Abstract
Male sex drive rhythm (MSDR) in Drosophila is a circadian behavior only observed in the social context of male-female pairs. In the presence of a female, males exhibit long periods of courtship activity with a pronounced rest phase at dusk, although isolated males exhibit an activity peak at dusk. The molecular mechanisms regulating the switch between these activity patterns are unknown. Here, we genetically manipulate the molecular clock in different subsets of neurons and find that proper oscillation of the molecular clock in ventral lateral neurons is essential for MSDR. These neurons express pigment-dispersing factor, the lack of which disrupts MSDR. Furthermore, we show that a cluster of dorsal neurons (DN1s) requires the molecular clock to synchronize the trough phase at dusk in MSDR and to establish the evening peak in single fly locomotor rhythm (SLR). Finally, we provide evidence that DN1s exert their roles in MSDR and SLR via distinct signaling pathways.
Keywords: circadian rhythm, courtship, molecular clock
Circadian rhythms regulate many cellular processes to optimize physiology, metabolism, and behavior to daily fluctuations of environmental conditions. WT Drosophilia melanogaster males show two locomotor activity peaks under standard light/dark (12:12 LD) cycles, one in the morning and a second one around dusk. This activity rhythm is sustained without external stimuli, such as in constant darkness (12:12 DD), by the intrinsic molecular clock in the brain. In Drosophila, the intrinsic molecular clock is controlled by two clock regulators, PERIOD (PER) and TIMELESS (TIM), both of which are expressed in small groups of neurons (clock neurons), where they oscillate in a 24-h fashion and, in turn, negatively regulate the activity of two transcription factors, CLOCK (CLK) and CYCLE (CYC) (1). CLK and CYC control expression of hundreds of genes believed to play specific roles in the circadian modulation of many behaviors and physiological processes (2, 3). The clock neurons can be subdivided into six distinct clusters: small and large ventral lateral neurons (sLNvs and lLNvs), dorsal lateral neurons (LNds), and three groups of dorsal neurons (DN1s, DN2s, and DN3s) (4). LNvs are essential for establishing the morning activity peak, whereas both LNds and LNvs, are required for the evening activity peak in DD (5, 6).
Although locomotor activity is the most investigated circadian behavior in Drosophila, other behaviors, including courtship and mating (7–9), are under strong circadian influence. We recently demonstrated that male courtship and locomotor activity exhibit a typical cycling pattern when males are paired with females. In contrast to single fly locomotor rhythm (SLR), this courtship rhythm, called male sex drive rhythm (MSDR), is characterized by a trough at subjective dusk, followed by a sharp increase of proximity encounters (i.e., courting) that peak during the subjective night (9). per0 and perS mutant males show arrhythmic and short-period (19.5 ± 0.4 h, n = 14) MSDR, respectively, when they are paired with WT females (9). Moreover, pairs in which males and females were entrained in different time zones exhibit MSDR synchronized with the male's internal clock (9). These findings suggest that MSDR depends on the intrinsic molecular clock of males, at least under the experimental conditions used in this study.
Results
Molecular Cycling (Clock) in a Subset of Clock Neurons Is Required for MSDR.
Asynchrony of MSDR and SLR is characterized by an activity peak and trough at dusk, respectively, and therefore raises intriguing questions. Are the same neural networks that control SLR also necessary for MSDR, and what are the genetic regulators and molecular mechanisms that switch the activity pattern of single males to that of males paired with females? To address these questions, we disrupted the molecular clock in different groups of neurons using the binary GAL4/UAS expression system (10) and by exploiting the dominant negative effect of the CYCΔ protein (11). Unlike effectors that induce cell death, CYCΔ specifically blocks the cycling of the molecular clock but appears not to affect other neuronal functions (6, 11). To facilitate comparison among different GAL4 drivers, we assigned a single numerical value to each genotype— the courtship rest index (CRI), defined as the ratio of the means of courtship during the whole day and around dusk [circadian time (CT) 10 to CT14].
We first tested the potency of the CYCΔ protein by combining UAS-CYCΔ with the panneuronal driver C155-elav-GAL4 (12) (Fig. 1A). Control (UAS-CYCΔ/+ and C155-elav-GAL/+) and WT (Oregon-R) males exhibit a CRI of ∼1.4, which is indicative of a trough of courtship activity at dusk that defines a robust MSDR, whereas C155-elav-GAL4;UAS-CYCΔ males have a CRI of ∼1.1 (P < 0.0001; Fig. 1B), which reflects an almost even distribution of courtship activity throughout the day. We then determined the CRI of 40 CNS- and peripheral nervous system-specific GAL4 drivers in males that also carry UAS-CYCΔ. Six drivers produced a significant (P < 0.01) reduction in the CRI when compared with control or WT males (Fig. 1C and Table S1). The largest reduction was observed with cry-GAL4_2 (Experimental Procedures and Fig. S1A) and tim-GAL4, which are expressed in the core clock neurons in the brain (4, 13). The other lines were c370, reported to be expressed in broad areas of the CNS, including the mushroom bodies (MBs); c772, which appears largely restricted to the MBs (14); and c319, which is expressed sparsely throughout the brain (Fig. S1B). Finally, the CRI was also strongly reduced with the fruGAL4 driver, which is expressed in a cellular network that constitutes a neural circuit for male courtship (15).
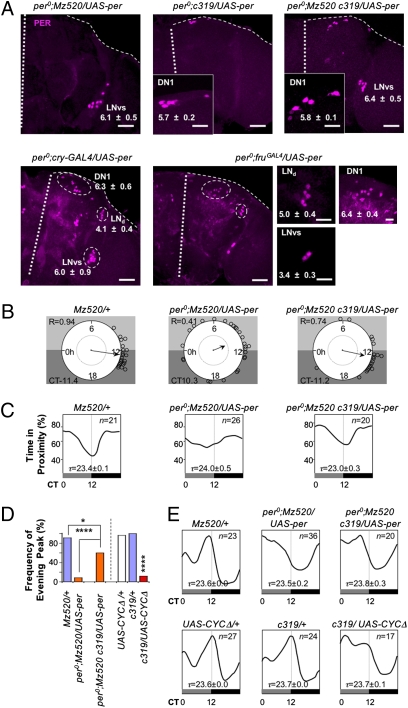
Fig. 1.
Expression of CYCΔ in clock neurons interferes with MSDR. (A) Ectopic expression of CYCΔ in all neurons (Right) abolishes the distinct trough of courtship activity around subjective dusk, while males containing only the driver or reporter alone (two graphs in the Center) show the same MSDR as WT males (Left; n = 16–48). Error bars denote SEM. Two consecutive 24-h periods of the data of time in proximity (time points 13–60) were pooled and are shown as a single 24-h interval. All behavioral tests were done under the DD condition in this study; thus, all black and gray bars indicate subjective night and subjective day, respectively. (B) Loss of MSDR results in a significant reduction of the CRI (****, P < 0.0001, Kruskal–Wallis ANOVA test). Error bars denote SEM. (C) Expression of CYCΔ by the indicated GAL4 drivers reduced the CRI. Levels of significance (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001) were determined in comparison to +/UAS-CYCΔ males, except for UAS-CYCΔ/c772-GAL4 (compared with +/c772-GAL4) (n = 16–55). Error bars denote SEM. Based on the results of Fig. 2D and Fig. S1C, the expression pattern of GAL4 drivers in each clock neuron cluster is indicated as +. (D) Brains of indicated genotypes were stained at ZT2 with anti-PER (magenta) and anti-GFP (green) antibodies. The number of GFP-positive cells in each cluster is indicated in the appropriate frame (n > 8 hemispheres). In this study, all brain hemispheres are oriented with the central brain on the left and optic lobe on the right unless otherwise stated. The images depicting lLNvs may include the PDF-negative fifth sLNv cell, and the DN1s may include some DN2 cells. (Scale bar: 10 μm.)
Except for cry-GAL4 and tim-GAL4, the expression of the other GAL4 drivers reducing the CRI when combined with CYCΔ has not been examined in clock neurons. We therefore determined whether these lines showed expression in some or all clock neurons by performing double-labeling experiments using anti-PER and anti-GFP antibodies on the brains of GAL4/UAS flies. c370 and c772 showed expression in all LNvs and LNds (Fig. S1C), whereas c319 showed strong expression in many DN1s and in one or two cells of the lLNv and LNd clusters (Fig. 1D). We note that another GAL4 driver exclusively expressed in DN1s (Clk4.4F-GAL4; refs. 16 and 17) caused the same phenotype as c319 when driving UAS-CYCΔ (Fig. S2, Table 1, and Table S2). Finally, fruGAL4 is strongly expressed in all sLNvs and LNds, in many DN1s, and occasionally in a single lLNv. These results indicate that all these GAL4 drivers are expressed in at least one of the six clock neuron clusters. However, there is no cluster common to all drivers, suggesting that the molecular clock in multiple groups of clock neurons is involved in establishing MSDR.
Table 1.
MSDRs for 5 days in DD
| Genotype | sLNv | lLNv | LNd | DN1 | Total flies, n | Rhythmic flies (%) | Period, h | Power | Trough phase | Phase concentration |
| Oregon-R | 15 | 11 (73) | 23.7 ± 0.1 | 124 ± 15 | −10.8 | 0.96 | ||||
| UAS-CYCΔ/+ | 17 | 12 (71) | 23.6 ± 0.2 | 88 ± 12 | 11.9 | 0.91 | ||||
| Mz520-GAL4/+ | 32 | 21 (66) | 23.4 ± 0.1 | 102 ± 12 | −11.4 | 0.94 | ||||
| c319-GAL4/+ | 32 | 25 (78) | 23.9 ± 0.1 | 126 ± 10 | −10.9 | 0.96 | ||||
| Clk4.4F-GAL4/+ | 32 | 25(78) | 23.2 ± 0.3 | 111 ± 11 | −10.5 | 0.88 | ||||
| cry-GAL4_2/+ | 17 | 13 (76) | 23.9 ± 0.2 | 91 ± 17 | −10.4 | 0.94 | ||||
| fruGAL4/+ | 26 | 17 (65) | 23.9 ± 0.4 | 80 ± 9 | −11.0 | 0.68 | ||||
| Pdf-GAL80/+;fruGAL4/+ | 18 | 11 (61) | 24.1 ± 0.4 | 109 ± 17 | −11.1 | 0.82 | ||||
| cry-GAL80 fruGAL4/+ | 14 | 10 (71) | 23.3 ± 0.2 | 124 ± 17 | −10.2 | 0.94 | ||||
| Pdf-GAL80/+ | 16 | 14 (88) | 23.7 ± 0.1 | 121 ± 14 | −11.1 | 0.99 | ||||
| Pdf01 | 27 | 4 (15) | 25.2 ± 1.4 | 51 ± 18 | 10.0 | 0.54† | ||||
| fruGAL4 | 30 | 2 (7) | 28.5 ± 2.8* | 43 ± 5 | 4.9 | 1.00† | ||||
| Mz520-GAL4/UAS-CYCΔ | + | + | 12 | 0 (0) | — | — | — | — | ||
| Mz520-GAL4 Pdf-GAL80/UAS-CYCΔ | 14 | 14 (100) | 23.2 ± 0.1* | 84 ± 13 | −10.9 | 0.95 | ||||
| c319-GAL4/UAS-CYCΔ | + | 27 | 6 (22) | 23.3 ± 0.9 | 51 ± 8 | −7.1 | 0.32† | |||
| UAS-CYCΔ/+;Clk4.4F-GAL4/+ | + | 27 | 4 (15) | 24.7 ± 1.9 | 62 ± 26 | −5.5 | 0.79† | |||
| UAS-CYCΔ/+;cry-GAL4_2/+ | + | + | + | + | 16 | 0 (0) | — | — | — | — |
| UAS-CYCΔ/+;fruGAL4/+ | + | + | + | 37 | 9 (24) | 23.0 ± 1.0 | 62 ± 13 | −9.0 | 0.62† | |
| Pdf-GAL80/UAS-CYCΔ;fruGAL4/+ | + | + | 27 | 8 (30) | 22.8 ± 0.7 | 51 ± 7 | −7.9 | 0.71† | ||
| UAS-CYCΔ/+;cry-GAL80 fruGAL4/+ | 26 | 13 (50) | 23.4 ± 0.3 | 117 ± 15 | −10.3 | 0.92 | ||||
| C155-elav-GAL4 per0 w;UAS-per2/+ | + | + | + | + | 24 | 19 (79) | 24.4 ± 0.3* | 93 ± 8 | −10.9 | 0.85 |
| C155-elav-GAL4 per0 w;Pdf-GAL80/+;UAS-per2/+ | + | + | 26 | 1 (4) | 21.7 | 38 | −2.6 | 1.00† | ||
| per0 w;UAS-per16/+ | 18 | 1 (6) | 30.0 | 25 | 0.5 | 1.00† | ||||
| per0 w;Mz520-GAL4/+;UAS-per16/+ | + | + | 43 | 26 (60) | 24.0 ± 0.5 | 78 ± 6 | 10.3 | 0.41† | ||
| per0 w;Mz520-GAL4 Pdf-GAL80/+;UAS-per16/+ | 7 | 1 (14) | 23.7 | 88 | 11.8 | 1.00† | ||||
| per0 w;c319-GAL4/+;UAS-per16/+ | + | 30 | 10 (33) | 23.8 ± 0.5 | 75 ± 12 | −8.9 | 0.82 | |||
| per0 w;Clk4.4F-GAL4/UAS-per16 | + | 22 | 4 (18) | 24.5 ± 0.3* | 57 ± 12 | −9.6 | 0.90† | |||
| per0 w;Mz520-GAL4 c319-GAL4/+;UAS-per16/+ | + | + | + | 36 | 20 (56) | 23.0 ± 0.3* | 87 ± 9 | −11.2 | 0.74 | |
| per0 w;UAS-per16/cry-GAL4_2 | + | + | + | + | 32 | 19 (59) | 22.5 ± 0.6* | 88 ± 11 | −10.2 | 0.69 |
| per0 w;UAS-per16/fruGAL4 | + | + | + | 33 | 20 (61) | 25.8 ± 0.4* | 82 ± 10 | −11.0 | 0.77 |
Genotypes in bold show significant (χ2 test, P < 0.05) decreases in the fraction of rhythmic flies compared with Oregon-R. Clock neurons expressing UAS-CYCΔ or UAS-per in each genotype are indicated by a +. All per0-rescue flies except C155-elav-GAL4 per0 w;Pdf-GAL80/+;UAS-per2/+, per0 w;Mz520-GAL4 Pdf-GAL80/+;UAS-per16/+, and per0 w;Clk4.4F-GAL4/UAS-per16 show significant (χ2 test, P < 0.05) increases in the fraction of rhythmic flies compared with per0 w;UAS-per16/+.
*Significant (Kruskal–Wallis ANOVA test, P < 0.05) period difference compared with Oregon-R.
†Low phase concentration (Rayleigh test, P > 0.01).
Pigment-Dispersing Factor Signaling Is Essential for MSDR.
LNvs, which express pigment-dispersing factor (PDF), play a critical role in SLR. For example, the genetic ablation of these cells leads to the loss of behavioral rhythms in DD (6, 18). Moreover, Pdf01 mutant flies lack SLR in DD (19). Although a Pdf-GAL4 driver, combined with UAS-CYCΔ, did not result in a significant reduction of the CRI in our initial screen (Table S1), a second and presumably more active LNv-specific driver [Mz520-GAL4 (5)] caused severe MSDR (Table 1) and SLR (Table S2) phenotypes, suggesting that LNvs are indeed necessary for generating MSDR. Although the Mz520-GAL4 driver appears to be expressed in one additional (nonclock) neuron per hemisphere when compared with Pdf-GAL4 (Fig. S2), suppression of CYCΔ in clock neurons by Pdf-GAL80 is sufficient to rescue both SLR and MSDR phenotypes in Mz520-GAL4;UAS-CYCΔ males (Table 1 and Table S2). A crucial role for the LNvs in MSDR is further supported by the finding that Pdf01 mutant males and males lacking PER expression in LNvs (C155-elav-GAL4 per0;Pdf-GAL80/UAS-per) also exhibited a strong MSDR phenotype (Table 1 and Table S2). Pdf-GAL80 is expressed in all PDF-positive LNvs (6) and represses transcription of the UAS reporter in these cells. Taken together, these results indicate that a molecular clock in LNvs plays not only a crucial role for SLR but represents a critical cellular component for proper MSDR.
The experiments described thus far address which cells are necessary to generate MSDR. To determine which clock neurons are sufficient for MSDR, we first combined UAS-CYCΔ and fruGAL4 with Pdf-GAL80 or cry-GAL80. cry-GAL80 is expressed in all LNvs and LNds and in some DN1s (6) (Fig. S3A). Restoration of the clock in a majority of clock neurons (UAS-CYCΔ/cry-GAL80 fruGAL4) rescued MSDR and SLR, whereas restoration of the molecular clock in PDF neurons (compare UAS-CYCΔ/Pdf-GAL80/fruGAL4 with UAS-CYCΔ/fruGAL4 controls) only rescued SLR (Table 1 and Table S2). We then carried out rescue experiments in per0 mutant males by expressing UAS-per under the control of various GAL4 drivers. Restriction of PER to the majority of clock (cry-GAL4_2) or FRUM-positive (fruGAL4) neurons showed robust MSDR, albeit altered period lengths (Table 1). PER nuclear accumulation at Zeitgeber time (ZT) 2 was observed in the clock neurons expressing these drivers (Fig. 2A). Surprisingly, PER expression restricted to LNvs (per0;Mz520-GAL4/UAS-per) restored MSDR well, with normal period length, albeit with a slightly advanced trough phase and lower phase concentration (Fig. 2 B and C and Table 1). This result indicates that the molecular clock in LNvs is sufficient for establishing the basic features of MSDR, whereas the timing of the courtship rest is poorly synchronized among different flies. When PER expression was restricted to DN1s (per0;c319-GAL4/UAS-per and per0;Clk4.4F-GAL4/UAS-per), the number of flies with MSDR increased but was still significantly lower than in WT males (P < 0.05; compare with per0;UAS-per/+ and Ore-R, respectively; Table 1).
Fig. 2.
Distinct functions of DN1s in MSDR and SLR. (A) Brains of the indicated genotype were stained at ZT2 with anti-PER. White thick and fine dotted lines indicate the midline and edge of the brain, respectively. The numbers of PER-positive cells in each cluster are indicated (n = 8–12 hemispheres). (Scale bars: large panels, 50 μm; small panels, 10 μm.) (B) Circular phase analysis (34) of MSDR for individual flies. Small circles represent the estimated trough phase of individual rhythmic flies (Table 1). Vector length and position indicate phase concentration (R) and averaged phase, respectively. An internal dotted circle represents a value of 0.5 in phase concentration. The trough phase is shown in CT hours. (C) Waveforms represent the averaged distribution of time in proximity through a circadian period (τ) window, for n rhythmic flies (Table 1). The τ-windows are normalized to 24 h. (D) Significant reduction in frequency of the evening peaks (%) of flies, which lack the molecular clock in a subset of DN1s. The frequency of rhythmic flies, which showed the highest activity peak between CT8 and CT16, was calculated (n = 17–36). *P < 0.05; ****P < 0.0001 (χ2 test). (E) Waveforms represent the averaged distribution of locomotor activity in SLR through a circadian period (τ) window for n rhythmic flies (Table S2). The τ-windows are normalized to 24 h. Activity levels are normalized independently for each genotype.
Because PER expression in LNvs or DN1s alone rescued MSDR partially in per0 males, we wondered whether combining PER expression in these neurons might rescue MSDR completely. We generated per0 males with both drivers and UAS-per (per0;Mz520-GAL4 c319-GAL4/UAS-per). Indeed, both the number of flies with robust MSDR and their phase concentration were restored to the level of WT males (Fig. 2 B and C and Table 1). We suggest that Pdf neurons are necessary and sufficient to produce the basic features of MSDR (i.e., large number of rhythmic flies), whereas the DN1 neurons are essential for phase concentration (i.e., synchronicity among males) and for setting the trough phase at dusk. Interestingly, per0;Mz520-GAL4 C319-GAL4/UAS-per flies also show restoration of the evening peak in SLR, albeit their phase is advanced by about 3.5 h (Fig. 2 D and E).
Discussion
DN1 Clock Neurons Have Distinct Roles in MSDR and SLR.
Our analyses revealed that LNvs play a crucial role in MSDR. This is not surprising, given the prominent role that these cells play in establishing SLR in DD and the central position they occupy in the neural circuitry of the clock cell network. However, we also find that DN1s provide critical but distinct functions for establishing more complex features of MSDR and SLR, respectively. Interestingly, both high (per0;Mz520-GAL4/UAS-per) and low (c319-GAL4/UAS-CYCΔ) CLK/CYC activity in DN1s of males leads to the same basic SLR phenotype, namely, the lack of a distinct evening peak (5) (Fig. 2E), but these manipulations affect MSDR differently. High CLK/CYC activity in DN1s leads to the spreading of the trough phase (Fig. 2 B and C), which exhibits tight coherence at dusk in WT males, whereas low CLK/CYC activity in these cells causes a loss of MSDR altogether. How can these CLK/CYC activity-dependent distinct phenotypes of MSDR be explained? We suggest two models, whereby signaling molecule(s) released from DN1s act on distinct neural targets that either trigger the evening peak or the courtship trough at dusk, respectively. Which of the two pathways is activated depends solely on whether or not external female stimuli are present. In the first model, we propose that DN1s secrete two distinct signaling molecules, SM1 and SM2, which mediate the specific motor output at dusk in SLR and MSDR, respectively (Fig. 3A). SM1 alone acts on neurons that control the evening peak in SLR. The absence of the evening peak observed in per0;Mz520-GAL4/UAS-per and c319-GAL4/UAS-CYCΔ males indicates that proper molecular cycling in DN1s is essential for timely SM1 release. SM2 is necessary for and acts on neurons suppressing courtship activity, generating the trough in MSDR precisely around dusk. Here, PDF signaling acts as a temporal trigger to induce SM2 production, regardless of proper molecular cycling in DN1s. For example, when CLK/CYC activity is constitutively high in DN1s (per0;Mz520-GAL4/UAS-per), SM2 may be released over an extended period, leading to variable placing of the trough in different males. To position the trough precisely at dusk (i.e., to release SM2 in a timely manner), a gating mechanism is necessary, which is implemented through cycling of CLK/CYC-dependent downstream genes in DN1s. However, CLK/CYC activity in DN1s is essential for releasing SM2, as evidenced by the lack of MSDR in c319-GAL4/UAS-CYCΔ males. In the second model, a single signaling molecule (SM2) generates both the trough at dusk in MSDR and the evening peak in SLR (Fig. 3B). The defining feature of this model is that different threshold concentrations of SM2 are required to activate the two different downstream targets. The target neurons that control evening peak activity in SLR require the maximum concentration of SM2, which is only achieved when the molecular clock in DN1s is intact. Release over an extended period occurs in the absence of a molecular clock in DN1s (e.g., in per0;Mz520-GAL4/UAS-per) and results in a lower SM2 concentration at the critical time, a level that is insufficient to trigger evening peak activity. However, the reduced concentration of SM2 is sufficient to induce the courtship trough, albeit less precisely (hence, the variable positioning).
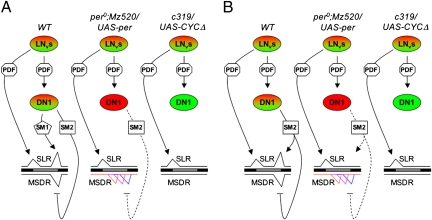
Fig. 3.
Models of DN1 function. (A) Model 1: Distinct signaling molecules generate a peak and trough in SLR and MSDR, respectively. WT males exhibit robust morning and evening activity peaks in SLR as well as precisely timed troughs at dusk in MSDR. Rhythmically released PDF (octagon) produces morning peaks (SLR) directly and synchronizes CLK/CYC activity in DN1s. SM1 (pentagon) and SM2 (square) are produced in and released from DN1s and are necessary to generate evening peaks (SLR) and troughs of courtship activity (MSDR) at dusk, respectively. Cells in red and green indicate high and low levels of CYC/CLK, respectively. Different colors of MSDR traces represent individual males. Enhancing and suppressing functions are indicated by arrows and blocked lines, respectively. The interrupted line indicates extended release of SM2 because of lack of CLK/CYC in DN1s. (B) Model 2: Distinct levels of a single signaling molecule are required to generate the evening peak and trough in SLR and MSDR, respectively. In WT males, tightly controlled release of SM2 acts on two distinct neuronal targets. In the absence of female stimuli, a high concentration of SM2 leads to an increase in locomotor activity at dusk (evening peak), whereas in the presence of female signals, SM2 suppresses directly or indirectly the courtship circuit, leading to the courtship trough. Lack of CLK/CYC cycling leads to extended release of SM2, and hence lower SM2 levels sufficient only to trigger a courtship trough but not an evening peak in SLR.
Cusumano et al. (20) have recently shown that PDF signaling from lLNvs to LNds is sufficient for establishing evening peak activity of single flies under LD conditions. This observation, along with our finding of PDF signaling from LNvs to DN1s to be sufficient for establishing evening peak activity in DD, suggests that parallel PDF signaling pathways are implemented, depending on external circumstances, to increase locomotion activity in anticipation of dusk.
FRUM Expression in Nonclock Neurons Might Be Necessary for Proper MSDR.
How do males alter their circadian activity pattern in response to external chemosensory (female-derived) signals? We demonstrated that many clock neurons in the sLNv, LNd, and DN1 clusters express FRUM (i.e., express fruGAL4) and are therefore sexually dimorphic in physiology (Fig. 1D and Fig. S4). Our finding broadly extends a previous observation, which reported FRUM-dependent male-specific neuropeptide F expression in three TIM-positive LNds (21). Unlike mutations in clock genes, fru mutations neither disrupt PER cycling in clock neurons (Table S3) nor SLR (Table S2), indicating that FRUM is not an essential component of the molecular clock. However, males with these fru mutations, which show robust courtship activity toward both males and females (15) (Fig. S3B), cause an arrhythmic MSDR phenotype (Table 1). Whether or not FRU function is required in clock neurons remains to be seen. The failure to induce arrhythmic MSDR as well as SLR in males with suppressed FRUM expression in clock neurons (UAS-fruMIR/cry-GAL4_2; Table S3 and Table S4) may indicate a requirement for FRUM in nonclock cells; alternatively, it may simply reflect insufficient interference with fru RNA in these cells.
FRUM is a central regulator of male courtship (20, 21) and sexual orientation (23, 24). Indeed, FRUM-expressing neurons can be viewed as defining the courtship circuitry itself. Thus, it will be of interest to identify those neurons that modulate courtship activity under the control of the intrinsic circadian clock. We suggest that the most recognizable feature of MSDR, courtship suppression at dusk, is established through SM2 signaling either by increasing the sensitivity of courtship inhibitory neurons or decreasing the sensitivity of courtship promoting neurons. However, elucidating the role of FRUM in the regulation of MSDR will require taking into account its complex role during development.
Experimental Procedures
Fly Strains.
Fly strains used in this study are as follows: Oregon-R, w1118, C155-elav-GAL4, fru3, and UAS-nucGFP (Bloomington Stock Center); UAS-CYCΔ (11); Pdf-GAL4 (6); tim-GAL4 (23); Clk4.4F-GAL4 (16,17) (P. Hardin, College Station, TX); fruGAL4 (15) (B. J. Dickson, Vienna, Austria); Pdf-GAL8096A and cry-GAL802e3m (6, 13); UAS-per2-4 (26) (M. Rosbash, Waltham, MA); Mz520-GAL4 (27); w per0;UAS-per16 (16) (F. Rouyer, Gif-sur-Yvette, France); UAS-fruMIR/CyO;UAS-fruMIR (which we describe as UAS-fruMIR in this report); P52a (28) (B. Baker, Stanford, CA); MJ63, MJ146, and MJ286 (29) (L. C. Griffith, Waltham, MA); Gr5a-GAL4, Gr22b-GAL4, Gr22e-GAL4, Gr28b.c-GAL4, Gr32-GAL4, Gr59b-GAL4, and Gr66a-GAL4 (30); Gr68a-GAL4 (31); Or47b-GAL4 (32) (L. B. Vosshall, New York, NY); Or83b-GAL4vp16 (33) (D. P. Smith, Dallas, TX); ok348-GAL4 (34) (T. Kitamoto, Iowa City, IA); and flytrap GAL4 lines 64Y, 104Y, 116Y, 187Y, 188Y, 201Y, 238Y, c182, c184, c319, c320, c343, c369, c370, c424, c500, c600, c739, c772, and c851 (http://www.fly-trap.org/). cry-GAL4_2 was an insertion in the third chromosome that was generated from cry-GAL4-24 (25) (J. Hall, Waltham, MA) by Δ2-3.
Behavioral Assays.
Assays for single fly locomotor activity and close proximity rhythms were performed as described previously (9), with the following modifications. We used virgin males of the indicated phenotypes as test subjects (5–12 days of age) and virgin w1118 females (4–8 days of age) as target objects. Males were entrained under 12:12 LD conditions in vials (about 20 animals per vial) from the time of hatching until they were used for the experiment. A single male and a single female were placed in the 15-mm diameter arena (24-well tissue culture plates with standard fly food) before lights-off time (ZT12, time 0). We videotaped (time lapse, one frame every 3 s) for 84 h under constant red dim light (<1 lux). Frequency of “close proximity encounters” (<1 mm) between the two flies was analyzed by EthoVison 3.1 (Noldus). The bin size was 60 min.
For longer recordings (5.5 days), we used apple juice media and infrared light instead of standard fly food and red dim light, respectively. Analysis of the longer recording data (Table 1 and Table S2) was done with FaasX software 1.6b (F. Rouyer). Rhythmic flies were defined by χ2 periodgram analysis with the following criteria (filter on): power ≥20, width ≥2 h, with selection on period = 24 ± 8 h. Phase concentration (R) indicates phase coherence among individual flies. The bin size was 20 min, and data length was 5 days (from CT0, time 12 h). Dead flies, which were determined by the video observation, were excluded.
CRI.
We defined the CRI as the mean of time in proximity over a 2-day period (48 h) divided by the mean of time in proximity during 2 4-h intervals on 2 consecutive days (CT10 to CT14). To calculate the CRI, we used the 48 h from time point 13 (subjective lights-on, CT0) to time point 60 (subjective lights-off, CT24).
Immunocytochemistry.
Brains were placed overnight in primary antisera solution at 4 °C. Rabbit anti-PER (33) and chicken anti-GFP (Molecular Probes) were diluted at 1:10,000 and 1:15,000 in PBS containing 0.1% Triton X-100 and 5% heat-inactivated goat serum, respectively. Brains were rinsed four times for 30 min and then placed in secondary antisera overnight at 4 °C. Dilutions (1:200) of Alexa555-conjugated goat anti-rabbit and Alexa488-conjugated goat anti-chicken (Molecular Probes) in PBS-TB were used. We included DN2s with DN1s because of their inconsistent separation from DN1s.
Statistical Analysis.
Effects on the CRI and averaged time in proximity were determined using the Student's t test (Table S1) for each genotype. If a significant difference was revealed between a genotype and its control, the Kruskal–Wallis ANOVA test (Fig. 1 B and C and Table S1) was applied. All analyses were done with JMP6 (SAS Institute) and FaasX software.
Supplementary Material
Acknowledgments
We thank P. Hardin, B. J. Dickson, M. Rosbash, F. Rouyer, B. Baker, L. C. Griffith, L. B. Vosshall, D. P. Smith, T. Kitamoto, J. Hall, and D. Tracy for various fly strains. We thank anonymous reviewers for insightful comments on this manuscript. This work was supported by National Institutes of Health Grant R01DC009014-03 (to H.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912457107/-/DCSupplemental.
References
- 1.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 2.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y, et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 6.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 7.Hardeland R. Species differences in the diurnal rhythmicity of courtship behaviour within the melanogaster group of the genus Drosophila. Anim Behav. 1972;20:170–174. doi: 10.1016/s0003-3472(72)80188-x. [DOI] [PubMed] [Google Scholar]
- 8.Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci USA. 2001;98:9221–9225. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 11.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 13.Emery P, et al. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 14.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 15.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchardon E, et al. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- 19.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 20.Cusumano P, et al. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci. 2009;12:1431–1437. doi: 10.1038/nn.2429. [DOI] [PubMed] [Google Scholar]
- 21.Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci USA. 2006;103:12580–12585. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito H, et al. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryner LC, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 24.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 27.Klarsfeld A, et al. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manoli DS, Baker BS. Median bundle neurons coordinate behaviours during Drosophila male courtship. Nature. 2004;430:564–569. doi: 10.1038/nature02713. [DOI] [PubMed] [Google Scholar]
- 29.Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and inhibitory switches for courtship in the brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. doi: 10.1016/j.cub.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 30.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39:1019–1029. doi: 10.1016/s0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- 32.Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- 33.Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, et al. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- 35.Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1–25. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.