Abstract
A longstanding question in centromere biology has been the organization of CENP-A–containing chromatin and its implications for kinetochore assembly. Here, we have combined genetic manipulations with deconvolution and super-resolution fluorescence microscopy for a detailed structural analysis of chicken kinetochores. Using fluorescence microscopy with subdiffraction spatial resolution and single molecule sensitivity to map protein localization in kinetochore chromatin unfolded by exposure to a low salt buffer, we observed robust amounts of H3K9me3, but only low levels of H3K4me2, between CENP-A subdomains in unfolded interphase prekinetochores. Constitutive centromere-associated network proteins CENP-C and CENP-H localize within CENP-A–rich subdomains (presumably on H3-containing nucleosomes) whereas CENP-T localizes in interspersed H3-rich blocks. Although interphase prekinetochores are relatively more resistant to unfolding than sur-rounding pericentromeric heterochromatin, mitotic kinetochores are significantly more stable, reflecting mitotic kinetochore maturation. Loss of CENP-H, CENP-N, or CENP-W had little or no effect on the unfolding of mitotic kinetochores. However, loss of CENP-C caused mitotic kinetochores to unfold to the same extent as their interphase counterparts. Based on our results we propose a new model for inner centromeric chromatin architecture in which chromatin is folded as a layered boustrophedon, with planar sinusoids containing interspersed CENP-A–rich and H3-rich subdomains oriented toward the outer kinetochore. In mitosis, a CENP-C–dependent mechanism crosslinks CENP-A blocks of different layers together, conferring extra stability to the kinetochore.
Keywords: CENP-A, centromere, super-resolution imaging, chromosome, boustrophedon
During mitosis, a condensed region of centromeric chromatin differentiates into the kinetochore, a complex multiprotein structure that directs chromosome segregation, mediates microtubule attachment, and signals improper attachments (1). Although centromeric DNA sequences are diverse within and between species, all centromeres contain the histone H3 variant CENP-A (2, 3). CENP-A replaces one or both copies of H3 in kinetochore nucleosomes (4, 5), but not every centromeric nucleosome contains CENP-A. In stretched human mitotic centromeres, CENP-A–containing blocks of chromatin are interspersed with H3 nucleosomes (6, 7) dimethylated on Lys4 (H3K4me2) (8). A similar pattern is also found in neocentromeres, rare chromosomal aberrations in which functional kinetochores assemble on unique sequence noncentromeric DNA (9). This pattern of H3 modifications is not universal, however, and a different pattern is observed at maize and rice centromeres (10–12).
It is unknown how the underlying chromatin fiber folds to bring several blocks of CENP-A chromatin together into a single compact kinetochore. One model proposes that kinetochore chromatin forms an “amphipathic”-like superhelix with CENP-A nucleosomes on its outer surface and H3 nucleosomes facing the interior (8). A recent immunoelectron microscopy study confirmed that CENP-A occupies the outer third of the centromeric chromatin (13). An alternative looping model has been suggested for the point centromere of budding yeast (14).
Here, we have used DT40 cells genetically depleted of condensin (15) and several constitutive centromere-associated network (CCAN) proteins (16–19) to map the pericentromeric region and to study chromatin organization in unfolded kinetochore fibers formed in vitro under conditions that preserve nonhistone protein attachments to chromatin (20). Examination of these unfolded fibers using super-resolution fluorescence microscopy with single molecule sensitivity allowed us to map the distribution of CENP-A relative to several histone modifications and other kinetochore proteins along the chromatin fiber with unprecedented resolution. Our studies also showed that CENP-C, but not the CCAN proteins CENP-H, CENP-N, or CENP-W, has an essential role in stabilizing kinetochore chromatin when cells enter mitosis.
Results
Mapping Pericentromeric Chromatin.
Centromeres are usually embedded in the condensed chromatin of the primary constriction. However, in condensin-depleted SMC2OFF metaphase cells, kinet-ochores undergo dramatic poleward “excursions” (15, 21), trailing behind them a thread of extended pericentromeric chromatin (Fig. 1B). Importantly, these kinetochores are functional, and after a slight delay, will subsequently direct sister chromatid segregation (15, 22). We used this system to map the distribution of histone modifications in the pericentromeric chromatin by indirect immunofluorescence and deconvolution microscopy.
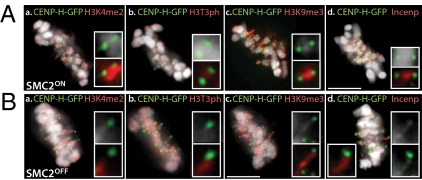
Fig. 1.
Mapping of pericentromeric chromatin. Localization of histone modifications and INCENP in pericentromeric region in SMC2ON (A) and SMC2OFF (B) metaphase cells expressing CENP-H-GFP: a, H3K4me2; b, H3T3ph; c, H3K9me3; and d, INCENP. (Scale bars in all figures: 5 μm, unless otherwise stated.)
H3K9me3, H3T3ph, and inner centromere protein INCENP were present between sister kinetochores in SMC2ON cells (Fig. 1A) and extended along the full length of the extended chromatin fiber trailing kinetochores undergoing excursions in SMC2OFF cells (Fig. 1B, b–d). In contrast, H3K4me2 was detected only in the chromosome-proximal portion of the stretched chromatin, with a distal nonstained region adjacent to the kinetochore (Fig. 1B, a). We conclude that DT40 kinetochore chromatin consists of compact CENP-A domains flanked at either side by pericentromeric heterochromatin.
Centromeric Region Unfolds to Higher Extents in Interphase than in Mitotic Cells.
We previously showed that exposure of chromosomes to low ionic strength TEEN buffer [a low-salt buffer in which chromatin higher-order structures are destabilized (20)] causes chromatin to unfold to beads-on-a-string nucleosomes while retaining kinetochore protein binding (15, 20). Although interphase prekinetochores are much more resistant to unfolding in this buffer than pericentromeric heterochromatin (15), they did unravel during longer incubations in TEEN buffer, producing fibers in which patches of CENP-A and H3-containing chromatin alternated (Fig. 2B). The intercalated H3-containing chromatin was rich in H3K9me3 (mean occupancy, 56 ± 20% of the CENP-A-containing region; n = 11). In other experiments, H3K4me2 appeared to be present at lower levels in the intercalated chromatin (mean occupancy, 18 ± 12%; n = 9; Fig. 2D). A similar distribution of histone modifications has been observed in centromeres in maize and rice (10, 12).
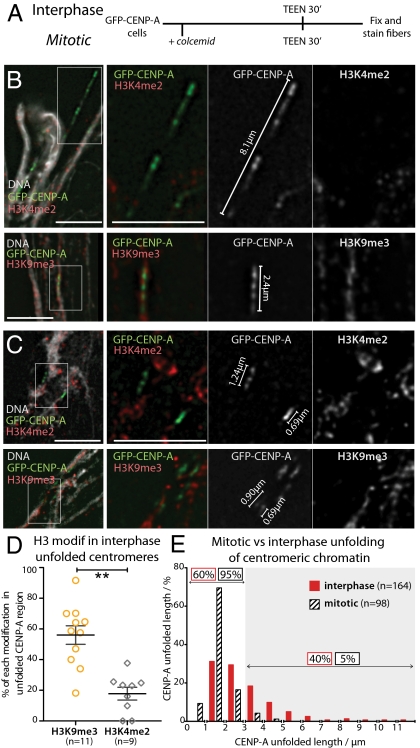
Fig. 2.
Mitotic kinetochores are more stable than interphase prekinetochores when subjected to unfolding induced by TEEN buffer. (A) Experimental procedure for unfolding centromeric region of mitotic and interphase cells. Examples of interphase (B) and mitotic (C) fibers immunostained for H3K9me3 (Upper) and H3K4me2 (Lower). (D) Quantification of the percentage of the total length of the CENP-A–containing fiber occupied by H3K9me3 (yellow) and H3K4me2 (gray). (E) Distribution of CENP-A fiber lengths measured for interphase (red) and mitotic (black) cells. Gray area indicates fibers larger than 3 μm, used to define a separation between the two groups.
Mitotic kinetochore chromatin is much more resistant to unfolding in TEEN buffer than its interphase counterpart, presumably because of structural maturation as cells enter mitosis (23, 24). The mean length (±SD) of unfolded kinetochores (determined by the borders of the GFP-CENP-A domains) derived from mitotic centromeres was 1.1 ± 0.44 μm (n = 98), compared with 2.47 ± 1.31 μm (n = 164) for interphase prekinetochores (Fig. 2 C and E). Considering that the kinetochore diameter determined by serial-sectioning electron microscopy is 145 ± 27 nm (n = 48; Fig. S1) in SMC2ON DT40 cells at metaphase, the average mitotic kinetochore fiber undergoes an eightfold extension in TEEN buffer. This contrasts with an average 17-fold extension of interphase prekinetochore fibers.
In mitosis, 95% of the extended CENP-A domains measured less than 3 μm (Fig. 2E). Similarly, most (60%) unfolded interphase prekinetochores yielded single CENP-A blocks less than 3 μm long. We refer to these interphase prekinetochores and normal mitotic kinetochores as having undergone “stage 1” unfolding. The remaining 40% of unfolded interphase kinetochores displayed multiple interspersed CENP-A subdomains stretching over more than 3 μm, up to a maximum of 13.4 μm. We refer to this as “stage 2” unfolding.
CENP-C Is Required for the Increased Structural Integrity of Mitotic Kinetochores.
In a genetic approach to identify proteins specifically required for the increased stability of mitotic kinetochores, we performed our kinetochore unfolding assay using mitotic cells depleted of CENP-C (18), CENP-H (16), CENP-N (17), and CENP-W (19). Remarkably, no significant difference was observed between unfolded WT kinetochores and those prepared from cells lacking CENP-H, CENP-N, or CENP-W (Fig. 3 A and B). Thus, we observed stage 2 CENP-A domain unfolding in 4%, 3%, and 8% of unfolded CENP-HOFF, CENP-NOFF, and CENP-WOFF kinetochores. This was not significantly different from the values obtained with CENP-HON, CENP-NON, and CENP-WON kinetochores in this assay, although following loss of CENP-W a few more unfolded prekinetochores were seen.
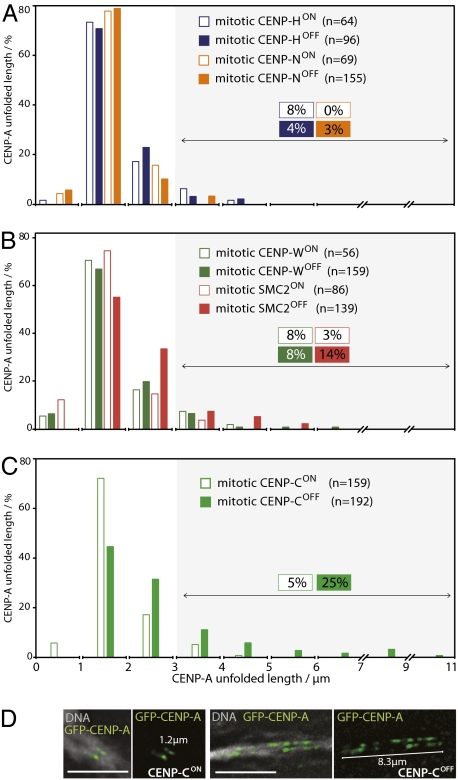
Fig. 3.
CENP-C is essential to confer extra stability to mitotic kinetochores. (A and B) Distribution of lengths of unfolded mitotic CENP-A chromatin fibers measured in the presence or absence of (A) CENP-N (orange) and CENP-H (blue), and (B) CENP-W (green) and condensin (pink). (C) Distribution of unfolded CENP-A chromatin fibers from mitotic CENP-CON and CENP-COFF cells. (D) In the absence of CENP-C (CENP-COFF), mitotic centromeres unravel to levels similar to those observed in interphase with CENP-CON.
In contrast, the loss of CENP-C caused a significant destabilization of the mitotic kinetochore chromatin in the TEEN assay, as the percentage of kinetochores undergoing stage 2 unfolding increased from 5% to 25% (Fig. 3 C and D). As with interphase prekinetochores, many CENP-A domains underwent only stage 1 unfolding following CENP-C depletion. However, the maximal lengths of stage 2–unfolded CENP-A domains achieved after CENP-C depletion approached those observed for interphase kinetochore fibers (Fig. 2E).
Depletion of condensin yielded intermediate results in this unfolding assay. The number of extended CENP-A domains larger than 3 μm increased from 4% to 14%, but domains larger than 6 μm were never observed (Fig. 3B). This suggests that condensin in the underlying heterochromatin may contribute in part to the structural integrity of kinetochore chromatin during mitosis, but that loss of condensin does not lead to full stage 2 unfolding.
Thus, of the four CCAN proteins tested, only CENP-C is required for the enhanced structural integrity of mitotic kinetochore chromatin.
Mapping Unfolded Interphase Prekinetochores Using Super-Resolution Microscopy.
Fluorescence microscopy with subdiffraction limit spatial resolution yielded further insights into the organization of kinetochore chromatin. Our method resembles (fluorescence) photoactivation-localization microscopy and stochastic optical reconstruction microscopy [reviewed by Lippincott-Schwartz and Manley (25)], which are based on single-molecule detection of switchable fluorophores and provide single-molecule sensitivity with a spatial resolution of tens of nanometers. Cycles of stochastic switching, detection, and localization of single molecules on a wide-field microscope are used to reconstruct super-resolution images (Materials and Methods). A related technique was recently used to localize chromosomal proteins in fixed cells with a precision of 20 to 30 nm (26).
We tagged CENP-A with the reversible fluorescent protein Dronpa, which photo-switches between dark and bright states after irradiation at 488 and 405 nm, respectively (27, 28). Chromatin from cells stably expressing Dronpa-CENP-A was unfolded as described earlier and immunostained for detection of different histone modifications using Alexa Fluor 647-labeled secondary antibodies. In the presence of a “switching buffer” containing an oxygen scavenger and reducing agent (Materials and Methods), Alexa Fluor 647 cycles reversibly between fluorescent and dark metastable states (29, 30). By sequential imaging with 488-nm and 633-nm irradiation, a two-color super-resolution map revealing the distribution of CENP-A relative to other kinetochore components could be constructed.
Fig. 4A shows an extended centromeric region stretching over 13 μm in which alternating CENP-A and H3K9me3 domains are readily apparent. Super-resolution microscopy also detected H3K4me2 between CENP-A domains, but whereas H3K9me3 was present as discrete blocks, H3K4me2 showed a more scattered distribution (Fig. 4B). Chromatin regions unlabeled with anti-H3K9me3 or anti-H3K4me2 may contain nucleosomes with other (or no) modifications.
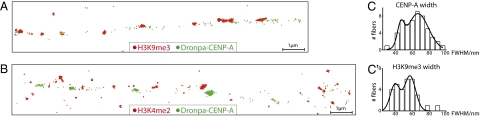
Fig. 4.
Characterization of unfolded prekinetochores using fluorescence microscopy with subdiffraction spatial resolution. (A) Example of a 13.4-μm interphase fiber in which H3K9me3 blocks are clearly observed between CENP-A arrays. (B) Example of a 15.1-μm fiber in which H3K4me2 is also detected between CENP-A arrays, but with a more diffuse distribution. Dronpa-CENP-A is represented as green, H3K9me3 and H3K4me2 labeled with Alexa647 are represented as red. Yellow represents colocalization. Each dot corresponds to the localization of a single molecule switching event. (C) Frequency distribution histogram of stretched fiber widths measured from super-resolution reconstructed images of (C) Dronpa-labeled CENP-A and (C′) Alexa Fluor 647–labeled H3K9me3. The solid lines represent the best fit to bimodal Gaussian distributions centered at 46 and 67 nm for CENP-A (n = 53) and 40 and 57 nm for H3K9me3 (n = 30).
A rough estimate of the amount of CENP-A in the fiber in Fig. 4A can be obtained from the number of localizations (a total of 123). Assuming that, on average, each Dronpa molecule switches approximately three to five times (31), the number of labeled CENP-A molecules is approximately 25 to 40. This value should be taken with caution, as Dronpa can switch as many as 170 times (28). For this estimation, we also assume that most CENP-A in the fibers is labeled with Dronpa and that the amount of endogenous CENP-A is not significant (Fig. S2A).
By reconstructing super-resolution images of the density of single-molecule localizations (Materials and Methods), the width could be estimated for CENP-A– and H3K9me3-labeled fibers (Fig. 4 C and C′). Both exhibited a bimodal distribution centered at approximately 40 nm and 60 nm. Taking into account that the values are convolved with a 37-nm error (Fig. S2 B–D), these measurements likely correspond to two populations of chromatin fibers that differ in diameter by 20 nm. For chromatin fibers in this size range, the most likely candidates are the canonical 10- and 30-nm fibers. This important result suggests the possibility to distinguishing between different levels of chromatin organization by super-resolution fluorescence microscopy.
Localization of CCAN Proteins in Stretched CENP-A–Containing Chromatin Using Super-Resolution Microscopy.
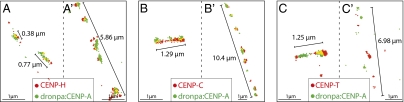
To determine whether components of the CCAN are associated with CENP-A or H3 blocks in kinetochore chromatin, we localized CENP-C, CENP-H, and CENP-T by super-resolution microscopy of unfolded kinetochores from cells expressing Dronpa-CENP-A (Fig. 5). All three CCAN components colocalized with CENP-A in shorter compact fibers (Fig. 5 A–C). However, when longer fibers were unfolded to reveal multiple CENP-A subdomains CENP-C and CENP-H remained closely associated with the CENP-A–rich subdomains, but CENP-T clearly localized between those subdomains (Fig. 5 A′–C′). Thus, the distribution of CENP-T resembles that of H3K9me3 and H3K4me2 in unfolded prekinetochores, and suggests the presence of blocks of canonical histone H3 in the outer surface of kinetochore chromatin.
Fig. 5.
Localization of CCAN components in interphase CENP-A chromatin fibers. CENP-H (A) and CENP-C (B) in red colocalize with CENP-A (green) both in short (A, B) and more extended (A′, B′) fibers. (C) CENP-T (red) colocalizes with CENP-A (green) in short fibers (C); however, in more extended fibers (C′), CENP-T is interspersed between CENP-A domains.
Discussion
We have combined gene targeting with biochemical manipulation of chromatin higher-order structure and super-resolution microscopy to characterize a vertebrate kinetochore. Kinetochores persist during interphase as locally condensed chromatin domains known as prekinetochores (32) that undergo a program of structural (23) and biochemical (24) maturation as cells enter mitosis. By using a protocol in which kinetochore chromatin is unfolded in vitro with TEEN buffer (20), we show that this maturation renders mitotic kinetochore chromatin considerably more robust than that of interphase prekinetochores. This presumably helps to give kinetochores the structural rigidity required to withstand pulling forces within the mitotic spindle.
Genetic analysis using DT40 conditional knockouts reveals that this mitotic stabilization of kinetochore chromatin requires CENP-C but not CENP-H, CENP-N, or CENP-W. This was surprising, as CENP-H is required for CENP-C accumulation at interphase prekinetochores (16, 18). However, a role for CENP-C in stabilization of the mitotic kinetochore is consistent with previous observations that CENP-C determines the size and continuity of the kinetochore plate (24, 33). CENP-C could perform a scaffolding role by interacting directly with DNA (19, 34) or RNA (35, 36) or with proteins such as CENP-L or Pcs1 as shown in Schizosaccharomyces pombe (37).
Interphase prekinetochore chromatin unfolded with TEEN buffer consists of extended fibers in which multiple CENP-A subdomains alternate with subdomains containing H3K9me3. This heterochromatin-associated modification was also seen to abut CENP-A domains in stretched pericentromeric chromatin trailing behind kinetochores undergoing poleward excursions in condensin-depleted cells. Furthermore, the pericentromeric chromatin also consistently displayed a gap between H3K4me2 staining and the kinetochore. However, further analysis of the histone modification pattern of unraveled centromeric fibers, using both deconvolution and super-resolution imaging, suggested that, as in rice centromeres (10), both histone H3 modifications (detected in independent experiments) are present in the centromeric region. Comparison of the two sets of experiments suggested that H3K4me2 may be present at lower levels than H3K9me3 in this region.
Although it is now accepted that CENP-A domains alternate with canonical H3 blocks within kinetochore chromatin (6, 7), the modification pattern of the canonical histones blocks seems to be less conserved. In human and Drosophila interphase prekinetochores, H3K4me2, but not H3K9me3, was found to be intercalated between CENP-A subdomains (8). H3K4me2 and lower levels of H3K9me3 were readily detected within the kinetochore of a human artificial chromosome by ChIP (38). In contrast, levels of H3K4me2 were much lower in one human neocentromere (39) and in maize centromeres (11, 12). Clearly, more work is required to understand the role of particular histone modifications in kinetochore chromatin structure and function.
Here we examined the distribution of members of the CCAN relative to CENP-A along the chromatin fiber. CENP-C, CENP-H, CENP-I, CENP-K-U, and CENP-W all coimmunoprecipitate with CENP-A following partial digestion of chromatin by micrococcal nuclease (40, 41), and ChIP studies showed colocalization of CENP-C and CENP-H with discontinuous domains of CENP-A in human neocentromeres (39, 42, 43). CENP-N is the only CCAN protein that has been shown to interact directly with CENP-A nucleosomes (44). CENP-C, CENP-T, and CENP-W coimmunoprecipitate with H3 nucleosomes after extensive nuclease digestion (19, 45). This suggests that some canonical H3 nucleosomes must be close to CENP-A–rich subdomains in the inner kinetochore. We confirmed this by super-resolution light microscopy, finding that CENP-C and CENP-H colocalized with CENP-A–rich subdomains in unfolded kinetochores. This suggests the presence of some canonical H3 nucleosomes within the CENP-A–rich subdomains. In contrast, CENP-T, H3K4me2, and H3K9me3 were interspersed between the CENP-A subdomains.
Detailed maps of kinetochore proteins in fixed chromosomes derived from measurements using two-color fluorescence light microscopy place CENP-C external, but very close to, CENP-A, with CENP-T slightly external to CENP-C (46, 47). Current kinetochore chromatin folding models based on data available from localization of canonical histones and CENP-A on unfolded chromatin fibers propose that CENP-A and H3 coexist in the same fiber and are sorted on different faces of an “amphipathic” superhelix, in which CENP-A faces the outer kinetochore and the H3-containing blocks are embedded in the centromere (7, 13). The data here presented extend this mapping of CENP-A relative to the CCAN proteins (most of which were still unidentified at the time the existent models were proposed) and require changes to the original solenoid model to take into account the fact that H3 domains with associated CENP-T are present on the outer face of the kinetochore chromatin.
Our data could be explained by a modification of the “amphipathic” superhelix model if helical segments were oriented radially with their long axes perpendicular to the chromosome axis. Such an orientation would expose some CENP-A and H3 on the outer surface of the chromosome, but is difficult to reconcile with immunoelectron microscopy and super-resolution colocalization of other kinetochore proteins with CENP-A by fluorescence microscopy, all of which indicate that CENP-A seems to provide a basal layer to the kinetochore that does not penetrate significantly into the chromosome interior (13, 46, 47).
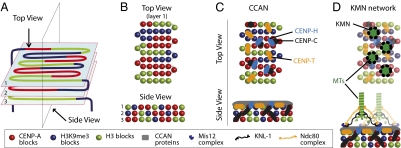
Based on our data, we suggest a simple alternative model for the topology of chromatin fiber folding in regional kinetochores. We propose that alternating CENP-A and H3 domains fold into a planar sinusoidal patch, or boustrophedon (Greek: “ox-turning”; Fig. 6). Such a topology would allow kinetochore size to vary according to the number of microtubules bound with minimal perturbation of local packing. Each kinetochore could be composed of several such patches stacked on top of one another as shown in Fig. 6A.
Fig. 6.
Model showing proposed boustrophedon arrangement of kinetochore chromatin. (A) A single continuous chromatin segment is arranged in a sinusoidal wave in a series of layers linked at both ends to heterochromatin. (B) Top and side view as indicated in A. (C) CCAN protein distribution in the kinetochore. (D) The KMN network assembles in mitosis on top of the CCAN and may confer stability to the mitotic kinetochore by crosslinking the CENP-C either directly or indirectly (see text for details).
This organization can explain the two stages of kinetochore chromatin unfolding observed in the present study. Stage 1 unfolding might correspond to the “straightening out” of the boustrophedon folds, with the different layers remaining held together laterally by structural crosslinks dependent at least in part on CENP-C. Stage 2 unfolding would entail the loss of connections between layers and stretching out of the whole kinetochore into a single contiguous linear segment. As we have shown, interphase prekinetochores frequently undergo stage 2 unfolding in TEEN buffer, whereas this is seldom observed for mitotic kinetochores. Given that CENP-C is required for this stability of the mitotic kinetochores, it is interesting to note that the genetic requirements for stable association of CENP-C with kinetochores differ between interphase and mitosis, with the former, but not the latter, requiring CENP-H (16, 18). This suggests that at least one aspect of kinetochore maturation may involve a change in the detailed mechanism of CENP-C association with kinetochore chromatin.
In surface view, our proposed model fits well with the recent proposed patterning of kinetochore proteins based on known protein associations in a “horizontal view” of the outer kinetochore (1). That model did not suggest a topological path for the chromatin fiber, which we propose here. In both models, CENP-A and H3 nucleosomes face the external surface, enabling the binding of all CCAN proteins. CENP-C could bind to the more internal CENP-A blocks, crosslinking several layers and explaining the similar oscillations undergone by CENP-A and CENP-C when kinetochores are under tension exerted by microtubules (47). The KMN network assembles in mitosis on top of the CCAN and binds microtubules. KMN binding may confer stability to the kinetochore by crosslinking the CENP-C chromatin either directly or indirectly.
Materials and Methods
DT40 conditional knockout cell lines were cultured as described before (22). Transfection of cells with a construct expressing GFP-CENP-A, antibody staining conditions, the quantification of DNA amounts in CENP-A and CENP-H kinetochore domains and detailed electron microscopy analysis of kinetochore sizes are described in SI Materials and Methods.
Super-Resolution Microscopy with Single Molecule Sensitivity.
Cells expressing Dronpa-CENP-A were plated on clean 22 × 22 mm coverslips for 20 min. Dronpa cDNA was provided by J. Lippincott-Schwartz (Bethesda, MD). The TEEN assay and antibody staining were performed as described in SI Materials and Methods using Alexa Fluor 647 (Invitrogen)-coupled secondary antibodies at 1:200 dilution. Coverslips were then attached to a CoverWell imaging chamber (Grace Bio Labs), containing “switching buffer” that promotes photo-induced blinking of Alexa Fluor 647: 10 mM PBS solution (pH 7.4) with an oxygen scavenger (0.5 mg/mL glucose oxidase; Sigma), 40 μg/mL catalase (Sigma), 10% wt/vol glucose (Fisher Scientific), and 50 mM β-mercaptoethylamine (Fluka) (29). Single-molecule fluorescence imaging was performed on a Nikon Eclipse TE2000 inverted microscope, equipped with a total internal reflection fluorescence oil-immersion objective (apochromat, magnification ×60; N.A., 1.49; Nikon). Wide-field illumination was achieved by focusing the expanded and collimated laser beam onto the back-focal plane of the objective. The resulting illuminated area was approximately 60 μm in diameter. Excitation was provided by a 488 nm CW Ar+ laser (163-C, 0.5 kW/cm2; Spectra-Physics) or 633-nm He/Ne CW laser (model 31–2140-000, 1 kW/cm2 at the sample; Coherent) passing through appropriate band-pass filters (Chroma Technology). Pulses for Dronpa photoactivation (2 Hz, 5 ms, 1 W/cm2) were provided by a CW 405-nm laser (Cube; Coherent) passing through an electronic shutter (Newport), controlled by a function generator (USB-6218; National Instruments).
Emitted fluorescence was collected by the same objective and imaged by an Andor Luca S electron-multiplying CCD camera after passing through a dichroic mirror (z488rdc or z633rdc; Chroma Technology), additional spectral filters (HQ500LP and HQ530/50, or HQ645LP and HQ700/75; Chroma Technology), and lenses resulting in a final pixel size of 74 nm. Integration time per frame was 100 ms. Typically 500 to 1,000 frames were collected. Two-color imaging of Dronpa and Alexa Fluor 647 was performed sequentially. Chromatic shifts were corrected by localizing immobilized 0.1 μm Tetraspeck beads (Invitrogen) with both colors.
Density super-resolution images like that in Fig. S2 were reconstructed by dividing each pixel into 16 subpixels and assigning each localization to a subpixel. The image brightness thus represents the density of localizations in a subpixel. The FWHM values were estimated from line cross-sections of density super-resolution images of fibers drawn with ImageJ by fitting a Gaussian function in GraphPad Prism. A spatial resolution of 37 nm was estimated by multiple localizations of the same single molecule of AlexaFluor647 analyzed under identical conditions, and fitting a Gaussian function to its cross section in a density reconstructed image (Fig. S2).
Movies were analyzed with Igor Pro by fitting Gaussian functions to individual molecules and localizing their centers.
Supplementary Material
Acknowledgments
We thank Dr. Peter Dedecker (Katholieke Universiteit, Louvain, Belgium) for the IgorPro routines. This work was supported by the Wellcome Trust (W.C.E.), Engineering and Physical Sciences Research Council Life Sciences Interface Program Grant EP/F042248/1 (to C.F.), the Darwin Trust of Edinburgh (S.A.R.), National Institutes of Health Grant R01GM06627 (to B.F.M.), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (T.F.). W.C.E. is a Principal Research Fellow of the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002325107/-/DCSupplemental.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma (Berl) 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 3.Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 5.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Sart D, et al. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, et al. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–1583. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin W, et al. Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 2008;16:1203–1214. doi: 10.1007/s10577-008-1269-8. [DOI] [PubMed] [Google Scholar]
- 13.Marshall OJ, Marshall AT, Choo KH. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribeiro SA, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukagawa T, et al. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 18.Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. J Cell Biol. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 22.Vagnarelli P, et al. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma (Berl) 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- 24.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunkel M, et al. Dual color localization microscopy of cellular nanostructures. Biotechnol J. 2009;4:927–938. doi: 10.1002/biot.200900005. [DOI] [PubMed] [Google Scholar]
- 27.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 28.Habuchi S, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 30.Steinhauer C, Forthmann C, Vogelsang J, Tinnefeld P. Superresolution micro-scopy on the basis of engineered dark states. J Am Chem Soc. 2008;130:16840–16841. doi: 10.1021/ja806590m. [DOI] [PubMed] [Google Scholar]
- 31.Flors C, et al. A stroboscopic approach for fast photoactivation-localization microscopy with Dronpa mutants. J Am Chem Soc. 2007;129:13970–13977. doi: 10.1021/ja074704l. [DOI] [PubMed] [Google Scholar]
- 32.Moroi Y, Hartman AL, Nakane PK, Tan EM. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981;90:254–259. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomkiel JE, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong LH, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obuse C, et al. A conserved Mis12 centromere complex is linked to hetero-chromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol. 2004;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- 41.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 42.Alonso A, et al. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007;8:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capozzi O, et al. Evolutionary and clinical neocentromeres: Two faces of the same coin? Chromosoma. 2008;117:339–344. doi: 10.1007/s00412-008-0150-z. [DOI] [PubMed] [Google Scholar]
- 44.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schittenhelm RB, et al. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.