Natural killer (NK) cells are key players in the innate immune system that provide rapid responses through cytokine secretion and direct lysis of stressed, infected, or transformed target cells (1). This capacity to unleash a powerful response is kept in check by inhibitory receptors such as the killer cell Ig-like receptor (KIR) family on human NK cells that recognize major histocompatibility complex class I (MHC-I) ligands on most somatic cells (2). The importance of interactions between MHC-I and KIR loci is underscored by associations of human leukocyte antigen (HLA)–KIR compound genotypes with infectious diseases, cancer, and other disorders (3). Inhibitory signaling by KIR results in a complete and proximal block of NK activation signals through recruitment of tyrosine phosphatase SHP-1 to the immunoreceptor tyrosine-based inhibition motifs (ITIM) in their cytoplasmic tail (4). Recognition of MHC-I by KIR and the resulting inhibition is influenced by the amino acid sequence of the peptide bound to MHC-I (5). In PNAS, Fadda et al. (6) now show that NK inhibition by peptide-MHC (pMHC) complexes on target cells can be overcome by specific peptide sequence variants that function as antagonists (6).
Receptor antagonism signifies not simply a lack of response but dominant inhibition of responses to agonist ligands. For instance, T-cell antagonism occurs when substitutions in certain T-cell receptor (TCR) contact residues in the pMHC not only fail to induce responses on their own but inhibit responses to the original pMHC epitopes. A unique aspect of the study by Fadda et al. (6) is that it is the inhibition of activation signals in NK cells that becomes the target of antagonist pMHC, leading to NK cell activation. The major significance of their work is that NK cells have the potential to be activated by even small fluctuations in peptide repertoires, as could arise during infections.
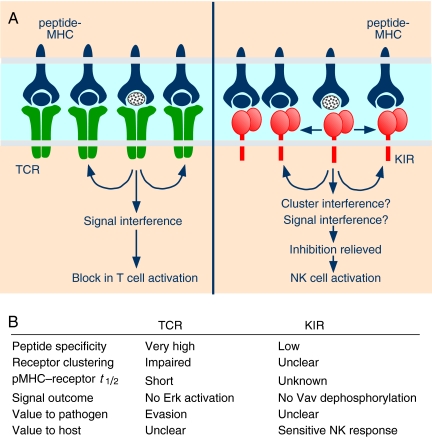
This unexpected activation of NK function by antagonist pMHC is not unlike T cell antagonism, which has been studied extensively (7–9). In the case of T cells, a shorter half-life of antagonist pMHC–TCR interactions favors a negative feedback loop, which becomes dominant and interferes with proper signaling by other TCRs (Fig. 1). Fundamental differences in the recognition of MHC-I by T cells and NK cells are worth reviewing before comparing peptide antagonism in T cells and NK cells. T-cell activation requires the productive engagement of antigen-specific TCR by pMHC on the surface of antigen-presenting cells. TCRs exhibit exquisite specificity for pMHC. In contrast, each germ-line-encoded KIR can accommodate many different peptides in the context of a large number of polymorphic MHC-I allotypes. Amino acid substitutions in the peptide side chains that point out of the MHC-I peptide binding site have the potential to interfere with KIR binding (10, 11). Although a given KIR can bind to MHC-I loaded with an array of peptides, recognition is sensitive to the nature of the side chains at positions P7 and P8 of the nonamer peptide (5). Thus, a wide array of peptides are competent to induce inhibition, and the requirements for productive peptide–MHC combinations are much less stringent for KIR than for TCR.
Fig. 1.
Peptide antagonism in T cells and NK cells. (A) MHC molecules loaded with different peptides, including an antagonist peptide (stippled), interact with TCR on T cells and with inhibitory KIR on NK cells. TCR engaged by antagonist pMHC favors feedback inhibition, which interferes with Erk activation by other TCRs. KIR engaged by antagonist pMHC accumulates at the immunological synapse and, by an unknown mechanism, interferes with Vav dephosphorylation. (B) Features of peptide antagonism in T cells and NK cells.
Using a series of peptide analogs, Fadda et al. show that some HLA-C binding peptides function as antagonists and relieve KIR-mediated inhibition of NK cell activation. They identified peptides that promote comparable HLA-C expression at the cell surface but differ in their ability to support recognition by KIR. (A cell line deficient in the transporter for antigen presentation was used to directly load defined, individual peptides onto HLA-C.) They further identified peptides that did not permit detectable KIR binding and yet relieved the inhibition mediated by a peptide that strongly supported KIR recognition of HLA-C. Such peptide antagonism is independent of KIR genotype, as there was no difference in antagonism among individuals with different KIR genotypes. Moreover, the relative concentrations of agonist and antagonist peptides determined the functional recognition of HLA-C by KIR. As seen with T cells, a binding threshold distinguishes pMHC with weak affinity for KIR from those that are antagonists. Surprisingly, antagonistic peptides were capable of promoting KIR clustering at the NK–target cell synapse. This is in contrast to T-cell antagonism, in which altered peptide ligands interfere with proper MHC clustering at the immune synapse (8). However, despite detectable KIR accumulation at the NK–target cell interface, inhibitory signaling failed, as shown by surface CD107a (a readout of NK degranulation), microtubule-organizing center (MTOC) polarization to the synapse, and the lack of dephosphorylation of the primary SHP-1 substrate Vav1.
How could KIR engaged by antagonist pMHC interfere with inhibitory signaling by neighboring KIRs at NK–target cell synapses? Perhaps SHP-1 recruited by antagonized KIR dephosphorylates ITIMs in other KIRs. Alternatively, antagonized KIR may transform into an activation receptor. However, in that case, it would have to induce signals that are resistant to standard KIR-mediated inhibition. Most of the evidence so far points to a dominant role of KIR inhibitory signaling over activation signals (4). A more appealing hypothesis may be that interference by antagonized KIR occurs at the level of regulated KIR clustering (Fig. 1). Such antagonism would be possible if inhibitory signaling by KIR requires the formation of well-organized multimers. It is not known whether inhibitory KIR microclusters (12) still form in the presence of antagonisticpeptides and whether the formation of qualitatively different microclusters would somehow hinder inhibitory signaling by KIR. Deciphering how antagonistic pMHC uncouples KIR clustering from inhibitory signaling may provide fascinating insights into the mechanism of inhibition
KIR antagonism would favor NK cell responsiveness in the host.
In T-cell antagonism, which offers pathogens an immune evasion strategy through mutation of epitopes into antagonist peptides, there is no obvious advantage to the host. In contrast, KIR antagonism would favor NK cell responsiveness in the host, as it could be a sensitive sensor of changes in the repertoire of peptides bound to MHC-I. However, such a system of recognition would have to occur by chance. The extensive polymorphism of HLA-C and the recognition of multiple HLA-C allotypes, which bind different peptides, by a single inhibitory KIR preclude self versus non-self discrimination. Therefore, antagonism by endogenous self peptides is inevitable. Antagonism, which results in weak inhibitory signaling, would also influence NK cell “licensing,” a process by which the intrinsic responsiveness of each NK cell is calibrated according to the strength of inhibitory signaling (13, 14). Nevertheless, the system of peptide antagonism of KIR inhibition, described here by Fadda et al., could represent a major strategy to mount rapid and sensitive NK responses to virus infections. Changes in the peptide repertoire of infected cells will cause either increased MHC-I occupancy by agonists (antagonist self peptides displaced by agonists), no change in the overall agonist–antagonist balance, or increased occupancy by antagonists (agonist self peptides displaced by antagonists). In the first two scenarios, inhibition by KIR is retained, and NK cell activation depends on MHC-I down-regulation or up-regulation of ligands for activation receptors. In the third, however, NK cells can attack infected cells even in the absence of MHC-I down-regulation. Such an ability to mount NK responses to small changes in peptide repertoire is a formidable weapon, albeit one that does not fire every time. Furthermore, as each NK cell expresses its own subset of inhibitory KIRs with distinct MHC-I specificity, antagonism of any one of those KIRs may suffice to mount an antiviral response. By exploiting the peptide selectivity of inhibitory receptors, NK cells have one more weapon in their arsenal to sense and respond quickly to alterations in their environment.
Footnotes
The authors declare no conflict of interest.
See companion article on page 10160 in issue 22 of volume 107.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 3.Kulkarni S, Martin MP, Carrington M. The yin and yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long EO. Negative signaling by inhibitory receptors: The NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long EO, Rajagopalan S. HLA class I recognition by killer cell Ig-like receptors. Semin Immunol. 2000;12:101–108. doi: 10.1006/smim.2000.0212. [DOI] [PubMed] [Google Scholar]
- 6.Fadda L, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci USA. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefanova I, Dorfman JR, Tsukamoto M, Germain RN. On the role of self-recognition in T cell responses to foreign antigen. Immunol Rev. 2003;191:97–106. doi: 10.1034/j.1600-065x.2003.00006.x. [DOI] [PubMed] [Google Scholar]
- 8.Sumen C, Dustin ML, Davis MM. T cell receptor antagonism interferes with MHC clustering and integrin patterning during immunological synapse formation. J Cell Biol. 2004;166:579–590. doi: 10.1083/jcb.200404059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wylie DC, Das J, Chakraborty AK. Sensitivity of T cells to antigen and antagonism emerges from differential regulation of the same molecular signaling module. Proc Natl Acad Sci USA. 2007;104:5533–5538. doi: 10.1073/pnas.0611482104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 11.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 12.Treanor B, et al. Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J Cell Biol. 2006;174:153–161. doi: 10.1083/jcb.200601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev. 2006;214:143–154. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 14.Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]