Abstract
We investigated two mitochondrial genes (cytb and cox1), one plastid gene (tufA), and one nuclear gene (ldh) in blood samples from 12 chimpanzees and two gorillas from Cameroon and one lemur from Madagascar. One gorilla sample is related to Plasmodium falciparum, thus confirming the recently reported presence in gorillas of this parasite. The second gorilla sample is more similar to the recently defined Plasmodium gaboni than to the P. falciparum–Plasmodium reichenowi clade, but distinct from both. Two chimpanzee samples are P. falciparum. A third sample is P. reichenowi and two others are P. gaboni. The other chimpanzee samples are different from those in the ape clade: two are Plasmodium ovale, and one is Plasmodium malariae. That is, we have found three human Plasmodium parasites in chimpanzees. Four chimpanzee samples were mixed: one species was P. reichenowi; the other species was P. gaboni in three samples and P. ovale in the fourth sample. The lemur sample, provisionally named Plasmodium malagasi, is a sister lineage to the large cluster of primate parasites that does not include P. falciparum or ape parasites, suggesting that the falciparum + ape parasite cluster (Laverania clade) may have evolved from a parasite present in hosts not ancestral to the primates. If malignant malaria were eradicated from human populations, chimpanzees, in addition to gorillas, might serve as a reservoir for P. falciparum.
Keywords: ape malaria, human malaria, Plasmodium malagasi, Plasmodium phylogeny
There is a revolution afoot concerning our understanding of human malaria. It was shown in 1994/1995 that the closest relative of Plasmodium falciparum, the agent of malignant malaria was Plasmodium reichenowi, a chimpanzee parasite, the only ape malaria parasite that had been molecularly characterized (1–4). The close phylogenetic relationship between P. falciparum and Plasmodium reichenowi, their distinctness from the three other known human malaria parasites (Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae), as well as from other primate parasites, and their remoteness from bird or lizard parasites, was soon confirmed by other studies (5–7). It was assumed, as a working hypothesis, that P. falciparum and P. reichenowi had evolved from a common ancestor parasite, independently in their respective hosts, humans and chimpanzees, as these two lineages gradually diverged from one another over the last 5–7 million years—the cospeciation hypothesis. Two alternative hypotheses—either (i) a human origin (P. reichenowi evolved from an introduction of P. falciparum into chimpanzee hosts) or (ii) a chimpanzee origin (P. falciparum evolved from the introduction of P. reichenowi into the human lineage)—could not be tested against each other or against the cospeciation hypothesis, because only one P. reichenowi isolate was available, which had been isolated from a captive chimpanzee.
It was soon demonstrated by Rich et al. (8), Rich and Ayala (9), and Ayala and Rich (10) that P. falciparum has very low levels of neutral genetic polymorphism, a result that was subsequently confirmed by other investigators (11–15). The scarcity of neutral polymorphism was explained as the result of a recent world expansion of P. falciparum—the Malaria's Eve hypothesis (8)—which was estimated to have happened starting a few thousand years ago, rather than millions of years ago, as expected according to the cospeciation hypothesis. Martin et al. (16) suggested a mechanism compatible with a relatively recent origin of P. falciparum from P. reichenowi based on the differential expression of host sialic acid (Sia) ligands that populate the surface of erythrocytes and other cells, and on differences in Sia-binding preferences among parasite receptors. A mutation in the human lineage inactivated the CMAH gene rendering human ancestors unable to generate the Sia Neu5Gc from its precursor NeuAc, thus making humans resistant to P. reichenowi. A later mutation in the dominant invasion receptor EBA175 provided the P. falciparum lineage with preference for the overabundant Neu5Ac precursor, accounting for its extreme human pathogenicity (16, 17).
Rich et al. (18) have recently analyzed, in eight new isolates of P. reichenowi, gene fragments from the three principal genomes of Plasmodium parasites, mitochondrial (cytB), apicoplast (clpC), and nuclear (18S rDNA). The isolates were obtained from blood samples of wild and wild-born captive chimpanzees in Cameroon and Côte d'Ivoire. The genetic analysis shows that P. reichenowi is a geographically widespread and genetically very diverse chimpanzee parasite. The genetic lineage comprising the totality of global falciparum is fully included within the much broader genetic diversity of P. reichenowi. The genetic analysis supports the hypothesis proposed by the authors that all extant populations of P. falciparum originated from P. reichenowi, likely by a single host transfer, which may have occurred as early as 2–3 million years ago or as recently as 10,000 years ago (18). This hypothesis is consistent with the hypothesis proposed by Martin et al. (16) and Varki and Gagneux (17) of the two successive mutational events, first in our human ancestors and then in the parasite, which account for the extreme pathogenicity of P. falciparum.
Other recent developments in the malaria story include the discovery of new Plasmodium species parasitic to apes, and the presence in chimpanzees and gorillas of Plasmodium parasites previously thought to be human specific. Moreover, a cercopithecoid parasite, Plasmodium knowlesi, has now been found in humans (19–22).
Duval et al. (23) screened DNA isolated from blood samples in captivity from 113 chimpanzees and 17 gorillas from Cameroon, initially kept as pets for variable time periods and then brought to local zoos or sanctuaries or confiscated by the Ministry of Environment and Forestry. Two chimpanzees from the subspecies Pan troglodytes troglodytes presented with single infections of Plasmodium ovale, molecularly characterized by means of partial fragments from three genes, the mitochondrial genes cytochrome b and cytochrome c oxidase and the nuclear gene lactate dehydrogenase.
Hayakawa et al. (24, 25) screened the blood of 60 captive chimpanzees, imported to Japan 30 years earlier, and obtained two isolates of Plasmodium malariae, molecularly characterized by a nearly complete sequence of the nuclear small subunit rRNA and the complete mitochondrial genome. Indigenous malaria disappeared in Japan more than 50 years ago. Thus, the two chimpanzees most likely were infected in Africa and the P. malariae parasites brought into Japan from Africa. One chimpanzee was a male from Sierra Leone, assumed to have been born in 1978, and exported to Japan in April 1980. The other chimpanzee is a female, assumed to have been born in 1976, and exported to Japan in March 1977 from an unrecorded African country of origin. Both chimpanzees have been asymptomatic to the present.
As pointed out above, P. reichenowi was identified in the 1990s as the sister lineage of P. falciparum (1–3) and had remained since then, until very recently, as the only known ape parasite that had been molecularly characterized. Ollomo et al. (26) obtained blood samples from 17 chimpanzees kept as pets by villagers, collected from different parts of Gabon. Two chimpanzees were infected by a new species named by the authors Plasmodium gaboni. Analysis of its complete mitochondrial genome showed that the new species is much more closely related to P. falciparum and P. reichenowi than to any other known Plasmodium species. Indeed, the three species make up a distinctive clade, considerably separated from the clade that includes all other known primate parasites, with P. gaboni about equally different from P. falciparum as from P. reichenowi. The time of divergence of P. gaboni from the clade making up P. falciparum and P. reichenowi, assuming that the divergence of the later two species occurred 4–7 million years ago, was estimated at ~21 ± 9 Mya, a time congruent with the radiation of the hominoids, suggesting that the Plasmodium lineage encompassing the three species may have been present in early hominoids and may have diversified simultaneously with their hosts (26).
The recent discovery that Plasmodium can be genotyped from urine, saliva, and fecal primate samples (5, 27, 28) has opened up considerably the opportunities to explore the presence of Plasmodium in apes, as well as in other primates and in all other sorts of animals, by noninvasive methods. Prugnolle et al. (29) have analyzed partial Cytb sequences (704 nucleotides) from five chimpanzee and seven gorilla DNA samples, obtained from fecal samples from wild chimpanzees (n = 125) and gorillas (n = 84) from Cameroon, plus three blood samples from captive wild-born gorillas from Gabon. The chimpanzee isolates clustered with either P. reichenowi or P. gaboni, congruent with previous observations. The gorilla isolates, however, clustered into three different groups. Two isolates identify new lineages and potentially new species. One lineage, labeled Plasmodium GorA, was a sister lineage to P. gaboni; the second lineage, Plasmodium GorB, clustered with the clade made up of P. reichenowi plus P. falciparum. Surprisingly, three gorilla samples, two fecal Cameroon samples and one blood sample from a captive gorilla from Gabon, turned out to be P. falciparum. The presence of P. falciparum in nonhuman primates had not been previously reported.
Also recently, Krief et al. (30) found four sequences related to P. falciparum and two related to P. malariae in blood samples from bonobos (Pan paniscus). In chimpanzees (Pan troglodytes), the investigators found, in addition to P. reichenowi, two sequences related to P. vivax, as well as two newly named species, Plasmodium billbrayi (four sequences) and Plasmodium billcollinsi (three sequences), which form a monophyletic clade with the previously discovered P. gaboni (see below).
Results
We investigated two mitochondrial genes (cytB and cox1), one plastid gene (tufA) and one nuclear gene (ldh) in blood samples positive for Plasmodium from 12 chimpanzees and two gorillas from Cameroon and one lemur from Madagascar. One gorilla sample (GGcam10) belongs within the P. falciparum cluster, thus confirming the presence of P. falciparum-related parasites in gorillas, as reported by Prugnolle et al. (29). The second gorilla sample (GGcam13) appears to be closest to P. gaboni, although statistically not closer to gaboni than to the falciparum–reichenowi clade. The chimpanzee samples proved to be extremely diversified. Two samples (CPZcam46 and CPZcam137) cluster with P. falciparum, suggesting that this parasite is present not only in gorillas, as already shown by Prugnolle et al. (29) and as observed by us, and in bonobos (30), but also in chimpanzees. A third chimpanzee sample (CPZcam61) appears to be P. reichenowi, and two others (CPZcam80 and CPZcam155) associate with P. gaboni. The other chimpanzee samples are very different from those in the ape cluster. Two samples (CPZcam89 and CPZcam91) are P. ovale; these are the same samples already reported (23). Another sample (CPZcam83) is similar to P. malariae. That is, at least three human Plasmodium parasite species have also been found in chimpanzees. Four chimpanzee samples manifested mixed infections: three samples (CPZcam72, CPZcam86, and CPZcam103) had mixed infections of P. reichenowi and P. gaboni, and one sample (CPZcam63) of P. reichenowi and P. ovale.
The lemur Plasmodium sample appears in our tree as a sister lineage to a large cluster of cercopithecoid and hominid primates, which does not include falciparum or the ape parasites, thus suggesting that this cluster must have originated from a parasite present in hosts that were not ancestral to the primates at large.
We screened a total of 113 chimpanzee, 17 gorilla, and 55 lemur DNA samples for Plasmodium parasite infection by PCR using two mitochondrial partial genes, cytochrome b (cyt b) and cytochrome c oxidase 1 (cox1). Two additional partial genes were sequenced for the purpose of confirming certain results, the nuclear gene lactate dehydrogenase (ldh) and the apicoplast elongator gene tufA. Twelve chimpanzees (7 Pan t. troglodytes and 5 Pan t. vellerosus) and two gorillas (Gorilla gorilla) from Cameroon and one lemur (Propithecus verreauxi) from Madagascar were infected with Plasmodium parasites (Table 1). As shown in Table 1, four chimpanzees had mixed infections consisting of P. reichenowi and either P. gaboni (three cases) or P. ovale (one case). The mixed-infection sequences were not used in the phylogenetic reconstruction.
Table 1.
Gorilla, chimpanzee, and lemur samples analyzed for cytb and cox1 partial gene sequences: Sample code, host species, sample collection date, gender, age, Plasmodium strain, and GenBank accession number
| GenBank accession no. | ||||||
| Sample code | Host | Sample collection date | Gender, age | Plasmodium strain | cyt b | cox1 |
| GGcam10 | Gorilla gorilla | March 2000 | M, <6 mo | P. falciparum | HM000105 | HM000114 |
| GGcam13 | Gorilla gorilla | April 1999 | M, 1 y | P. GorB | HM000106 | HM000115 |
| CPZcam46 | Pan t. vellerosus | February 2002 | M, 3 y | P. falciparum | HM000107 | HM000116 |
| CPZcam61 | Pan t. troglodytes | February 1999 | F, NA | P. reichenowi | HM000108 | HM000117 |
| CPZcam63 | Pan t. vellerosus | September 1998 | M, adult | P. reichenowi/P. ovale* | AJ251941/ FJ409564 | AJ251941/ FJ409568 |
| CPZcam72 | Pan t. troglodytes | February 2000 | F, 2 y | P. reichenowi/P. gaboni* | AJ251941/ FJ895307 | AJ251941/ FJ895307 |
| CPZcam80 | Pan t. troglodytes | July 2001 | M, 1 y | P. gaboni | HM000109 | HM000118 |
| CPZcam83 | Pan t. vellerosus | April 1999 | F, juvenile | P. malariae | HM000110 | HM000119 |
| CPZcam86 | Pan t. vellerosus | February 2000 | M, 1 y | P. reichenowi/P. gaboni* | AJ251941/ FJ895307 | AJ251941/ FJ895307 |
| CPZcam89 | Pan t. troglodytes | February 2000 | F, juvenile | P. ovale | FJ409565 | FJ409569 |
| CPZcam91 | Pan t. troglodytes | February 2001 | M, adult | P. ovale | FJ409564 | FJ409568 |
| CPZcam103 | Pan t. vellerosus | April 2000 | M, 1 y | P. reichenowi/P. gaboni* | AJ251941/ FJ895307 | AJ251941/ FJ895307 |
| CPZcam137 | Pan t. troglodytes | February 2003 | F, NA | P. falciparum | HM000111 | HM000120 |
| CPZcam155 | Pan t. troglodytes | March 2004 | M, 1.5 y | P. gaboni | HM000112 | HM000121 |
| Lemur | Propithecus verreauxi | November 2002 | M, adult | Plasmodium malagasi | HM000113 | HM000122 |
M, male; F, female; NA, not available.
*Mixed infections.
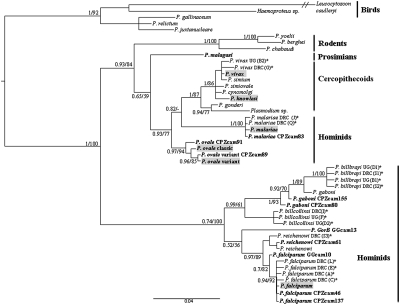
Figure 1 shows the phylogeny of the remaining newly obtained sequences (eight from chimpanzees, two from gorillas, and one from a lemur) together with relevant sequences from other Plasmodium species, listed in Table 2. The phylogenetic analysis was carried out using a concatenate sequence of 1672 bp, consisting of partial cyt b (708 bp) and cox1 (964 bp) genes. Maximum likelihood and Bayesian methods were used. Identical topologies were obtained by both methods. Bayesian posterior probabilities and bootstrap values are both given in Fig. 1. The topology of the reference sequences (Table 2) is consistent with previously published phylogenies (5, 24).
Fig. 1.
Phylogeny of Plasmodium species inferred from cyt b and cox1 nucleotide sequences. Values are Bayesian posterior probabilities (left of slash) and bootstrap percentages obtained by maximum likelihood (right of slash). Values less than 0.55 or 55% are not shown. CPZcam, chimpanzee from Cameroon; GGcam, gorilla from Cameroon. Boldface type indicates species and strains discovered in this work; gray overlay represents human parasites. *Sequences from Krief et al. (30). DRC, Democratic Republic of Congo; UC, Uganda. P. simium is a parasite of New World monkeys that is genetically identical to P. vivax. Hosts are shown on the right. P. knowlesi is a simian parasite recently found in humans.
Table 2.
Cytb and cox1 DNA GenBank sequences used in the phylogeny presented in Fig. 1, with their natural hosts and accession numbers
| GenBank accession no. | |||
| Plasmodium parasites species | Natural hosts | cyt b | cox1 |
| P. falciparum | Homo sapiens | M76611 | M76611 |
| P. falciparum DRC (A)* | Pan paniscus | GQ355474 | GQ355474 |
| P. falciparum DRC (C)* | Pan paniscus | GQ355473 | GQ355473 |
| P. falciparum DRC (E)* | Pan paniscus | GQ355472 | GQ355472 |
| P. falciparum DRC (L)* | Pan paniscus | GQ355475 | GQ355475 |
| P. vivax | Homo sapiens | AY598139 | AY598139 |
| P. vivax DRC (G)* | Pan t. troglodytes | GQ355480 | GQ355480 |
| P. vivax UG (B2)* | Pan t. schweinfurthii | GQ355481 | GQ355481 |
| P. ovale classical | Homo sapiens | FJ409567 | FJ409571 |
| P. ovale variant | Homo sapiens | FJ409566 | FJ409570 |
| P. malariae | Homo sapiens | AB489194 | AB489194 |
| P. malariae DRC (J)* | Pan paniscus | GQ345486 | GQ355486 |
| P. malariae DRC (Q)* | Pan paniscus | GQ355485 | GQ355485 |
| P. reichenowi | Pan t. troglodytes | AJ251941 | AJ251941 |
| P. reichenowi DRC (S3)* | Pan t. troglodytes | GQ355476 | GQ355476 |
| P. gaboni | Pan t. troglodytes | FJ895307 | FJ895307 |
| P. gaboni DRC (S1)* | Pan t. troglodytes | GQ355468 | GQ355468 |
| P. gaboni DRC (S2)* | Pan t. troglodytes | GQ355469 | GQ355469 |
| P. gaboni UG (B1)* | Pan t. schweinfurthii | GQ355470 | GQ355470 |
| P. gaboni UG (D1)* | Pan t. schweinfurthii | GQ355471 | GQ355471 |
| P. gaboni DRC (I)* | Pan t. troglodytes | GQ355479 | GQ355479 |
| P. gaboni UG (D2)* | Pan t. schweinfurthii | GQ355477 | GQ355477 |
| P. gaboni UG (F)* | Pan t. schweinfurthii | GQ355478 | GQ355478 |
| P. knowlesi | Old World monkeys† | AY598141 | AY598141 |
| P. simiovale | Old World monkeys | AY800109 | AY800109 |
| P. cynomolgi | Old World monkeys | AY800108 | AY800108 |
| P. gonderi | Old World monkeys | AY800111 | AY800111 |
| P. simium | New World monkeys | AY800110 | AY800110 |
| Plasmodium sp. | Old World monkeys | AY800112 | AY800112 |
| P. yoelii | Thamnomys rutilans | M29000 | M29000 |
| P. berghei | Grammomys surdaster | AF014115 | AF014115 |
| P. chabaudi | Thamnomys rutilans | AF014116 | AF014116 |
| P. gallinaceum | Gallus gallus | AB250690 | AB250690 |
| P. relictum | Birds | AY099032 | EU254593 |
| P. juxtanucleare | Gallus gallus | AB250415 | AB250415 |
| Haemoproteus sp. | Lichenostomus frenatus | AY733087 | AY733087 |
| Leucocytozoon caulleryi | Birds | AB302215 | AB302215 |
Five chimpanzees, CPZcam61 plus the four chimpanzees with mixed infection (CPZcam63, CPZcam72, CPZcam86, and CPZcam103), were infected with P. reichenowi, the parasite previously considered chimpanzee specific but recently shown to infect gorillas as well (29). Five chimpanzees are infected with P. gaboni, the recently identified chimpanzee parasite (26, 29): CPZcam80, CPZcam155, and the doubly infected CPZcam72, CPZcam86, and CPZcam103. One gorilla, GGcam13, is infected with a strain identical to the recently discovered gorilla parasite named P.GorB by Prugnolle et al. (29).
Six chimpanzees and one gorilla were infected with parasites previously thought to be human specific. One gorilla sample (GGcam10) belongs within the P. falciparum cluster, thus confirming the presence of P. falciparum-related parasites in gorillas, as reported by Prugnolle et al. (29). However we found two chimpanzees, CPZcam46 and CPZcam137, that were also infected with P. falciparum, and a chimpanzee, CPZcam83, infected with P. malariae. Krief et al. (30) found four bonobos also infected with P. falciparum and one bonobo infected with P. malariae. In addition, three chimpanzees are shown in Table 1 to be infected with P. ovale; one of the chimpanzees, CPZcam63, in a mixed infection (with P. reichenowi) and two more, CPZcam89 and CPZcam91, previously reported by Duval et al. (23). Of these two chimpanzees, CPZcam 89 was infected with “classic” P. ovale. The parasites from CPZcam91 are more similar to the “variant” P. ovale strain, although somewhat different from it, which might suggest the presence of a distinct strain of P. ovale (23).
The identification of strains GGcam10, CPZcam46, and CPZcam137 as P. falciparum-related parasites was confirmed by two additional partial gene sequences obtained from GenBank (Table 2): nuclear ldh (368 bp) and plasmid tufA(677 bp). The identification of CPZcam61 as P. reichenowi was confirmed by the ldh partial sequence (no tufA sequence of P. reichenowi was available from GenBank). The monophyly of the set of strains made up of the three P. falciparum and one P. reichenowi strains plus the P. gaboni CPZcam155 and CPZcam80, plus the P. GorB GGcam13 was confirmed by both ldh and tufA (as well as cytb and cox1). The monophyly has 100% bootstrap support and 1.00 Bayesian posterior probability (Fig. 1). All tufA sequences of this monophyletic group (Laverania clade, discussed below) show a three-nucleotide insertion when compared with the tufA sequences of other Plasmodium parasites from GenBank. These three nucleotides coding for either a proline (CCT) or a serine (TCT) at position 263 in the protein sequence are specific to the Laverania parasites.
Of the 55 Madagascar lemur samples examined, one Plasmodium strain was isolated from Propithecus verreauxi. We provisionally name this strain Plasmodium malagasi. This parasite forms a monophyletic clade with the clade that includes all Cercopithecoid parasites plus P. malariae and P. ovale (Fig. 1). Notably, this larger monophyletic clade does not include the clade encompassing P. falciparum, P. reichenowi, P. gaboni, P. GorA, P. GorB, and the newly named (30) P. billbrayi and P. billcollinsi (29) (Fig. 1). Thus, the latter clade (Laverania, discussed below) evolved from parasites acquired by host transfer from an evolutionary lineage that had diverged from the primate lineage before the divergence between lemurs (prosimians) and primates, which occurred before the Cretaceous/Tertiary boundary, ~75–80 Mya (31, 32).
Discussion
Apes as Reservoirs for Human Malaria Parasites.
Prugnolle et al. (29) identified P. falciparum in fecal samples form two wild gorillas in Cameroon (two distant localities) and in one blood sample from a captive gorilla from Gabon, and Krief et al. (30) found P. falciparum in blood samples from bonobos. The likely success of the campaign to eradicate malaria is hardly in sight. Were it to succeed, one consideration to keep in mind, as pointed out by Prugnolle et al. (29), is that gorillas might serve as a reservoir for malignant malaria and the possibility that humans might acquire P. falciparum by host transfer from gorillas, or from bonobos (30). We confirm the presence of P. falciparum–related parasites in gorillas, sample GGcam10 (Table 1 and Fig. 1). Moreover, we have found P. falciparum in blood samples from two chimpanzees belonging to two different subspecies, Pan troglodytes vellerosus (CPZcam46) and P.t. troglodytes (CPZcam137) (Table 1 and Fig. 1). The possibility that African apes might serve as a reservoir for malignant malaria is thereby much increased.
We have found the human parasite P. ovale in samples from two chimpanzees (CPZcam89 and CPZcam91) of the subspecies P.t. troglodytes and in one sample (CPZcam63) from P.t. vellerosus. Krief et al. (30) found the human parasite P. malariae in two blood samples from bonobos, and we have found it in one sample (CPZcam83) from P.t. vellerosus chimpanzees. These chimpanzees were held in captivity. Increased contact with humans, relative to wild chimpanzees, may have facilitated cross-species malaria transmission. There is no information on the capacity of the human Plasmodium parasites to produce viable gametocytes in chimpanzees that would be required for transmission by mosquitoes from chimpanzees or bonobos to humans or to other chimpanzees, gorillas, or different primate species.
The increased contact of humans with apes in the wild raises the question whether likely transmissions of P. falciparum (and other human Plasmodium parasites) may endanger the survival of the ape species in the wild. There are no definitive answers to this question. Experimental infections with human blood infected with P. falciparum fail to induce malaria in chimpanzees (reviewed in ref. 33). Nonsplenectomized chimpanzees experimentally infected with P. falciparum developed a low-grade parasitemia (up to 1,000/mm3) and maintained the infection without evidence of eliminating the parasites (34). Even after splenectomy to increase parasite survival, experimentally infected chimpanzees did not develop a parasitemia equivalent to that observed in humans (33, 34).
The strong human specificity of P. falciparum may be due to species-specific erythrocyte recognition profiles (16, 17). The mutational loss of the common primate Sia Neu5Gc may have protected our human ancestors from P. reichenowi. However, the major merozoite-binding protein (the erythrocyte-binding antigen-175, EBA-175) would have consequently evolved in P. falciparum to take advantage of the accumulated excess on human erythrocytes of the Neu5Gc precursor, the Sia Neu5Ac. These evolutionary changes would account for the reduced virulence of P. falciparum in apes and the presumed similar reduced virulence of P. reichenowi in humans. These two-way attenuations of virulence are consistent with the specific binding activities demonstrated by Martin et al. (16). The enhanced specificity of falciparum EBA-175 for the human Sia Neu5Ac (rather than for the ape Sia Neu5Gc) indicates that P. falciparum evolved in association with the hominid lineage, rather than in chimpanzees, bonobos, or gorillas. The presence of P. falciparum in these apes would therefore have resulted from host transfer from humans to apes. The older age and much greater genetic diversity of P. reichenowi compared with P. falciparum (18) also support the evolution of P. falciparum in the human lineage from ancestral P. reichenowi acquired in turn by host transfer from chimpanzees (18).
The Laverania Clade.
Plasmodium schwetzi was originally described by Reichenow in 1920 in the blood of apes in Cameroon (33). Experimental infections by P. schwetzi in humans have also been reported (35). In 1970, Contacos asserted its potential as a zoonosis in Africa (36). P. schwetzi is morphologically similar to the two human pathogens, P. vivax and P. ovale. Arguments have been advanced in favor of one or the other of these species as the most closely related to P. schwetzi (33). No isolate of this parasite is available from which molecular sequences could be obtained to decide its phylogenetic position.
The only known genetic relative of P. malariae is Plasmodium brasilianum (37), a parasite of several species of New World monkeys. The two species are indistinguishable at the molecular level, so that a very recent host transfer between humans and monkeys must have occurred (1, 3, 38). Plasmodium rodhaini was reported in 1920 from blood smears of chimpanzees and gorillas in Cameroon. It was morphologically described as being similar to the human parasite P. malariae (33). Transmission studies with quartan parasites isolated from chimpanzees have shown that P. rodhaini is actually P. malariae (39).
Plasmodium falciparum was discovered by Alphonse Laveran in 1880; P. falciparum has been classified with its sister species, P. reichenowi, a malaria parasite of chimpanzees, into the subgenus Laverania because of specific characters such as aspects of life cycle, high antigenic polymorphism, crescent-shaped gametocytes, and long gametocytogenesis duration as compared with that of other human malaria species placed in the subgenus Plasmodium (33, 40). Our results show that P. falciparum and P. reichenowi belong to the same clade as the recently discovered P. gaboni, P. GorA, and P. GorB (29), and P. billbrayi and P. billcollinsi (30). The set of these seven monophyletic species may appropriately be included in the Laverania subgenus, and be referred to as the Laverania clade (41).
The evolutionary origin of the Laverania clade predates the divergence of the prosimian P. malagasi from the Cercopithecoid clade, which may have occurred before 75–80 Mya. Currently, there seems to be no evidence of the Plasmodium parasites from which the Laverania clade evolved, or the nature of their hosts, or if and when a host transfer may have occurred from some remote ancestor to a more recent ancestor of the Plasmodium Laverania clade. What we know is that such a host transfer must have occurred before ~10 Mya, the time of divergence of the gorilla lineage from the human-plus-chimpanzee lineage. Be that as it may, our results confirm that (i) as asserted by Rich et al. (18), Ollomo et al. (26), Prugnolle et al. (29), and Krief et al. (30), the molecular diversity of the Laverania clade is much greater than previously thought, and (ii) chimpanzees, bonobos, and gorillas are a potential reservoir of P. falciparum, as well as of P. malariae and P. ovale.
Materials and Methods
Most of the chimpanzees and gorillas used in the study originated from different areas of Cameroon, and were initially kept as pets for different periods of time and then brought to local primate facilities or confiscated by the Ministry of Environment and Forestry. These animals had blood samples extracted for virological studies at the Virology Unit of Centre Pasteur du Cameroon (42, 43). A DNA bank was established between 1998 and 2004.
In total, we tested 130 DNA samples from great apes for Plasmodium infection, using primarily cytochrome b (cyt b) molecular tools (44); the apes included 105 chimpanzees from four subspecies (60 Pan t. troglodytes, 39 P. t. vellerosus, three P. t. schweinfurthii, and three P. t. verus), eight chimpanzees of undetermined subspecies, and 17 gorillas (Gorilla gorilla).
A total of 55 lemurs belonging to six genera (Hapalemur, Eulemur, Indri, Avahi, Varecia, and Propithecus) were caught between 1996 and 2002 in different areas of Madagascar. These animals were sampled for cytogenetic and molecular taxonomic studies by the Institut d'Embryologie of the University Louis Pasteur in France (45). Under an agreement with the Institut Pasteur de Madagascar, the University Louis Pasteur of Strasbourg provided blood samples from these different lemurs to the Institut Pasteur de Madagascar, where they were frozen and stored. DNA was obtained from lemur blood samples using a phenol/chloroform extraction technique and then screened for Plasmodium infection using cyt b molecular tools (44).
Isolated parasites from African great apes (chimpanzees and gorillas) and lemurs were molecularly characterized with the mitochondrial genes cytochrome b (cyt b) and cytochrome c oxidase 1 (cox1). The gene cox1 has been selected for biodiversity identification within the international barcoding program (46). Like cyt b gene, cox1 is a conserved gene and is useful for resolving phylogenetic relationships between parasite species that diverged over tens to hundreds of millions of years ago (47). In addition, we used the nuclear gene lactate dehydrogenase (ldh) gene and the tufA apicoplast elongation factor for additional characterization of parasites isolated from chimpanzees, gorillas and lemurs. All primers used are specifics of Haemosporidia parasites and do not amplify other Apicomplexa parasites or host DNA. All PCR products were sequenced by Macrogen in Korea.
Primers and PCR conditions used to amplify the cytb, cox1, ldh and tufA genes were as follows. Cytb gene fragments of 708 bp were amplified using published primers (44). The PCR and nested PCR reactions were carried out in a final volume of 25 μL under the following conditions: 2.5 μL of each primer, 2 mM of each dNTP, 2.5 U of Taq polymerase (Solis), 2 mM MgCl2, 5 min at 94 °C, 30 s at 94 °C, 30 s at 55 °C, and 90 s at 72 °C for 40 cycles, and a final 10-min extension at 72 °C. Cox1 gene fragments of 964 bp were obtained with the following primers: cox1a: 5′-CGCCTGACATGGATGGATAATAC -3′ and cox1b: 5′-CCATTTAAAGCGTCTGGATAATC -3′, and nested PCR primers: cox1c: 5′-GATTAACCGCTGTCGCTGGGACTG -3′ and cox1d: 5′-CGTCTAGGCATTACATTAAATCC -3′. The PCR and nested PCR reactions were carried out as for cytb, except 30 s at 94 °C, 30 s at 53 °C for PCR, and 30 s at 58 °C for nested PCR, and 2 min at 72 °C for 40 cycles and a final 10-min extension at 72 °C. Cox1c and cox1d were used for sequencing. We amplified ldh gene fragments of 350 bp with two primers for PCR: Ldh1 (5′-GGNTCDGGHATGATHGGAGG-3′) and Ldh2 (5′-GCCATTTCRATRATDGCAGC-3′), and two for nested PCR: Ldh7 (5′-TGTDATGGCWTAYTCVAATTGYMARGT-3′) and Ldh8 (5′-CCATYTTRTTNCCATGWGCWSCDACA-3′). PCR and nested PCR were carried out as for Cytb, except 10 pmol/μL of each primer; 0.2 mM of each dNTP, 2.5 mM MgCl2, 2.5 μL PCR buffer 10×, and 2 μL of DNA, heating for 5 min at 94 °C, 30 s at 94 °C, 30 s at 53 °C, and 30 s at 52 °C for nested PCR, and 1 min at 72 °C for 40 cycles and a final 10-min extension at 72 °C. Ldh7 and Ldh8 were used for sequencing. 677 bp of the tufA gene were amplified using two primers for PCR: TufA1 (5′-GGGCATGTASATCATGGGAAAAC-3′) and TufA2 (5′-CCTGCTCCTATWGTTTTWCCT-3′), and two for nested PCR reactions: TufA3 (5′-GGAGCTACACAAATGGATRTAGC-3′) and TufA4 (5′-GGTTTATGACGACCHCCYTC-3′). PCR and nested PCR were as for ldh, except 1 min at 53 °C for PCR and 90 s at 72 °C for 40 cycles and a final extension phase for 10 min at 72°C. TufA3 and TufA4 were used for sequencing. All primers used are specific for Haemosporidia parasites and do not amplify other Apicomplexa parasites or host DNA. All PCR products were sequenced by Macrogen in Korea.
The cytb, cox1, ldh, and tufA sequences were checked using chromatograms and CLUSTAL W alignment to ensure that no position was ambiguous (48). Mixed infections were discarded from the phylogenetic study. Phylogenetic analyses were based on the use of 708 bp cyt b and 964 bp cox1 concatenated sequences. Reference sequences without ambiguous positions for either cyt b or cox1 were obtained from GenBank (Table 2).
We performed a statistical analysis, based on the method of Xia and Xie (49), to ascertain whether the number of substitutions was saturated. In this method, both transitions and transversions are plotted against evolutionary distances calculated with the JC69 model. The relative rates at which transitions and transversions saturated at the third position were compared by counting substitutions in all pairwise comparisons between sequences. The analysis showed that the third base was saturated, and this base was therefore discarded for subsequent phylogenetic analyses.
The most appropriate nucleotide substitution model was identified, based on hierarchical likelihood ratio tests and the Akaike information criterion, using PHYML (50) and Garli (51). Model selection was also performed using MrBayes by computing the Bayes factor. The General Time Reversible model incorporating a proportion of invariant sites and a gamma correction for variable sites (52) was favored by the hierarchical likelihood ratio tests, Akaike information criterion, and Bayes factor. Maximum likelihood analysis was carried out with PHYML, with clade support evaluated by nonparametric bootstrapping (1,000 replicates). Bayesian analysis was performed with MrBayes (53), using two runs of 1 million generations sampled every 100 generations. Convergence was determined using the standard deviation of the split frequencies, and runs were terminated when a value of less than 0.01 was reached. The burn-in phase was defined as the first 250,000 generations. Partitioning the alignment using independent models for each of the two genes did not improve the likelihood estimates in MrBayes.
Acknowledgments
We acknowledge all zoos and primate-keeping institutions in Cameroon, the Cameroonian Wild Aid Fund, the Limbe Wildlife Foundation, and the Pandrillus Organization, which provided chimpanzee and gorilla primate blood specimens and information for this study. We acknowledge the Direction des Eaux et Forêts of Antananarivo and the Association Nationale pour la Gestion des Aires Protégées of Antananarivo (ANGAP) for permission to capture the animals and take samples.
Footnotes
References
- 1.Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escalante AA, Ayala FJ. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92:5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: Evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- 4.Di Fiore A, Disotell T, Gagneux P, Ayala FJ. In: Primate Parasite Ecology. The Dynamics and Study of Host-Parasite Relationships. Huffman MA, Chapman CA, editors. Cambridge, UK: Cambridge Univ Press; 2009. pp. 141–182. [Google Scholar]
- 5.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagner SC, Misof B, Maier WA, Kampen H. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 2007;101:493–503. doi: 10.1007/s00436-007-0499-6. [DOI] [PubMed] [Google Scholar]
- 7.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Rich SM, Licht MC, Hudson RR, Ayala FJ. Malaria's Eve: Evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rich SM, Ayala FJ. Population structure and recent evolution of Plasmodium falciparum. Proc Natl Acad Sci USA. 2000;97:6994–7001. doi: 10.1073/pnas.97.13.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayala FJ, Rich SM. Genetic variation and the recent worldwide expansion of Plasmodium falciparum. Gene. 2000;261:161–170. doi: 10.1016/s0378-1119(00)00478-9. [DOI] [PubMed] [Google Scholar]
- 11.Volkman SK, et al. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- 12.Tishkoff SA, et al. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 13.Hartl DL, et al. The paradoxical population genetics of Plasmodium falciparum. Trends Parasitol. 2002;18:266–272. doi: 10.1016/s1471-4922(02)02268-7. [DOI] [PubMed] [Google Scholar]
- 14.Tishkoff SA, Verrelli BC. In: Infectious Disease and Host-Pathogen Evolution. Dronamraju KR, editor. Cambridge, UK: Cambridge Univ Press; 2004. pp. 113–140. [Google Scholar]
- 15.Escalante AA, Cornejo OE, Rojas A, Udhayakumar V, Lal AA. Assessing the effect of natural selection in malaria parasites. Trends Parasitol. 2004;20:388–395. doi: 10.1016/j.pt.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human-chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varki A, Gagneux P. Human-specific evolution of sialic acid targets: Explaining the malignant malaria mystery? Proc Natl Acad Sci USA. 2009;106:14739–14740. doi: 10.1073/pnas.0908196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rich SM, et al. The origin of malignant malaria. Proc Natl Acad Sci USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 20.Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004;10:2211–2213. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vythilingam I, et al. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans R Soc Trop Med Hyg. 2006;100:1087–1088. doi: 10.1016/j.trstmh.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Kantele A, Marti H, Felger I, Müller D, Jokiranta TS. Monkey malaria in a European traveler returning from Malaysia. Emerg Infect Dis. 2008;14:1434–1436. doi: 10.3201/eid1409.080170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duval L, et al. Chimpanzee malaria parasites related to Plasmodium ovale in Africa. PLoS ONE. 2009;4(5):5520. doi: 10.1371/journal.pone.0005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K. Big bang in the evolution of extant malaria parasites. Mol Biol Evol. 2008;25:2233–2239. doi: 10.1093/molbev/msn171. [DOI] [PubMed] [Google Scholar]
- 25.Hayakawa T, et al. Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees. PLoS ONE. 2009;4:e7412. doi: 10.1371/journal.pone.0007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ollomo B, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5(5):1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mharakurwa S, Simoloka C, Thuma PE, Shiff CJ, Sullivan DJ. PCR detection of Plasmodium falciparum in human urine and saliva samples. Malar J. 2006;5:103. doi: 10.1186/1475-2875-5-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nwakanma DC, et al. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J Infect Dis. 2009;199:1567–1574. doi: 10.1086/598856. [DOI] [PubMed] [Google Scholar]
- 29.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krief S, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoder AD, Cartmill M, Ruvolo M, Smith K, Vilgalys R. Ancient single origin for Malagasy primates. Proc Natl Acad Sci USA. 1996;93:5122–5126. doi: 10.1073/pnas.93.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui A, Rakotondraparany F, Munechika I, Hasegawa M, Horai S. Molecular phylogeny and evolution of prosimians based on complete sequences of mitochondrial DNAs. Gene. 2009;441:53–66. doi: 10.1016/j.gene.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Coatney GR, Collins WE, Warren M, Contacos PG. The Primate malarias (illustrated). Washington, DC: US Government Printing Office; 1971. [Google Scholar]
- 34.Taylor DW, et al. Parasitologic and immunologic studies of experimental Plasmodium falciparum infection in nonsplenectomized chimpanzees (Pan troglodytes) Am J Trop Med Hyg. 1985;34:36–44. doi: 10.4269/ajtmh.1985.34.36. [DOI] [PubMed] [Google Scholar]
- 35.Rodhain J, Dellaert R. Contribution a l etude de Plasmodium schwetzi E. Brumpt (2eme note). Transmission de Plasmodium schwetzi a l'homme. Ann Soc Belg Med Trop. 1955;35:757–775. [PubMed] [Google Scholar]
- 36.Contacos PG, et al. Transmission of Plasmodium schwetzi from the chimpanzee to man by mosquito bite. Am J Trop Med Hyg. 1970;19:190–195. doi: 10.4269/ajtmh.1970.19.190. [DOI] [PubMed] [Google Scholar]
- 37.Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev. 2002;15:564–594. doi: 10.1128/CMR.15.4.564-594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayala FJ, Escalante AA, Rich SM. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia. 1999;41:55–68. [PubMed] [Google Scholar]
- 39.Collins WE, Jeffery GM. Plasmodium malariae: Parasite and disease. Clin Microbiol Rev. 2007;20:579–592. doi: 10.1128/CMR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray RS. Studies on malaria in chimpanzees. VI. Laverania falciparum. Am J Trop Med Hyg. 1958;7:20–24. doi: 10.4269/ajtmh.1958.7.20. [DOI] [PubMed] [Google Scholar]
- 41.Bray RS. The malaria parasites of anthropoids apes. J Parasitol. 1963;49:888–891. [PubMed] [Google Scholar]
- 42.MacFie TS, Nerrienet E, de Groot NG, Bontrop RE, Mundy NI. Patterns of diversity in HIV-related loci among subspecies of chimpanzee: Concordance at CCR5 and differences at CXCR4 and CX3CR1. Mol Biol Evol. 2009;26:719–727. doi: 10.1093/molbev/msp016. [DOI] [PubMed] [Google Scholar]
- 43.Calattini S, et al. Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies. J Med Primatol. 2006;35(2):59–66. doi: 10.1111/j.1600-0684.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 44.Duval L, et al. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 2007;6:157. doi: 10.1186/1475-2875-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rumpler Y, et al. Comparing chromosomal and mitochondrial phylogenies of sportive lemurs (Genus Lepilemur, Primates) Chromosome Res. 2008;16:1143–1158. doi: 10.1007/s10577-008-1265-z. [DOI] [PubMed] [Google Scholar]
- 46.Hajibabaei M, Singer GA, Hebert PD, Hickey DA. DNA barcoding: How it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007;23:167–172. doi: 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis. Austin, TX: University of Texas; 2006. [Google Scholar]
- 52.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 53.Huelsenbeck JP, Ronquist F. MrBayes: A program for the Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]