoding a marker to select for transformation of the recipient yeast strain. We fused these protoplasts with yeast spheroplasts prepared from a suitable [pin–][psi–] strain and determined the frequency of conversion to [PSI+] among the transformants. Similar to the results of the extract transformation experiments, the conversion frequency was ∼1% among the transformants that we obtained using protoplasts prepared from cells containing both fusion proteins (Fig. 3C). In contrast, we found no [PSI+] clones among the transformants that we obtained using protoplasts prepared from cells containing either the NM fusion protein only or the New1 fusion protein only (Fig. 3C). These findings reinforce our conclusion that the NM-YFP fusion protein can access the infectious prion conformation in E. coli cells that also contain a PIN factor provided by the New1-CFP fusion protein. Although the bacterial protoplast fusion experiment did not provide evidence for infectious material in cells containing the NM-YFP fusion protein only, we cannot exclude the possibility that such material was present at levels that were below the detection limit.
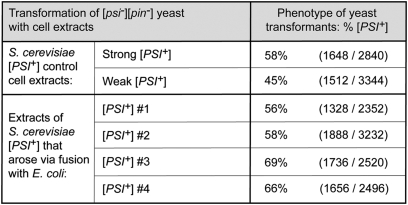
The introduction of bacterially derived material into [pin–][psi–] yeast cells resulted in relatively low [psi–] to [PSI+] conversion frequencies (Fig. 3 B and C). Work in yeast has revealed that Sup35 can adopt distinct amyloid conformations, giving rise to distinct prion strains that result in distinguishable in vivo phenotypes that are stably propagated (i.e., heritable) (32–34, 29, 30). We therefore considered the possibility that the specific amyloid conformation adopted by the NM-YFP fusion protein in E. coli limits its infectivity when transferred to yeast cells (see, for example, ref. 35). To test this, we prepared extracts from [PSI+] yeast cells that arose via bacterial protoplast fusion and from control [PSI+] yeast cells, and used these extracts to transform [pin–][psi–] yeast spheroplasts. The frequency of conversion to [PSI+] among the transformants was similar in the two cases (Fig. 4). We therefore do not favor the possibility that the bacterially derived amyloid consists of an inherently low-infectivity conformer. Furthermore, when we performed seeding reactions similar to those in Fig. 3A using either [PSI+] yeast extract or E. coli twisted ribbons extract as the seed, we found that the resulting material was comparably infectious (Fig. S5), also suggesting that the bacterially derived NM fusion protein has not adopted an alternative and less infectious amyloid conformation.
Fig. 4.
Material derived from [PSI+] yeast strains that arose via fusion with E. coli protoplasts is similarly infectious to material derived from control [PSI+] yeast strains. Extracts of four representative [PSI+] yeast strains obtained via fusion with E. coli protoplasts (labeled [PSI+] #1–4) as well as control extracts from both strong (SG862) and weak (SG863) [PSI+] yeast strains were used to transform [pin−] [psi−] yeast cells. Frequency of [PSI+] observed (as percentage of total transformants) is shown. Strain details are given in Table S1; colony phenotypes are shown in Fig. S4.
Discussion
Our results establish that the Sup35 NM moiety can access an infectious prion conformation in E. coli cells in a PIN-facilitated manner. Purified Sup35 (or NM-GFP) can form amyloid fibrils in vitro, and this in vitro generated material is infectious when introduced into yeast cells (36). Nevertheless, in yeast cells, spontaneous conversion of Sup35 to the prion form is dependent on the presence of a PIN factor (10, 11, 24). Furthermore, cellular chaperones, which are required for the stable propagation of the prion state (15, 16), have also been implicated in the de novo formation of [PSI+] (refs. 37 and references therein, 38). It is striking that the stimulatory effect of PIN has been recapitulated in bacterial cells, which are thought to have diverged from eukaryotes 2.2 billion years ago (39), and it will be interesting to learn how the bacterial chaperone environment influences the formation of infectious amyloid.
As noted above, transfer of protein from E. coli cells containing both the New1 and NM fusion proteins to [pin–][psi–] yeast cells resulted in relatively low [psi–] to [PSI+] conversion frequencies (Fig. 3 B and C). We suggest that the ribbon-like aggregates that are formed in E. coli cells containing both the New1 and NM fusion proteins may provide relatively few free ends to nucleate the polymerization of soluble Sup35, leading to a low seeding efficiency. Consistent with this suggestion, when high-molecular-weight aggregates were isolated from the bacterial extracts by centrifugation and subsequently fragmented by sonication, the infectivity of the resulting material increased, and the average NM polymer size decreased (Fig. S6). In yeast cells the fragmentation of prion fibers into seeding-competent oligomeric species (known as propagons), which is mediated principally by the Hsp104 chaperone system, is thought to be essential for the stable propagation of [PSI+] through multiple cell divisions (21, 40–43). Whether the E. coli chaperone environment can be altered to increase the infectivity of material extracted from cells containing both the New1 and the NM fusion proteins remains to be investigated.
The mechanistic basis for the ability of [PIN+] to facilitate the conversion of Sup35 to the prion form has not been fully elucidated; however, several lines of evidence are consistent with a direct cross-seeding mechanism (ref. 44 and references therein), which is also supported by our findings that (i) [PIN+] can function in a heterologous cellular environment lacking other yeast factors, and (ii) New1-CFP and NM-YFP aggregates appear to colocalize before the formation of twisted ribbons consisting of NM-YFP but no detectable New1-CFP. In addition to providing a heterologous system in which to study prion biology, the demonstration that the E. coli cytoplasm can support the formation of infectious amyloid raises the possibility that endogenous bacterial proteins exist that might take advantage of this capacity to function as epigenetic switches.
Materials and Methods
Plasmids, Strains, and Cell Growth.
Overnight cultures of bacterial strains were diluted to OD600 0.02 in LB supplemented with the appropriate antibiotics (carbenicillin 100 μg/mL; chloramphenicol 25 μg/mL), grown for 30 min at 30 °C and induced with the appropriate inducers (L-Arabinose at 0.02% wt/vol; IPTG at 1 mM). Additional details on plasmids and strains are given in SI Materials and Methods and Table S1.
Fluorescence Microscopy.
Cells were harvested at 3,000 × g, resuspended in PBS, and spotted onto agarose pads (1% wt/vol in PBS; Seakem LE Agarose, Lonza) for visualization with a UplanFLN 100× phase contrast objective on an Olympus BX61 microscope as described elsewhere (45). Images were captured with a CoolSnapHQ camera (Photometrics) and the Metamorph software package, version 6.1 (Universal Imaging). Exposures were typically 50–200 ms. For the comparison in Fig. 2B, a total of 701 cells that produced detectable levels of both fluorescent fusion proteins (NM-YFP and New1-CFP-3×HA) were counted.
Bacterial Extract Preparation.
Cultures (50 mL) were grown to the indicated times and the cell densities were recorded (OD600). The cultures were centrifuged 10 min at 3,000 × g, supernatants removed, and the pellets frozen on dry ice. Pellets were thawed and resuspended in buffer STC (1 M sorbitol, 10 mM Tris pH 7.5, 10 mM CaCl2) based on 1 mL buffer for 50 mL culture (OD600 = 1.0). rLysozme (EMD Biosciences) was added to 3,000 units/mL and the mixture was incubated, rocking, at room temperature (RT) for 30 min. The lysozyme-treated cell suspension was frozen on dry ice and thawed on ice water to complete lysis. Cell debris was removed from the lysate by two sequential rounds of centrifugation, each at 500 × g for 15 min at 4 °C. Clarified lysates were normalized for total protein as assayed by Bradford or bicinchoninic acid (ThermoFisher).
Yeast Extract Preparation.
[psi−] or [PSI+] yeast strains producing NMWT-GFP under the control of the CUP1 promoter (Table S1) were grown at 30 °C in SD–URA media and induced with 50 μM CuSO4 and grown for an additional 4 h. All other yeast strains (Table S1) were grown at 30 °C in YPD media. Log-phase cells were pelleted, and resuspended in 1/100th volume STC buffer with 2× Complete EDTA-free protease inhibitor (Roche). The resuspension was combined with 1 volume acid-washed glass beads (400 μm) and subjected to vortexing for 5 min at 4 °C. Unlysed yeast cells and cell debris were removed by two sequential rounds of centrifugation, each at 500 × g for 15 min at 4°C. The supernatant was taken and normalized for total protein concentration as determined by Bradford assay or bicinchoninic acid (ThermoFisher).
Semidenaturing Detergent Agarose Gel Electrophoresis.
SDD-AGE was performed as previously described (23) using 1.5% agarose. Blots were probed with anti-GFP, anti-HA (clone 3F10; Roche), or anti-Sup35 yS-20 (Santa Cruz Biotechnology). Secondary antibodies were HRP-conjugated antimouse IgG (Cell Signaling 7076), antirat IgG (Abcam ab6734), and antigoat IgG (Santa Cruz Biotechnology sc-2354). Blots were detected using an ECLplus Western Blotting Detection System (GE Heathcare).
Extract Seeding and Filter Retention Assay.
Bacterial cell extract containing NM-GFP was adjusted with buffer STC to 2 mg/mL total protein, whereas extracts that were used as seed were adjusted to 1 mg/mL total protein. The indicated amounts of the various seeds were added to extracts of NM-GFP, and the reactions were incubated at room temperature without agitation. Samples were removed at the indicated times, treated with 2% SDS, and frozen on dry ice. After the final time point, samples were thawed and applied to a 0.22-μm cellulose acetate membrane (GE Osmonics Labstore) as described elsewhere (46). The experimental protocol is further outlined in SI Materials and Methods.
Protein Extract Transformations.
A 25-μL quantity of bacterial or yeast extract (1 mg/mL total protein in Fig. 3 and 2 mg/mL in Fig. 4) was used to transform 100 μL of [pin–] [psi–] yeast spheroplasts as previously described (28), with the exception that yeast spheroplasts were formed using lyticase (Sigma L5263) in buffer ST (1 M sorbitol, 10 mM Tris, pH 7.5) under conditions that caused more than 95% of yeast cells to form spheroplasts in 30–60 min. [PSI+] transformants were scored as described (28).
Bacteria and Yeast Fusions.
Fusion of bacterial protoplasts to [pin–][psi–] yeast spheroplasts was performed as described elsewhere (31), with the following alterations. Protoplasts of bacteria containing yeast shuttle vector pSG378 were formed using the PeriPreps Kit (Epicentre) as described by the manufacturer, pelleted at 3,000 × g, and resuspended in STC buffer. Bacterial protoplasts and yeast spheroplasts were mixed and incubated for 5 min at RT. Fusion was induced by the addition of 9 volumes PEG buffer (20% wt/vol PEG 8,000, 10 mM Tris, pH 7.5, and 10 mM CaCl2) and allowed to proceed for 30 min at RT. The fusion reactions were pelleted for 1.5 min at 200 × g, gently resuspended in SOS buffer (1 M sorbitol, 7 mM CaCl2, 0.25% wt/vol yeast extract, 0.5% wt/vol bacto-peptone), and allowed to recover for 30 min at 30 °C. The recovered reactions were plated in SD-URA top agar with 10 mg/mL adenine. Putative [PSI+] URA+ transformants were identified as previously described (28). [PSI+] was confirmed by examining colony color on 1/4 YPD plates and by curing via passage on 3 mM GuHCl (28). Additional details are given in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Tanaka and J. Weissman (University of California, San Francisco, CA) for reagents and guidance on protein transformations, R. Cohen-Kupiec for preliminary work with Sup35NM-GFP, B. Bukau and P. Deighan for valuable discussion, R. Halfmann for helpful advice, and S. Dove for comments on the manuscript. This work was supported by a National Institutes of Health Pioneer Award OD003806 (to A.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913280107/-/DCSupplemental.
References
- 1.Aguzzi A, Polymenidou M. Mammalian prion biology: One century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 2.Wickner RB, Liebman SW, Saupe SJ. Prions of yeast and filamentous fungi: [URE3+], [PSI+], [PIN+], and [Het-s] In: Prusiner SB, editor. Prion Biology and Diseases. Cold Spring Harbor Lab Press: Cold Spring Harbor, NY; 2004. pp. 305–372. [Google Scholar]
- 3.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 4.Pan KM, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura Y, Koitabashi S, Fujita T. Analysis of yeast prion aggregates with amyloid-staining compound in vivo. Cell Struct Funct. 2003;28:187–193. doi: 10.1247/csf.28.187. [DOI] [PubMed] [Google Scholar]
- 6.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 7.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 8.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 11.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 16.True HL. The battle of the fold: Chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 18.Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 20.Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 21.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 22.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 23.Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. July 16, 2008;(17) doi: 10.3791/838. 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 26.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:0442–0451. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka M, Weissman JS. An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 2006;412:185–200. doi: 10.1016/S0076-6879(06)12012-1. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 30.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 31.Gyuris J, Duda EG. High-efficiency transformation of Saccharomyces cerevisiae cells by bacterial minicell protoplast fusion. Mol Cell Biol. 1986;6:3295–3297. doi: 10.1128/mcb.6.9.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 33.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 35.Krammer C, et al. The yeast Sup35NM domain propagates as a prion in mammalian cells. Proc Natl Acad Sci USA. 2009;106:462–467. doi: 10.1073/pnas.0811571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparrer HE, Santoso A, Szoka FCJ, Jr, Weissman JS. Evidence for the prion hypothesis: Induction of the yeast [PSI+] factor by in vitro-converted Sup35 protein. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- 37.Tuite M, Stojanovski K, Ness F, Merritt G, Koloteva-Levine N. Cellular factors important for the de novo formation of yeast prions. Biochem Soc Trans. 2008;36:1083–1087. doi: 10.1042/BST0361083. [DOI] [PubMed] [Google Scholar]
- 38.Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andréasson C, Lindquist S, Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One. 2007;3:e1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng DF, Cho G, Doolittle RF. Determining divergence times with a protein clock: Update and reevaluation. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007 doi: 10.1371/journal.pbio.0050024. 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derkatch IL, Liebman SW. Prion-prion interactions. Prion. 2007;1:161–169. doi: 10.4161/pri.1.3.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 46.Boyé-Harnasch M, Cullin C. A novel in vitro filter trap assay identifies tannic acid as an amyloid aggregation inducer for HET-s. J Biotechnol. 2006;125:222–230. doi: 10.1016/j.jbiotec.2006.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.