Abstract
The fundamental chemistry of trace elements dictates the molecular speciation and reactivity both within cells and the environment at large. Using protein structure and comparative genomics, we elucidate several major influences this chemistry has had upon biology. All of life exhibits the same proteome size-dependent scaling for the number of metal-binding proteins within a proteome. This fundamental evolutionary constant shows that the selection of one element occurs at the exclusion of another, with the eschewal of Fe for Zn and Ca being a defining feature of eukaryotic proteomes. Early life lacked both the structures required to control intracellular metal concentrations and the metal-binding proteins that catalyze electron transport and redox transformations. The development of protein structures for metal homeostasis coincided with the emergence of metal-specific structures, which predominantly bound metals abundant in the Archean ocean. Potentially, this promoted the diversification of emerging lineages of Archaea and Bacteria through the establishment of biogeochemical cycles. In contrast, structures binding Cu and Zn evolved much later, providing further evidence that environmental availability influenced the selection of the elements. The late evolving Zn-binding proteins are fundamental to eukaryotic cellular biology, and Zn bioavailability may have been a limiting factor in eukaryotic evolution. The results presented here provide an evolutionary timeline based on genomic characteristics, and key hypotheses can be tested by alternative geochemical methods.
Keywords: Archean-Proterozoic, biogeochemistry, bioinorganic chemistry, evolution, metal homeostasis
Metalloproteins contain one or more ions of an inorganic element in their 3D structure and are said to comprise 30% of all proteins (1). Many biological pathways contain at least one metalloenzyme (2) and consequently require Mg, K, Ca, Fe, Mn, and Zn to sustain life (3). Other elements, like Cu, Mo, Ni, Se, and Co, are required by many—though not all—organismal lineages, and the utilization of trace elements varies greatly between species (3). The different elements have a range of affinities for most coordinating environments in the order Mg+2/Ca+2 < Mn+2 < Fe+2 < Co+2 < Ni+2 < Cu+2 ∼Zn+2, an equilibrium series known as the Irving-Williams Series (4). This array of outer-sphere chemistry provides significant catalytic diversity, yet has consequences for both biological and environmental chemistry. Within the cell, metals compete for protein binding sites; hence, extensive protein networks involving transporters and metal-sensing regulatory proteins are required to maintain the proper subcellular concentrations of each element, and in some cases directly shuttle each metal to its requisite metalloprotein in the proper cellular compartment (1, 5, 6). It has been hypothesized that the establishment of a metal homeostasis system is required for distinct phylotypes of cells to develop (7).
Environmentally, for similar fundamental chemical reasons, the shifts in global redox state hypothesized to have occurred over the past 4.5 billion years (3, 7, 8) greatly influenced trace-metal abundance. Specifically, the anoxic and reducing Archean ocean would have been enriched in Fe, Mn, and Co, yet low in Cu, Zn, and Mo (8, 9). Following the invention of oxygenic photosynthesis and the resulting increase in atmospheric O2 levels around 2.4 billion years ago (GYA) (10), increased continental weathering and oceanic sulfate reduction promoted a transition to a euxinic (anoxic and sulfidic) Proterozoic ocean (11, 12). In this high-sulfide, reducing ocean, Fe, Mn, and Co concentrations would have declined yet remained greatly elevated relative to the modern oxic ocean, whereas Cu and Zn would have decreased several orders of magnitude (9). This transition was likely not ocean-wide, with sporadic returns to anoxia and high spatial variability (13, 14), although coastal surface waters perhaps exhibited conditions similar to those of the modern oxic ocean. The transition to a generally oxic and oxidizing ocean (∼0.8–0.5 GYA) prompted large increases in Cu, Zn, and Mo, with concomitant decreases in Fe, Mn, and Co (9). The enormous redox shifts temporally encompass the evolution of modern life, and it has been suggested that this influenced the selection of the elements for biological utilization (15), although there is sparse evidence (16).
The advent of large-scale whole-genome sequencing can help elucidate how trace-metal chemistry influenced the evolution of protein macromolecules and how the different superkingdoms of life incorporated this machinery. Both specific amino acid residues and an appropriate 3D arrangement of those residues are required for proper metal binding by a protein, implying that an analytical approach using protein structure is more appropriate than a purely sequence-based methodology. A recent survey of Fe-, Zn-, Mn-, and Co-binding structures in over 300 genome-encoded proteomes revealed that the abundance of proteins binding a specific metal within a proteome scales to proteome size as a power law with distinct slopes (α) for each superkingdom and metal (17). Specifically, the proteomes of akaryotes (organisms in superkingdoms Archaea and Bacteria lacking a nucleus) were described by power laws with values of α > 1 for Fe-, Mn-, and Co-binding structures, while the eukaryotic proteomes have values of α > 1 for Zn-binding structures. The different power law slopes for Fe, Mn, Zn, and Co parallels the hypothetical chemical environment in which each superkingdom supposedly evolved, providing full-genome evidence for the influence of trace metal geochemistry on biological evolution.
In that study, the lack of knowledge about the order of metalloenzyme evolution precluded any conclusions about how the chemistry of the different trace metals have impacted the emergence of diversified life. Although the power-law scaling revealed a consistent evolutionary trajectory, it lacked direction, impeding interpretation over evolutionary history. Here, we expand upon this previous study by examining a larger set of elements and introduce a temporal scale. The relative timing for the evolution of metal-binding protein structures, both those exclusive and promiscuous in metal choice, was determined using a phylogeny of protein architectures that has been developed and refined over the past 7 years (18, 19). The occurrences and abundances of all of the diverse Fe-, Zn-, Mn-, Co-, Ni-, Cu-, Mo-, and Ca-binding protein families in 313 genomes were also determined. By examining the intersections of these datasets, we identify several genomic fossils that illuminate how trace-metal chemistry has constrained evolution. Environmental concentrations of trace metals influenced what type of metal-binding proteins evolve. The invention of the protein machinery needed to solve the equilibrium puzzle of the Irving-Williams Series coincided with the birth of metal-binding electron-transfer proteins. Finally, based upon proteome content, protein-domain evolution, and cell biology, it seems likely that low Zn, Mo, and Cu concentrations prevented the widespread emergence and diversification of the eukaryotic superkingdom until the advent of a planet-wide shift in redox state.
Results
Overview.
The evolutionary units in our study are protein structural domains: more specifically, compact and independently folding 3D architectures. The structural classification of proteins (SCOP) uses structural similarity to group domain fold-structures into fold superfamilies (FSF), which are one or more evolutionarily related protein fold-families (FF) with little sequence similarity. FFs are sequence clusters nested within each FSF (20). SCOP (20) provides a hierarchical classification of all protein domains published in the Protein Data Bank (PDB) (21) and version 1.69 of SCOP sorts 70,800 domains into 945 defined folds that are assigned to 1,539 FSFs, which are further subdivided into 2,845 FFs. In the simplest scenario, a protein contains one domain belonging to a FF and its parent FSF. More complex proteins contain multiple domains from one or more FF and FSF. Here, we examine the distribution of metal-binding FFs in extant genomes, simultaneously examining the temporal evolution of the parent category, the FSFs.
Trends of Metallome Composition in the Different Superkingdoms.
The metallomes, defined here as the proteomic complement of Ca-, Zn-, Fe-, Mn-, Cu-, Co-, Mo-, and Ni-binding FFs, of 313 genome-encoded proteomes representing the three superkingdoms, were examined relative to the total number of protein domains assigned to a genome, which is directly proportional to genome size. In terms of sheer proteome proportion, Fe-, Zn-, and Ca-binding domains are the most abundant, followed by Cu and Mn, then finally Co, Ni, and Mo (Table S1). However, as reported previously, the abundances of Fe-, Zn-, Mn-, and Co-binding protein domains scale to proteome size as a power law, with different slopes for each metal and superkingdom (Fig. S1 and Table S1). This scaling indicates that a raw percentage is a misrepresentation of proteome abundance. The power-law scaling is weak (Fig. S1), yet Ni- and Mo-binding structures are clearly more abundant in akaryotic relative to eukaryotic proteomes, although both only comprise a minuscule portion of most proteomes (Table S1). Cu-binding domains are in similar abundance in all three superkingdoms (Table S1), a result consistent with a previous survey conducted using sequence-based methods (22). Many akaryotic proteomes contain only one or no Cu-binding protein domains, preventing a robust estimation of a power-law slope, whereas eukaryotic proteomes exhibit a preferential accumulation of Cu-binding domains with increasing proteome size (Fig. S1 and Table S1). Ca-binding domains within a proteome scale with power-law slopes of greater than one for all three superkingdoms (Fig. S1 and Table S1), although Archaea and Eukarya have significantly (P < 0.01) greater slopes than Bacteria. Large-scale expansions of Ca-binding protein families have been reported in the larger eukaryotic proteomes (23), and the α >1 power-law scaling extends those trends to both akaryotic superkingdoms.
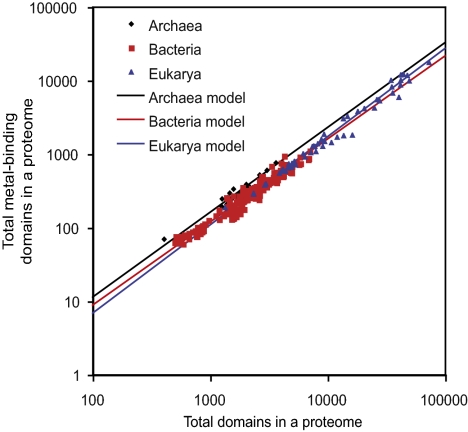
A comparison of the sum of all metal-binding domains and proteome size reveals a remarkable superkingdom-independent scaling (Fig. 1). This finding indicates that the overall gain and loss of metal-binding domains is a fundamental and constant rate for all of life. Furthermore , the slope of >1 indicates that the metal-binding proportion of a proteome increases with expansions in proteome size; only ∼8.5% of the small proteomes of parasitic Bacteria bind metals, whereas >25% of the largest mammalian proteomes is metal binding (Fig. 1). One caveat might be the lack of complete proteome annotation; 60 to 65% of a given proteome can be structurally characterized with the SUPERFAMILY HMMs, yet the structural genomics initiative suggests that the nonannotated portions of proteomes have a similar proportion of metal-binding domains (24).
Fig. 1.
Universal power-law scaling in metallomes. The total number of metal-binding domains relative to the total number of structural domains assigned to a given proteome is shown in log-log form, as are the fitted power laws from Table S1.
Diversity of Metal-Binding Compromises.
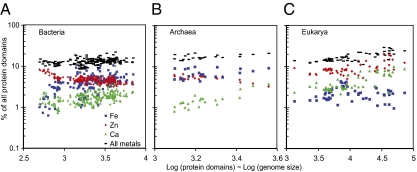
The different trends for each metal and superkingdom add up to a shared universal trend, indicating the need for diverse compromises. To visualize these compromises, we plotted the percent of a proteome that binds the three most abundantly used metals (Fe, Zn, and Ca) against the total number of protein domains in a proteome for each superkingdom (Fig. 2). In general, Bacteria eschew Ca for Fe and Zn, but the choice between the latter two metals depends upon proteome size. Bacteria with small proteomes contain a higher proportion of Zn-binding domains, most of them involved in tRNA synthesis, transcription, and translation (17). The essentiality of many of these protein domains means that the retention in small genomes is not surprising, yet the concomitant exclusion of Fe-binding domains is striking. In contrast, larger Bacterial proteomes tend to contain a higher proportion of Fe-binding domains. Similar trends are observed for archaeal proteomes (Fig. 2). Eukaryotic proteomes always contain a greater proportion of Zn-binding structures relative to those that bind Fe. Ca-binding proteins are often more abundant than Fe-binding proteins, particularly in larger eukaryotic proteomes (Fig. 2). Despite these prevailing trends, a great diversity of compromises exists within each superkingdom. Essentially, life has chosen diversely from the pool of available metalloenzymes, but selecting one element requires the relinquishment of another.
Fig. 2.
Metallomic compromises. The percent of the proteome that binds Zn, Fe, Ca, and the sum of the three metals is shown for (A) Bacteria, (B) Archaea, and (C) Eukarya.
Emergence of Metal-Binding Protein Structures.
A genome-based phylogeny of protein architectures at the FSF level that described the evolution of the protein world was previously reconstructed by Wang et al. (19), and the relative age of each metal-binding architecture was determined from this phylogenomic tree. For visualization, the age of each FSF (node distance, nd) is displayed as the number of nodes from the hypothetical ancestor in the tree on a relative 0 to 1 scale, with nd = 0 representing the birth of the protein universe and nd = 1.0 representing the most recent structural innovation (Fig. 3). The exact order of closely positioned FSFs is potentially debatable in trees of this size, but trends across the phylogeny are certainly robust and informative (25). For example, Wang et al. examined the evolution of protein architectures specific to each superkingdom and delineated the entire phylogeny into three epochs (19). FSFs ubiquitous to life arose during the first epoch (architectural diversification; nd = 0–0.391), and these core architectures catalyze much of modern metabolism, at least in their modern manifestation (19). The second epoch (superkingdom specification; nd = 0.391–0.61) describes the evolution of protein structures at a time when first lineages emerged and superkingdoms diversified. The third and final epoch (organismal diversification; nd = 0.61–1.0) contains a host of protein architectures specific and ubiquitous to Eukarya and describes the rise of lineages in an emerging and complex organismal world.
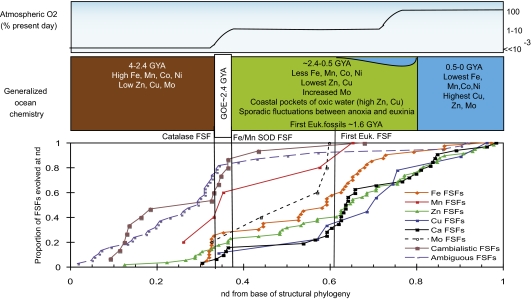
Fig. 3.
The appearance of metal-binding FSFs over the course of protein evolution and the correlation with oceanic geochemistry. (Top and Middle) A summary of the current consensus on the oxygen content of the atmosphere and the metal chemistry of the global oceans over geological time adapted from Lyon and Reinhard (14) and Saito et al. (9). (Bottom) The temporal evolution of several classes of metal-binding FSFs. Each point is a single metal-binding FSF, with the x axis indicating the time of appearance on a protein architecture phylogeny and the y axis indicating the total proportion of each category that has appeared up to that time. The three panels were aligned at two anchor points by a pair of transitional arguments detailed in the main text.
The nd increments are by nature a linear scale, yet equal nd steps do not necessarily correspond to equal time steps. To better interpret the structural evolution data, we aligned this structural phylogeny with a generalized geochemical and physical fossil record at two points. First, rising oxygen in the atmosphere at 2.4 GYA (14) would result in a physiological necessity for the oxidative defense-enzymes superoxide dismutase and catalase (Fig. 3), and the FSFs for these enzymes evolved at nd = 0.326 and 0.364, respectively. Second, the appearance of the first Eukarya-specific structure (nd = 0.614) was aligned with the first appearance of unambiguous physical fossils of eukaryotic life (1.6 GYA) (Fig. 3) (26). The anchor points allow a delineation of the structural phylogeny into gross geochemical epochs, which align remarkably well with the protein evolution epochs designated by Wang et al. (19). Metalloenzyme evolution was predominantly cladogenic, with numerous metal-binding structures appearing very close to each other within the phylogeny (Fig. 3). Over 80% of cambialistic (multiple metal binding; Methods) and ambiguous (may bind no or alternative metals; Methods) metal-binding architectures evolved during the first epoch, predating both the appearance of unambiguous metal-binding FSFs and the first loss of a FSF by a superkingdom (Fig. 1). The cambialistic FSFs contain an ensemble of metal-specific FFs. Essentially, these FSFs are 3D scaffolds that have achieved diversified metal-binding through alternations to the predominantly catalytic metal-binding active site. The cambialistic FSFs contain FFs that bind Fe, Zn, Mn, or Ni, with a few binding Mo, Ca, or Co (Table S2). The ambiguous FSFs contain FFs that either bind or lack metals.
Around nd = 0.3, two cambialistic FSFs prominently involved in metal homeostasis evolved: the helical backbone metal receptor FSF (nd = 0.34, SCOP ccs c.92.2, involved in Fe, Zn, and Mn uptake and sensing) and the heavy metal-associated domain FSF (nd = 0.321, SCOP ccs d.58.17, involved in Cu and Zn homeostasis). These FSFs contain multiple metal-specific FFs, are central to nearly all characterized metal-homeostasis systems, and are found in nearly all akaryotic proteomes in high copy number. The emergence of these metal homeostasis FSFs coincides with the appearance of many metal-binding FSFs, including those unambiguous and cambialistic in metal choice (Fig. 3). This appearance occurs just before the predominant period of diversification of the akaryotic superkingdoms (19). The majority of the emerging architectures bind metals abundant in the Archean ocean (9), and most Fe (>60%) and Mn-binding (100%) structures appear between 0.3 and 0.6 nd (Fig. 3). Structures that bind Fe in mixed Fe-S clusters evolve before those that bind Fe through porphyrin rings or direct amino acid bonds (Fig. S2A). In contrast, despite their early appearance, most (>60%) Zn-binding FSFs evolve after 0.6 nd, a pattern shared by Cu- and Ca-binding FSFs (Fig.3). Most of the late evolving Zn-FSFs use Zn as a structural molecule (Fig S2B).
Relative Age of the Metallome in the Three Superkingdoms.
We determined the percent of proteomes in each superkingdom that contain each Fe-, Zn-, and Ca-binding FSF and compared it to the age of the structure. The earliest evolving metal-binding protein structures are found within most proteomes of Archaea, Bacteria, and Eukarya, regardless of the metal-bound (Fig. S3). Structures that evolved during the second epoch appear in a smaller percentage of modern proteomes in each superkingdom, again in metal-independent fashion (Fig. S3). To a large extent, extant Bacteria eschew the architectures that evolved during the third epoch. Archaea share with Eukarya a late-evolving set of six Zn-binding architectures involved in transcription or translation (17), but otherwise contain few metal-binding structures that evolved after 0.6 nd. In contrast, eukaryotes increasingly incorporate metal-binding structures that evolved during the third epoch, particularly those that bind Ca and Zn (Fig. S3). Most of the Fe-binding structures that evolve after 0.6 nd are also found predominantly in Eukarya (Fig. S3). In summary, although the modern akaryotes still predominantly use ancient metal-binding protein structures, most eukaryotes use both early- and late-evolving structures.
Discussion and Conclusions
Influence of Ocean Chemistry on the Selection of the Elements.
Of the various metals, Fe, Mn, and Mo were preferentially selected by early biological life (Fig. 3), with many of the FSFs binding these metals appearing before the Great Oxidation Event (GOE). In contrast Cu and Zn utilization evolved after the GOE when these metals were available in at least some environments. The implication is that the geochemistry of the Archean ocean influenced both the evolution of metal-binding architectures and the selection of elements by the ancestors of Archaea and Bacteria. The early evolution of the low redox-state Fe-S-containing structures (Fig. S2) and delayed emergence of structures binding Fe directly using amino acid side chains, bolsters this conclusion. The analysis of metal-binding protein distribution and evolution presented here provides the first extensive temporal support for previous transitional argument hypotheses (15–17).
Emergence of the Akaryotes, Metal Homeostasis, and Biogeochemical Cycles.
Cambialistic metal-binding protein architectures capable of binding multiple metals appeared earlier than metal-specific counterparts, and the trend from promiscuity to specificity in metal-binding parallels general patterns observed in evolution of modern metabolism (27). The hypothesis that a functional metal homeostasis system would be necessary for the first steps in akaryotic evolution has been argued using transitional arguments (3), and the concomitant appearances of structures involved in metal homeostasis, metal-specific architectures, and the diversification of the akaryotic lineages provides the first genomic evidence of this. Before this coordinated evolution, organic cofactors able to transport protons had already evolved (28), yet when coupled with metal-binding electron transport proteins, redox transformations become possible. The earliest evolving Fe-, Mo-, and Cu-binding protein architectures are intrinsically involved in electron transfer and the transformations of C, N, S, and O (Table S3). In the environment, organisms metabolize the waste products of other organisms through interconnected biogeochemical cycles that attain a steady state resistant to further change (29–31). The coordinated appearance of many of the metal-binding electron transfer proteins provided the potential for the emergence of a nascent biogeochemical network, increasing the number of ecological niches for emergent lineages of Bacteria and Eukarya. If the evolution of metal-binding electron-transfer proteins and a biogeochemical network were indeed closely linked, parallel measurements of the isotopic fractionation of trace metals (Fe) and biogeochemical elements (N, S) of fossils of ancient microbial communities should reveal punctuated and concomitant variations.
Zinc and the Evolution of Eukarya.
The Zn power-law scaling, particularly the intercept, shows that the last common ancestor of Eukarya certainly contained more Zn-binding proteins than an equivalent akaryote (Table S1). Even modern Eukarya that inhabit anoxic environments, such as Giardia, have substantially more Zn-binding proteins than akaryotes (7.3% of the proteome). The eukaryotic proteomes contain most of the early evolving Zn proteins, likely acquired through endosymbiotic gene transfer (19, 32), yet are the sole carriers of late-evolving Zn-binding proteins (Fig. S3), where the metal is predominantly structural in nature (Fig. S2). An examination of the subcellular localization of metal-binding proteins within eukaryotic cells reveals an enrichment of Zn-binding domains targeted to the nucleus (Fig. S4), a result corroborated by the observation of elevated concentrations of Zn in the nuclei of marine flagellates (33). This eukaryote-specific utilization of Zn even came at the expense of Fe utilization (Fig. 2). In the geologic record, the eukaryotic fossils from the Proterozoic ocean were found in regions that were likely coastal (26), where a combination of photosynthesis in shallow water and O2-induced continental weathering likely resulted in increased Zn and Cu concentrations. The dependence of eukaryotes on Zn and the presumably low bioavailability of Zn during the Proterozoic (9) suggests that low environmental Zn concentrations may have limited eukaryotic diversification. The other biogeochemical features of the Proterozoic ocean invoked as limiting factors to eukaryotic evolution are low Mo and high sulfide concentrations (12, 34). The former would place a limit on global nitrogen fixation; the latter is presumed to have been toxic and either may have spatially confined the eukaryotic evolution to what was coincidentally high Zn environment, with a subsequent bias for Zn during structural innovations.
Currently, the timing of eukaryotic evolution is contentious and the unique utilization of Zn by Eukarya might provide a rich avenue of future research. Concrete fossilized remains are only found 1.6 GYA, and organic biomarkers placing an emergence at 2.7 GYA (35) have recently been called into doubt (36). In the structural phylogeny, the first eukaryotic-specific FSF appears much later than superoxide dismutases and catalases (Fig. 3), enzymes that would have been necessary for life to tolerate increased oxygen. This finding supports a temporal lag between the GOE and eukaryotic emergence, although molecular clock studies will be required to verify this. Eukaryotes are known to fractionate Zn during high affinity uptake (37), and an examination of the isotopic record of Zn may be a powerful ancillary approach to polarizing this fundamental argument.
Ca, Co, Mo, and the Evolution of Eukarya.
Ca concentrations are relatively unaffected by redox chemistry, seeming to remove a geochemical explanation for the observed late evolution of Ca-binding domains (Fig. 3). Like the Zn-binding proteins, many of the late-evolving Ca-binding FSFs are eukaryotic-specific (Fig. S3) and may have evolved to address a eukaryotic cellular need. Specifically, Ca, with its high coordination number and affinity for hard ligands, is ideally suited for signaling between organelles or cells (3, 38). Supporting this hypothesis, the most abundant Ca-binding domains in Eukarya are predominantly involved in signaling within or between cells (Table S4). The proteomes of the akaryotes that do have multiple organelles, Planctomycetes, are also enriched in Ca-binding domains (10% of the proteome), bolstering the hypothesis that this utilization is related to cellular complexity and not environment.
Cu and Mo bioavailability in the oceans would have paralleled that of Zn (9) and also likely influenced eukaryotic evolution. The Cu-binding cupredoxin FSF (b.6.1, nd = 0.34) evolved early and, although necessary for a complete global nitrogen cycle, only select akaryotic lineages contain this FSF. In contrast, this FSF is ubiquitous in eukaryotic proteomes, suggesting a preference for Cu in electron transport. Therefore, to a lesser extent, environmental Cu concentrations may have impeded eukaryotic evolution. Mo concentrations increased following the transition to an oxic ocean (39); however, in direct contrast to Zn, akaryotes dedicate a larger proportion of their proteome to binding Mo relative to Eukarya (Table S1). All of the Mo-binding structures emerged before the first eukaryotic FSF (Fig. 3) and catalyze biogeochemical transformations of N, such as nitrogen fixation and nitrate reduction, reactions that are the provenance of akaryotes (30). In contrast, many eukaryotes acquire nitrogen though ingestion of other organisms or by establishing symbiotic relationships. The influence of Mo on evolution is likely indirect; the overall passage of nitrogen through biogeochemical systems and thus ecosystem productivity depends upon Mo bioavailability (12, 40). On a global scale, increased Mo concentrations could have facilitated higher rates of N-fixation, releasing nutrient limitation of phytoplankton, increasing global carbon flow, and thus allowing for the energetically expensive eukaryotic cell.
Influence of Changing Zn on Metal Utilization.
Recently, Mulkidjanian and Galperin suggested that the Zn-binding proteins and RNA structures found in all of modern life must have been present in the last universal common ancestor (41). Invoking the hypothesized Zn concentrations in the Archean ocean (9), they concluded that the last universal common ancestor must have evolved near metal-rich hydrothermal vents (41). Our results do not support this hypothesis. Within our structural phylogeny, most of the early-evolving Zn-binding protein domains are in cambialistic and ambiguous FSFs (Fig. 3), and the progenitor structures may have bound a different metal or no metal at all. After the discovery of metal homeostasis, life predominantly turned to Fe and Mn (Fig. 3). Three early-evolving unambiguous Zn-binding FSFs are found in almost all extant akaryotic organisms: the Zn-dependent exopeptidases (c.56.5, nd = 0.125), the Zn-β ribbon (g.41.3, nd = 0.29), and RNA polymerase (e.29.1, nd = 0.27), yet the physiological ramifications of this appear to be slight. Modern akaryotes, like marine cyanobacteria, can grow continuously at concentrations of Zn approaching those of the Archean ocean, whereas eukaryotes cannot (9, 42, 43).
A more important question is how the progenitors of diversified life coped with a dramatic four to five order-of-magnitude decline in Zn concentrations accompanying the onset of oceanic euxinia at the Archean–Proterozoic transition. Chemical modeling studies suggest that, although uncomplexed Zn was likely very low, aqueous Zn-S species were certainly present (9) and microbial Zn acquisition might have proceeded through a ligand production and scavenging strategy similar to that employed for Fe acquisition in the modern world. Alternatively, early life might have turned to cambialism, using a different metal in what are modern-day Zn metalloenzymes. In the exopeptidases, Mn or Co can act as a substitute (44, 45). Cobalt can substitute for Zn in RNA polymerase without a dramatic loss in functionality, as shown in ref. 46. Structural proteins like Zn-β ribbons and Zn fingers also bind Co, although the DNA binding activity of a Co-bound Zn finger is much less efficient relative to the Zn-bound form (47). In each of these scenarios, the Irving-Williams series dictates that with increased Zn bioavailability, the other metal would be displaced. The utility of Zn fingers likely increased following the change in global Zn bioavailability, possibly prompting a burst in the innovation of proteins like Zn fingers (48) and quickening the diversification and rise of Eukarya. Essentially, the eukaryotic strategy of substituting Co for a majority of Zn requirements (42, 49) and the scavenging of metals using ligand export and retrieval may be vestigial abilities from the high sulfide Proterozoic, as suggested by Saito et al. (9).
Methods
Data Sources.
Libraries of hidden Markov models built from structural alignments of the FSFs and FFs are available from the Superfamily database and allow the conversion of a proteome sequence into a complement of structural protein architectures (50, 51). Release 1.69 of Superfamily covers 313 complete genomes (23 Archaea, 233 Bacteria, and 57 Eukarya) and was the source for all domain assignments. The Superfamily database employs a hybrid approach; a hidden Markov model-searching protocol and subsequent pairwise comparisons with a probability cutoff of E = 2 × 10−2 for identifying likely members of a group; it also provides a confidence level (in the form of an E value) for every candidate identified. As was done in Yang et al. (52), a more stringent E value cutoff of 10−4 was employed here for domain assignments.
Annotation of Metal Binding.
Each FSF in the SCOP database was manually examined for structures containing a covalently bound metal ion. This process requires examining FFs, domains, and specific PDB structures within each FSF. If all of the representative structures in a FSF or FF bind the same metal, then the family is considered metal specific or “unambiguous.” The manual curation of the SCOP revealed that most (∼80–90%) of the metal-binding FFs and their cognate domains bind a specific metal unambiguously (17), but the same cannot be said for the FSFs and the introduction of some descriptive terminology is required before proceeding further. Approximately 65% of the FSFs contain FFs binding the same metal, but the other 35% exhibit one of two deviations from this behavior (17). “Ambiguous” refers to FSFs where not all of the FFs bind a metal, whereas cambialistic FSFs have FFs binding different metals. Some of the cambialistic and ambiguous FSFs are the largest in the SCOP in terms of the number of FFs they contain (Table S2). Cambialistic and ambiguous FSFs and the metals they bind are listed in Table S2; all unambiguous metal-binding FFs and FSFs are listed in Table S5.
Analysis of Metal-Binding Domain Abundance.
Abundance matrices for each specific metal-binding FF in the individual proteomes were constructed, with rows representing a specific proteome and columns representing architectures. To determine the power-law fitting, the data were log-transformed and the linear fit and curve statistics were calculated using a geometric mean least-squares fitting technique. The slopes for each power law were compared using ANOCOVA by testing the hypothesis that the slopes were equal. The ubiquity of a FSF or FF is defined here as the percentage of proteomes within a superkingdom that contain a specific FF or FSF. All computations were conducted in Matlab.
Phylogenies of Protein Architectures.
A phylogenomic tree of protein architecture reconstructed using a global genomic census of protein-domain structure at the FSF level in 185 genomes spanning 19 Archaea, 129 Bacteria, and 37 Eukarya (19) was used to generate a timeline of architectural discovery. The tree was generated from FSF architectural abundance and was rooted by polarizing characters with a model based on two fundamental premises: (i) that protein structure is far more conserved than sequence and carries a strong phylogenetic signal, and (ii) that architectures that are popular in proteomes are more ancient. As with any phylogenetic method, the validity of character argumentation can be contentious but this has been discussed in detail elsewhere (25). The age (nd) of metal-binding FSFs relative to the rest of the protein universe was obtained directly from the tree of all FSFs. We chose this tree and not updated versions to allow direct comparison of evolutionary inferences of metal-binding and proteome diversification and because FSF functions are well annotated (25).
Supplementary Material
Acknowledgments
We thank Minglei Wang for help with tree reconstructions and Ken Nealson for a critical reading of the manuscript. This work was supported by a director's discretionary fund grant from the National Aeronautics and Space Administration National Astrobiology Institute (to C.L.D.) and National Science Foundation Grant MCB-0749836 and US Department of Agriculture-Cooperative State Research Education and Extension Service Grant Hatch Illu-802-314 (to G.C.-A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912491107/-/DCSupplemental.
References
- 1.Rosenzweig AC. Metallochaperones: Bind and deliver. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: From enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 3.Williams RJP, Frausto da Silva JJR. The Chemistry of Evolution: The Development of Our Ecosystem. Amsterdam: Elsevier; 2006. [Google Scholar]
- 4.Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747. [Google Scholar]
- 5.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 6.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 7.Williams RJP, Fraústo Da Silva JJ. Evolution was chemically constrained. J Theor Biol. 2003;220:323–343. doi: 10.1006/jtbi.2003.3152. [DOI] [PubMed] [Google Scholar]
- 8.Anbar AD. Oceans. Elements and evolution. Science. 2008;322:1481–1483. doi: 10.1126/science.1163100. [DOI] [PubMed] [Google Scholar]
- 9.Saito MA, Sigman DM, Morel FMM. The bioinorganic chemistry of the ancient ocean: The co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean-Proterozoic boundary? Inorganica Chemica Acta. 2003;356:308–318. [Google Scholar]
- 10.Bekker A, et al. Dating the rise of atmospheric oxygen. Nature. 2004;427:117–120. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- 11.Arnold GL, Anbar AD, Barling J, Lyons TW. Molybdenum isotope evidence for widespread anoxia in mid-Proterozoic oceans. Science. 2004;304:87–90. doi: 10.1126/science.1091785. [DOI] [PubMed] [Google Scholar]
- 12.Anbar AD, Knoll AH. Proterozoic ocean chemistry and evolution: A bioinorganic bridge? Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. [DOI] [PubMed] [Google Scholar]
- 13.Reinhard CT, Raiswell R, Scott C, Anbar AD, Lyons TW. A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science. 2009;326:713–716. doi: 10.1126/science.1176711. [DOI] [PubMed] [Google Scholar]
- 14.Lyons TW, Reinhard CT. Early Earth: Oxygen for heavy-metal fans. Nature. 2009;461:179–181. doi: 10.1038/461179a. [DOI] [PubMed] [Google Scholar]
- 15.Williams RJP. The natural selection of the chemical elements. Cell Mol Life Sci. 1997;53:816–829. doi: 10.1007/s000180050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji H-F, Chen L, Jiang Y-Y, Zhang H-Y. Evolutionary formation of new protein folds is linked to metallic cofactor recruitment. Bioessays. 2009;31:975–980. doi: 10.1002/bies.200800201. [DOI] [PubMed] [Google Scholar]
- 17.Dupont CL, Yang S, Palenik B, Bourne PE. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc Natl Acad Sci USA. 2006;103:17822–17827. doi: 10.1073/pnas.0605798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caetano-Anollés G, Caetano-Anollés D. An evolutionarily structured universe of protein architecture. Genome Res. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Yafremava LS, Caetano-Anollés D, Mittenthal JE, Caetano-Anollés G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007;17:1572–1585. doi: 10.1101/gr.6454307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreini C, Banci L, Bertini I, Rosato A. Occurrence of copper proteins through the three domains of life: A bioinformatic approach. J Proteome Res. 2008;7:209–216. doi: 10.1021/pr070480u. [DOI] [PubMed] [Google Scholar]
- 23.Morgan RO, Martin-Almedina S, Iglesias JM, Gonzalez-Florez MI, Fernandez MP. Evolutionary perspective on annexin calcium-binding domains. Biochim Biophys Acta. 2004;1742:133–140. doi: 10.1016/j.bbamcr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Shi W, et al. Metalloproteomics: High-throughput structural and functional annotation of proteins in structural genomics. Structure. 2005;13:1473–1486. doi: 10.1016/j.str.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Caetano-Anollés G, Wang M, Caetano-Anollés D, Mittenthal JE. The origin, evolution and structure of the protein world. Biochem J. 2009;417:621–637. doi: 10.1042/BJ20082063. [DOI] [PubMed] [Google Scholar]
- 26.Knoll AH, Javaux EJ, Hewitt D, Cohen P. Euykaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caetano-Anollés G, et al. The origin and evolution of modern metabolism. Int J Biochem Cell Biol. 2009;41:285–297. doi: 10.1016/j.biocel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Ji H-F, Chen L, Zhang H-Y. Organic cofactors participated more frequently than transition metals in redox reactions of primitive proteins. Bioessays. 2008;30:766–771. doi: 10.1002/bies.20788. [DOI] [PubMed] [Google Scholar]
- 29.Kump LR. In: Scientists Debate Gaia. Schneider SH, Miller JR, Crist E, Boston PJ, editors. Cambridge: MIT press; 2004. pp. 93–100. [Google Scholar]
- 30.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 31.Volk T. In: Scientists Debate Gaia. Schneider SH, Miller JR, Crist E, Boston PJ, editors. Cambridge: MIT press; 2004. pp. 27–36. [Google Scholar]
- 32.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 33.Twining BS, Baines SB, Vogt S, de Jonge MD. Exploring ocean biogeochemistry by single-cell microprobe analysis of protist elemental composition. J Eukaryot Microbiol. 2008;55:151–162. doi: 10.1111/j.1550-7408.2008.00320.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnston DT, Wolfe-Simon F, Pearson A, Knoll AH. Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth's middle age. Proc Natl Acad Sci USA. 2009;106:16925–16929. doi: 10.1073/pnas.0909248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature. 2008;455:1101–1104. doi: 10.1038/nature07381. [DOI] [PubMed] [Google Scholar]
- 37.John SG, Geis RW, Saito MA, Boyle EA. Zinc isotopic fractionation during high-affinity and low-affinity zinc transport by the marine diatom Thalassiosira oceanica. Limnol Oceanogr. 2007;52:2710–2714. [Google Scholar]
- 38.Vardi A, et al. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006;4:e60. doi: 10.1371/journal.pbio.0040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott C, et al. Tracing the stepwise oxygenation of the Proterozoic ocean. Nature. 2008;452:456–459. doi: 10.1038/nature06811. [DOI] [PubMed] [Google Scholar]
- 40.Glass JB, Wolfe-Simon F, Anbar AD. Coevolution of metal availability and nitrogen assimilation in cyanobacteria and algae. Geobiology. 2009;7:100–123. doi: 10.1111/j.1472-4669.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 41.Mulkidjanian A, Galperin M. On the origin of life in the Zinc world. 2. Validation of the hypothesis on the photosynthesizing zinc sulfide edifices as cradles of life on Earth. Biol Direct. 2009;4:27. doi: 10.1186/1745-6150-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sunda WG, Huntsman SA. Co and Zn interreplacement in marine phytoplankton: Evolutionary and ecological implications. Limnol Oceanogr. 1995;40:1404–1417. [Google Scholar]
- 43.Saito MA, Moffett JW, Chisholm SW, Waterbury JB. Cobalt limitation and uptake in Prochlorococcus. Limnol Oceanogr. 2002;47:1629–1636. [Google Scholar]
- 44.Heese D, Berger S, Röhm K-H. Nuclear magnetic relaxation studies of the role of the metal ion in Mn2(+)-substituted aminoacylase I. Eur J Biochem. 1990;188:175–180. doi: 10.1111/j.1432-1033.1990.tb15385.x. [DOI] [PubMed] [Google Scholar]
- 45.Maric S, et al. The M17 leucine aminopeptidase of the malaria parasite Plasmodium falciparum: Importance of active site metal ions in the binding of substrates and inhibitors. Biochemistry. 2009;48:5435–5439. doi: 10.1021/bi9003638. [DOI] [PubMed] [Google Scholar]
- 46.Speckhard DC, Wu FY-H, Wu C-W. Role of the intrinsic metal in RNA polymerase from Escherichia coli. In vivo substitution of tightly bound zinc with cobalt. Biochemistry. 1977;16:5228–5234. doi: 10.1021/bi00643a011. [DOI] [PubMed] [Google Scholar]
- 47.Predki PF, Sarkar B. Metal replacement in “zinc finger” and its effect on DNA binding. Environ Health Perspect. 1994;102(Suppl 3):195–198. doi: 10.1289/ehp.94102s3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravind L, Iyer LM, Koonin EV. Comparative genomics and structural biology of the molecular innovations of eukaryotes. Curr Opin Struct Biol. 2006;16:409–419. doi: 10.1016/j.sbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Price NM, Morel FMM. Cadmium and cobalt substitution for zinc in a marine diatom. Nature. 1990;344:658–660. [Google Scholar]
- 50.Gough J. Genomic scale sub-family assignment of protein domains. Nucleic Acids Res. 2006;34:3625–3633. doi: 10.1093/nar/gkl484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 52.Yang S, Doolittle RF, Bourne PE. Phylogeny determined by protein domain content. Proc Natl Acad Sci USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.