Abstract
Small heat-shock proteins (sHsps) are molecular chaperones that play an important protective role against cellular protein misfolding by interacting with partially unfolded proteins on their off-folding pathway, preventing their aggregation. Polyglutamine (polyQ) repeat expansion leads to the formation of fibrillar protein aggregates and neuronal cell death in nine diseases, including Huntington disease and the spinocerebellar ataxias (SCAs). There is evidence that sHsps have a role in suppression of polyQ-induced neurodegeneration; for example, the sHsp alphaB-crystallin (αB-c) has been identified as a suppressor of SCA3 toxicity in a Drosophila model. However, the molecular mechanism for this suppression is unknown. In this study we tested the ability of αB-c to suppress the aggregation of a polyQ protein. We found that αB-c does not inhibit the formation of SDS-insoluble polyQ fibrils. We further tested the effect of αB-c on the aggregation of ataxin-3, a polyQ protein that aggregates via a two-stage aggregation mechanism. The first stage involves association of the N-terminal Josephin domain followed by polyQ-mediated interactions and the formation of SDS-resistant mature fibrils. Our data show that αB-c potently inhibits the first stage of ataxin-3 aggregation; however, the second polyQ-dependent stage can still proceed. By using NMR spectroscopy, we have determined that αB-c interacts with an extensive region on the surface of the Josephin domain. These data provide an example of a domain/region flanking an amyloidogenic sequence that has a critical role in modulating aggregation of a polypeptide and plays a role in the interaction with molecular chaperones to prevent this aggregation.
Keywords: fibrillogenesis, α-crystallin
Small heat-shock proteins (sHsps) are important in the maintenance of cellular homeostasis. sHsps are induced under stress conditions and interact with partially unfolded proteins on their off-folding pathways, thereby providing a protective mechanism against protein misfolding and aggregation (1). They are present in many species, and 10 have been identified in the human proteome (see reviews in refs. 1 and 2). Alpha-crystallin (α-crystallin) is the most well characterized sHsp. The two isoforms of α-crystallin, αA-crystallin (αA-c) and αB-crystallin (αB-c), are a large component of the human eye lens, and αB-c is expressed in many other cell types including neurons (3, 4). Monomeric αB-c is approximately 20 kDa in mass, and under native conditions it forms a heterogeneous array of multimeric complexes ranging from 160 to 1,000 kDa in mass (2). The activity of αB-c is thought to involve hydrophobic interactions between αB-c and the partially folded protein. Under stress conditions, such as elevated temperature, the activity of αB-c is enhanced because of a conformational change within the multimeric complex (5, 6). Cellular stress is induced in several neurodegenerative diseases, such as Alzheimer’s disease (7), prion diseases (8), Parkinson disease (4), and Huntington disease (HD) (4), and there is evidence of increased αB-c activity (4). In addition, αB-c reduces neurotoxicity in a number of disease models (9, 10). In vitro the interaction between αB-c and a number of aggregating proteins has been characterized. Most substrates aggregate through a nucleation-dependent kinetic mechanism; however, the mode of interaction with αB-c appears to be dependent upon the specific target protein and the type of aggregation—i.e., amorphous or fibrillar (11–13). Complex stability is influenced by properties of the target protein, for example, the fibril-forming proteins apolipoprotein-CII (11) and amyloid-β (14) transiently interact with αB-c whereas α-synuclein (6) and κ-casein (15) form stable noncovalent complexes. Determining the mechanism by which αB-c interacts with specific aggregation-prone substrates is therefore important and may provide insight into the misfolding pathways leading to toxicity.
Polyglutamine (polyQ) expansion leads to the formation of fibrillar protein aggregates and neuronal cell death in nine diseases, including HD and the spinocerebellar ataxias (SCAs) 1, 2, 3, 6, 7, and 17. PolyQ peptides and proteins form fibrillar aggregates by a nucleation-dependent mechanism that is initiated by a monomeric nucleus (16–18). Recent evidence suggests that polyQ protein misfolding involves not only the polyQ tract but other aggregation-prone regions within the polyQ proteins (19–21). There is experimental evidence that the proteins ataxin-3 (in SCA3), ataxin-1 (in SCA1), and huntingtin (in HD) form fibrillar aggregates by a multidomain misfolding mechanism in which non-polyQ regions of the protein self-associate before the polyQ tract (19, 20, 22). Our laboratory has previously shown that ataxin-3 has a minimal two-stage aggregation mechanism (19). The first stage involves aggregation of the globular N-terminal Josephin domain that precedes the self-association of expanded polyQ segments (19).
sHsp overexpression decreases neuronal toxicity in HD by nonchaperone mechanisms, including suppression of reactive oxygen species (23) and stimulation of autophagy (24). In disease models overexpression of αB-c modulates toxicity in a context-dependent manner. αB-c did not alter the HD phenotype in a Drosophila model (25); however, it suppressed SCA3 toxicity in a Drosophila model (9). Enhanced suppression was observed when αB-c was coexpressed with full-length ataxin-3 in comparison with a C-terminal fragment not containing the Josephin domain (9). These data suggest that non-polyQ protein regions influence sHsp chaperone activity.
In this study we tested the chaperone ability of αB-c for a polyQ protein (SpAcQ52) that forms fibrillar aggregates by a mechanism involving only the polyQ region. We found that αB-c does not inhibit fibrillogenesis in this system. We then tested the effect of αB-c on the aggregation of ataxin-3 harboring a pathological length polyQ tract [at3(Q64)] and a truncated variant comprising only the Josephin domain. Our data show that αB-c potently inhibits the initial Josephin-dependent stage of ataxin-3 aggregation. By using NMR spectroscopy, we determined that αB-c interacts with specific regions of Josephin. These data provide an example and characterization of a domain/region flanking an amyloidogenic sequence that has a critical role in modulation of aggregation by interacting with molecular chaperones.
Results
αB-c Does Not Inhibit PolyQ-Mediated Aggregation.
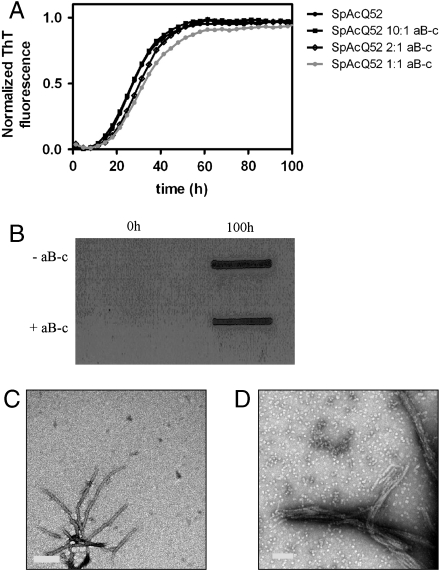
We first tested whether αB-c can prevent polyQ tract-mediated aggregation. We coincubated αB-c with a polyQ protein (SpAcQ52), which forms SDS-insoluble aggregates by a mechanism involving only the polyQ tract (26). The rate at which SpAcQ52 forms ThT-reactive aggregates does not change when incubated with increasing concentrations of αB-c (Fig. 1A). It has been previously shown that molecular chaperones can redirect the aggregation pathway of a protein from amyloid to amorphous aggregation (27, 28). Amorphous aggregates can be differentiated from amyloid by their sensitivity to SDS and appearance when visualized by transmission electron microscopy (TEM). So we tested whether SpAcQ52 incubated with αB-c forms SDS-insoluble aggregates by using a membrane filter-trap assay (29). These data demonstrate that the aggregates formed in the presence or absence of αB-c are insoluble in SDS (Fig. 1B). In addition, TEM analysis of the aggregates indicated that αB-c does not affect their fibrillar morphology. Fibrils ranging from 15 to 8 nm in width and 300 to 400 nm in length were observed in both the presence and absence of αB-c (Fig. 1C). Together, these data demonstrate that αB-c does not inhibit or modify the fibrillar aggregation of SpAcQ52.
Fig. 1.
The effect of αB-c on poly(Q) aggregation. (A) SpAcQ52 aggregation in the presence of increasing concentrations of αB-c monitored by ThT fluorescence. (B) Analysis of the SDS solubility of endpoint aggregates (100 h) in the presence and absence of αB-c. (C and D) TEM analysis of SpAcQ52 aggregates in the absence (C) and presence of αB-c. Ratios of SpAcQ52:αB-c are given on a molar basis.
αB-c Inhibits the Josephin-Dependent Stage of the Two-Stage Ataxin-3 Aggregation Pathway.
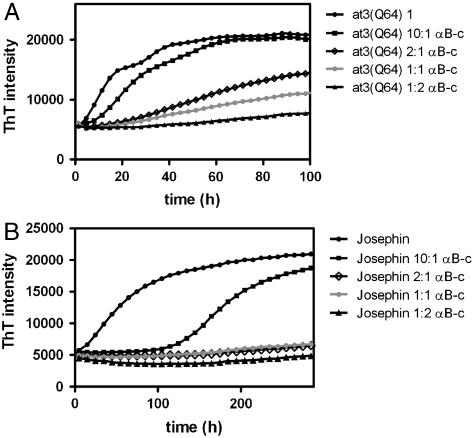
We have previously reported that full-length ataxin-3 forms aggregates by a two-stage mechanism (18). The aggregates formed in both stages are ThT-reactive, and in the absence of glycerol the two stages are characterized by a biphasic time course (Fig. 2A). The first stage is dominated by interactions of the N-terminal Josephin domain, which is an independently aggregation-prone domain (30). Aggregates formed during the first stage appear as short curvilinear structures and can be solubilized by SDS (19). These aggregates are stabilized in the second stage by interactions of the polyQ regions. Stage-two aggregates can be distinguished from stage-one aggregates because they are SDS-insoluble and morphologically appear as larger fibrillar structures. We determined the effect of αB-c on the two-stage aggregation pathway of at3(Q64). αB-c significantly slows the formation of ThT-reactive at3(Q64) aggregates in a concentration-dependent manner (Fig. 2A and Table 1).
Fig. 2.
The effect of αB-c on at3 aggregation. (A) At3(Q64) and (B) Josephin aggregation in the presence of increasing concentrations of αB-c monitored by ThT-fluorescence. Ratios of at3 variant:αB-c are given on a molar basis.
Table 1.
Half-lives (in hours) of Ataxin-3(Q64) aggregation followed by ThT and SDS insolubility in the presence and absence of αB-c
| ThT | SDS insolubility | ||
|
Stage 1 |
Stage 2 |
||
| Ataxin-3(Q64) | 9.3 ± 0.2 | 31.7 ± 0.9 | 25.5 ± 1.1 |
| Ataxin-3(Q64) 10∶1 αB-c | 19.7 ± 0.6 | 49 ± 4 | 45.1 ± 1.3 |
| Ataxin-3(Q64) 1∶1 αB-c | * | * | * |
| Josephin | 59.1 ± 3 | † | † |
| Josephin 10∶1 αB-c | 174 ± 0.3 | † | † |
| Josephin 1∶1 αB-c | ‡ | † | † |
*Aggregation incomplete after 300 h; therefore, data could not be fitted.
†Josephin does not undergo stage 2 of the ataxin-3 aggregation pathway and therefore does not form SDS-insoluble fibrils.
‡An equimolar amount of αB-c completely inhibited Josephin aggregation over 300 h.
The Josephin domain forms short aggregates with a ribbon-like morphology (30); however, the aggregation kinetics have not been described. Isolated Josephin forms ThT-reactive aggregates at a much slower rate than at3(Q64) (Fig. 2B and Table 1). The half-life of the first ThT-reactive phase of at3(Q64) aggregation represents the formation of Josephin-dependent fibrils (19). The half-life shifts from approximately 9 h in the full-length protein to around 60 h for the isolated Josephin domain (Table 1). These data indicate that the C-terminal polyQ tail in the context of the full-length protein accelerates Josephin domain aggregation.
αB-c has potent inhibitory activity on the aggregation of Josephin (Fig. 2B and Table 1), compared to the full-length at3(Q64) variant. Thus, an equimolar concentration of αB-c completely suppresses Josephin aggregation, but at3(Q64) still forms some ThT-reactive aggregates, even when αB-c is present in a 2∶1 molar excess. The reaction time course is too long to determine if the biphasic ThT pattern is altered at stoichiometric concentrations of αB-c. The potent inhibitory effect of αB-c on Josephin aggregation, combined with the data in Fig. 1 showing that αB-c does not inhibit polyQ-dependent aggregation, suggests that the slow ThT phase in the presence of high αB-c concentrations is dominated by polyQ intermolecular interactions.
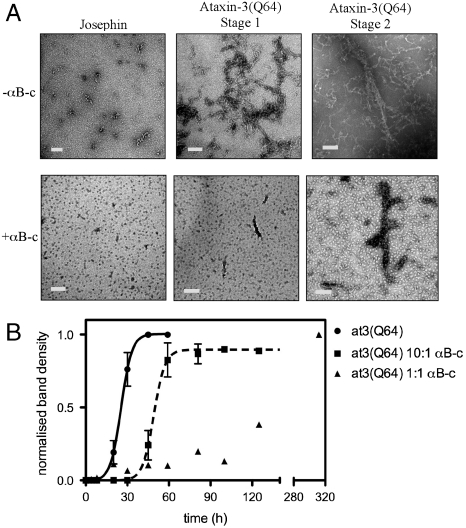
We analyzed the Josephin and at3(Q64) fibrils by TEM (Fig. 3A). Josephin forms short curvilinear fibrils, 5–10 nm in width and 10–100 nm long, when incubated alone. However, no fibrils were observed in samples incubated with equimolar concentrations of αB-c. Small, spherical aggregates characteristic of oligomeric αB-c are observed (Fig. 3A). After 7 h of incubation, curvilinear worm-like aggregates, 5–15 nm in width, consistent with the previously described morphology of stage-one aggregates (19), were observed in samples of at3(Q64) incubated alone. In the presence of αB-c, there were very few small aggregates present after 7 h. After 24 h, at3(Q64) was beginning to form larger fibrillar structures, ranging from 200 to 300 nm in length and 12 to 30 nm in width. These are consistent with the morphology of stage-two/endpoint fibrils. There were very few stage-two fibrils present in the samples incubated with αB-c.
Fig. 3.
The effect of αB-c on the formation of at3(Q64) SDS-insoluble fibrils. (A) TEM analysis of Josephin and ataxin-3 fibrils. (B) The poly(Q)-dependent stage of At3(Q64) aggregation was monitored by detection of SDS-insoluble fibrils using the membrane filter-trap assay. Membranes were analyzed by densitometry and data normalized.
At3(Q64) Forms SDS-Insoluble Fibrils at a Rate Modulated by Increasing αB-c Concentrations.
We tested the effect of αB-c on the formation of SDS-resistant at3(Q64) fibrils by using the filter-trap assay. Analysis of the membranes by densitometry showed that αB-c retards the formation of SDS-insoluble fibrils, as demonstrated by a twofold increase in half-life for at3(Q64) aggregation occurring in the presence of substoichiometric concentrations of αB-c [at3(Q64) 10∶1 αB-c] (Fig. 3B and Table 1). The inhibitory effect of αB-c is more pronounced at stoichiometric concentrations where SDS-insoluble fibrils were still formed but at much lower amounts relative to at3(Q64) incubated alone. The equimolar data could not be fit to the classic nucleation-dependent model because aggregation is incomplete over the 300-h time scale.
The Presence of αB-c Retains Ataxin-3 in a Monomeric Conformation.
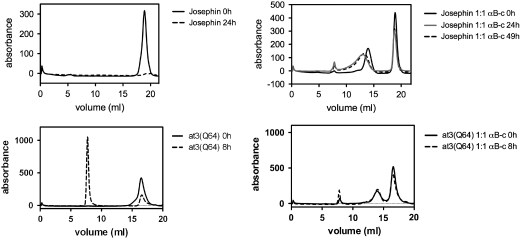
αB-c exists as a dynamic oligomeric complex (2). Depending on the target protein, it can irreversibly or transiently bind to target proteins, thereby inhibiting aggregation. To determine the stability of the αB-c:at3 association we performed size-exclusion chromatography (SEC) of Josephin and at3(Q64) incubated with equimolar concentrations of αB-c (Fig. 4).
Fig. 4.
Ataxin-3 aggregation in the presence and absence of αB-c monitored by SEC. Samples of Josephin and At3(Q64) incubated in the presence and absence of αB-c were removed at indicated time points and analyzed by SEC using a Superose 6 column. In the samples containing αB-c, equimolar amounts of substrate: αB-c were used.
Monomeric Josephin eluted from a Superose 6 column at approximately 18.9 mL. After 24-h incubation, Josephin formed insoluble aggregates that were removed by centrifugation prior to injection onto the column; therefore, the monomeric protein peak eluting at 18.9 mL was diminished (Fig. 4, Top Left). αB-c displayed a broad peak centered around 14 mL, consistent with αB-c being present as a polydisperse oligomer. In the presence of αB-c, most of the Josephin is retained in a monomeric conformation at 49 h, with some recruited into the αB-c complex, observed by a distinct shift and broadening of the αB-c peak (Fig. 4, Top Right). We performed SDS-PAGE to examine the composition of the peaks eluting at various times in the SEC experiments (Fig. S1A). A small amount of Josephin coelutes with the multimeric αB-c complex after a 48-h incubation period, and this is consistent with the shift and broadening of the peak corresponding to multimeric αB-c (Fig. 4).
At3(Q64) eluted as a single peak with a retention volume of approximately 17.8 mL. After 8 h, most of the at3(Q64) had aggregated, indicated by the presence of a large peak in the void volume (approximately 8 mL) of the Superose 6 column (Fig. 4, Bottom Left). After 8 h in the presence of αB-c, most of the at3(Q64) is retained in a monomeric conformation, with little change in the elution profile after this time (Fig. 4, Bottom Right). No at3(Q64) coeluted with the multimeric αB-c (Fig. S1B), consistent with the observation that the αB-c peak did not shift in the experiments with at3(Q64) (Fig. 4). The SEC data suggest that αB-c does not form a stable complex with at3(Q64)/Josephin maintaining the proteins in a monomeric conformation.
Addition of αB-c Affects Josephin/at3(Q64) Fibril Propagation.
The observation that αB-c retains Josephin and at3(Q64) in a monomeric conformation suggests that αB-c inhibits the conformational changes involved in nucleation of aggregation. αB-c has also been shown to inhibit fibril propagation in a number of systems (6, 11, 14, 27). We tested the effect of αB-c on Josephin and at3(Q64) fibril propagation by adding αB-c at various intervals after the commencement of aggregation.
When αB-c was added to isolated Josephin at 48 and 96 h, there was no further increase in ThT intensity, suggesting that αB-c prevents the further propagation of Josephin fibrils (Fig. S2A). Similar behavior is observed when αB-c is added at time points along the aggregation pathway of α-synuclein (6). When αB-c was added to the at3(Q64) reactions at various intervals, there was a flattening of the curve for a period of time and then a second slower aggregation phase (Fig. S2B). Because of the absence of this phase in the isolated Josephin reactions, we predict that the second phase is because of polyQ-dependent aggregation. Interestingly, the length of the flattened period was longer when αB-c was added at earlier time points in comparison to later time points, supporting the hypothesis that the rate at which poly(Q) interactions occurs is accelerated by the initial formation of stage-one Josephin-dependent fibrils.
αB-c Interacts with an Extensive Region of Josephin.
The ability of αB-c to inhibit ataxin-3 aggregation appears to be derived from inhibition of the Josephin-dependent stage (stage one) of the multidomain aggregation pathway. We used NMR spectroscopy to probe the molecular details of the Josephin:αB-c interaction surface.
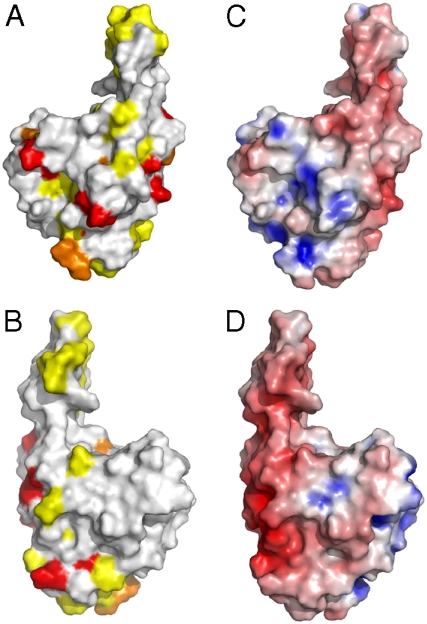
The NMR data show that αB-c interacts with an extensive region of Josephin (Fig. 5). A number of backbone amide cross-peaks in the 15N-HSQC (heteronuclear single quantum coherence) spectrum of Josephin were significantly broadened upon titration of αB-c up to a fivefold stoichiometric excess (Fig. S3). The broadening is consistent with exchange between the free and αB-c bound states of Josephin. Exchange broadening occurs when the time scale of the interaction is commensurate with the difference in NMR frequencies between the bound and free states, which is consistent with the transient interaction mechanism observed by SEC (Fig. 4). The Josephin residues perturbed to the largest degree were Q16, L19, N21, E26, S29, S35, I36, I77, W87, Q100, C114, V123, G127, F131, S135, L137, T138, L148, L155,Q156, E158, I162, Q176, and R182. These residues are confined to the globular subdomain of Josephin and include the active site residue C114.
Fig. 5.
αB-c interacts with specific regions of Josephin. (A and B) The residues perturbed upon addition of αB-c were mapped onto the Josephin domain (Protein Data Bank ID code 1YZB). Residues in intermediate exchange in the presence of αB-c (Red), residues shielded from broadening by Gd(DTPA-BMA) in the presence of αB-c (Yellow), and orange shows the overlapping residues. A and B show opposing faces of the Josephin structure. (C and D) The electrostatic surface of Josephin. Blue represents positive charge and red indicates negatively charged regions. C and D show opposing faces of the Josephin structure.
Because chemical shift perturbations can arise through indirect mechanisms (rather than a direct interaction between αB-c and Josephin), we used the paramagnetic relaxation agent Gd-diethylenetriamine pentaacetic acid-bismethylamide [Gd(DTPA-BMA)] to independently confirm the interaction surface (31, 32). The presence of Gd(DTPA-BMA) in solution causes paramagnetic relaxation enhancement, broadening the protein resonances. By performing these 15N-HSQC experiments in the presence and absence of αB-c, we could determine the αB-c interaction surface of Josephin, because the regions that interact with αB-c were partially shielded from paramagnetic relaxation (Fig. S4). Josephin residues that had > 20% stronger signals in the presence of Gd(DTPA-BMA) and a twofold excess of αB-c were S3, G11, L13, Q24, F28, A49, E50, G51, G52, T54, E56, D57, R59, T60, F61, S66, D71, G73, F75, S76, E90, N115, K128, N132, G139, L142, F151, G159, D168, E173, L178, M180, and I181. The identity of these residues correlates well with those broadened in the αB-c-bound state of Josephin (see above, Fig. 5). When mapped to the Josephin structure all of the residues that appeared to be most shielded from the paramagnetic probe by the presence of αB-c were found to be surface-exposed, validating these regions as the αB-c binding interface.
Discussion
sHsps Do Not Inhibit PolyQ Aggregation.
sHsps have a critical role in cellular homeostasis and can provide a barrier to the aggregation of partially unfolded proteins that are central to many neurodegenerative diseases. We investigated the effect of αB-c on the aggregation of a polyQ-containing model protein, finding that αB-c does not affect the formation of SDS-resistant polyQ fibrils when fibril formation is entirely polyQ-dependent (Fig. 1). Typically, sHsps (along with many other molecular chaperones) interact with exposed hydrophobic patches on target proteins. Glutamine is a hydrophilic amino acid; therefore, the surface hydrophobicity of polyQ would not increase during early aggregation stages. In addition, the polyQ region is thought to be in a condensed disordered state in the native ensemble (33, 34), therefore decreasing the accessibility of the side chains to sHsp interactions (35). In a manner akin to our findings with sHsps, molecular chaperones from other classes, e.g., Hsp70 and the chaperonin TRiC, which recognize surface hydrophobic patches of amyloidogenic intermediates, only weakly associate with polyQ proteins (28, 36). Therefore, the physiochemical properties of the polyQ tract may lead to its evasion by some chaperone protective mechanisms.
αB-c Inhibits the Josephin-Dependent Stage of Ataxin-3 Aggregation.
Pathological polyQ tracts are located within specific host proteins, and properties of the host protein are known to modulate aggregation and cellular toxicity (37, 38). Although molecular chaperones do not alter the misfolding of pure polyQ, genetic screens in Drosophila and Caenorhabditis elegans have identified several chaperones that modulate toxicity induced by ataxin-3, ataxin-1, and Htt exon 1, proteins that contain well-structured domain(s) coupled to a polyQ repeat (9, 39). As a result, we hypothesize that non-polyQ aggregation mechanisms play a role in pathogenesis. A previous study from our group has described a two-stage aggregation mechanism for pathological variants of ataxin-3 (19). Our data show that αB-c inhibits nucleation of the Josephin-dependent stage of ataxin-3 aggregation, subsequently having an indirect effect on the rate of poly(Q) intermolecular interactions. The chaperonin TRiC was recently shown to have a similar effect on the aggregation of Htt exon 1; TRiC suppresses aggregation by binding to the N-terminal domain adjacent to the polyQ tract (40).
αB-c had a more potent inhibitory effect on the aggregation of Josephin alone compared with at3(Q64). The Josephin aggregation stage is enhanced when this domain is part of at3(Q64), suggesting that the presence of the polyQ-containing C-terminal tail increases the rate of Josephin aggregation. The presence of the C-terminal polyQ tail leads to a slight decrease in the thermodynamic stability of Josephin (41), and we speculate that the tail alters the dynamic properties of the core domain enhancing the rate of conformational rearrangements on the aggregation pathway. The observation that αB-c has a greater inhibitory effect on the aggregation of isolated Josephin can be rationalized by a previous study demonstrating that larger target proteins require more αB-c (on a molar basis) for complete suppression of aggregation (42). The altered potency is also consistent with data showing that αB-c has a preference for target proteins that aggregate more slowly (43).
For the full-length protein, the interaction of αB-c with Josephin significantly slows the subsequent poly(Q)-dependent aggregation (Fig. 1 and Table 1). These data suggest that Josephin self-interactions lower the kinetic barrier to poly(Q)-dependent aggregation. The multidomain aggregation mechanisms described for ataxin-3 and Htt exon 1 suggest that a pool of small stage-one species can enhance the rate of polyQ-dependent aggregation by increasing the local polyQ concentration (19, 20). This suggestion is consistent with our data (Fig. S2B), which demonstrated that the formation of small Josephin fibrils is necessary for efficient formation of poly(Q)-dependent SDS-insoluble fibrils.
The significant effect of αB-c on ataxin-3 aggregation is consistent with a Drosophila model in which overexpression of αB-c decreased SCA3-induced toxicity (9), suggesting that inhibition of aggregation by chaperones is relevant in vivo. Another Drosophila study demonstrated that chaperone-induced retention of ataxin-3 in a monomeric conformation suppresses toxicity (44). Conversely, αB-c overexpression had no effect on the HD phenotype (25). Htt exon 1 has been proposed to have a similar two-stage aggregation mechanism compared to ataxin-3 (20). The alternative effects of αB-c in the ataxin-3 and Htt systems demonstrate how specific differences in multidomain misfolding pathways could lead to variable cellular responses.
αB-c Transiently Interacts with Monomeric Josephin.
The aggregation data clearly show that αB-c suppresses aggregation by associating with species on the Josephin aggregation pathway. Our SEC and NMR data show that inhibition mechanistically occurs by transient association of αB-c with a defined region on the Josephin surface that is partially coincident with the ubiquitin and HHR23B interacting surfaces (45–47). Interestingly, the perturbed residues are not all hydrophobic in character, and it is possible that some surface polar residues appear perturbed in our NMR studies because their amide groups are neighboring interacting hydrophobic groups. Whereas hydrophobic interactions are clearly important for chaperone action, it is also being increasingly recognized that substrate specificity is not purely driven by hydrophobic contacts (48–50).
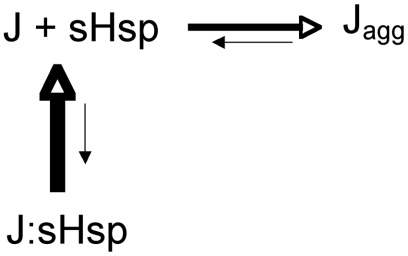
The SEC data show that the proteins are retained in a monomeric conformation in the presence of αB-c (Fig. 4). These data suggest that the rate at which αB-c interacts with Josephin is more rapid than the rate of misfolding. The NMR data indicate that this is a low affinity interaction; therefore, during this interaction, most of the Josephin is in a monomeric uncomplexed state. These data are consistent with a mechanism whereby αB-c binds and sequesters the free pool of monomeric proteins, thereby preventing fibril growth (Fig. 6).
Fig. 6.
A schematic model describing the effect of αB-c on at3 aggregation. Multimeric αB-c (a sHsp) transiently binds to at3 molecules in which Josephin is in an aggregation-prone conformation (Jagg). The J:sHsp complex is unstable, and monomeric Josephin is released from the sHsp.
The interaction between αB-c and monomeric Josephin, encompassing a large region of the target protein, is also observed, in the interaction of αB-c with the fibril-forming proteins α-synuclein (27) and κ-casein (6).
Conclusions
Our data demonstrate that inhibition of a non-polyQ aggregation stage by a chaperone can significantly impede protein aggregation involved in polyQ diseases. These data support the two-stage model as a plausible scheme for ataxin-3 aggregation in vitro. Therapeutic up-regulation of chaperone activity may be beneficial in SCA3 and other polyQ diseases that have a multidomain aggregation mechanism.
Experimental Procedures
Preparation of Proteins.
SpAcQ52 and the ataxin-3 variants used in this study were expressed and purified as previously described (26, 51). αB-c was expressed and purified as described in ref. 52.
Protein Aggregation Assays.
Aggregation assays were performed for SpAcQ52 and ataxin-3 variants at a protein concentration of 20 μM in Tris-buffered saline (TBS) (100 mM Tris, 80 mM NaCl, pH 7.4) containing 2 mM PMSF, 5 mM EDTA, and 15 mM ß-mercaptoethanol. Variable concentrations of αB-c were added to the samples, ranging from 2 to 40 μM. Samples were incubated at 37 °C.
ThT Measurements, Membrane Filter-Trap Assay, and TEM.
ThT measurements, the membrane filter-trap assay, and TEM analysis were performed as described in refs. 19 and 26.
SEC.
SEC of at3(Q64) and Josephin in the presence and absence of equimolar concentrations of αB-c was performed on an Akta-FPLC equipped with a Superose 6 10/300 GL column (Pharmacia). Prior to injection onto the column, samples were centrifuged for 5 min at 13,000 rpm to remove insoluble aggregated material. The absorbance at 214 nm was measured as proteins were eluted in TBS buffer at a constant flow rate of 0.5 mL/ min.
NMR Spectroscopy.
NMR methods are described in SI Text.
Supplementary Material
Acknowledgments.
This work was supported by the Australian Research Council and the National Health and Medical Research Council (NHMRC). S.P.B. is a NHMRC Research Fellow. H.E. was a NHMRC Peter Doherty Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914773107/-/DCSupplemental.
References
- 1.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12(10):842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 2.Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66(1):62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76(2):145–153. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 4.Iwaki T, et al. Accumulation of alpha B-crystallin in central nervous system glia and neurons in pathologic conditions. Am J Pathol. 1992;140(2):345–356. [PMC free article] [PubMed] [Google Scholar]
- 5.Das KP, Surewicz WK. Temperature-induced exposure of hydrophobic surfaces and its effect on the chaperone activity of alpha-crystallin. FEBS Lett. 1995;369(2–3):321–325. doi: 10.1016/0014-5793(95)00775-5. [DOI] [PubMed] [Google Scholar]
- 6.Rekas A, Jankova L, Thorn DC, Cappai R, Carver JA. Monitoring the prevention of amyloid fibril formation by alpha-crystallin. Temperature dependence and the nature of the aggregating species. FEBS J. 2007;274(24):6290–6304. doi: 10.1111/j.1742-4658.2007.06144.x. [DOI] [PubMed] [Google Scholar]
- 7.Lowe J, et al. alpha B crystallin expression in non-lenticular tissues and selective presence in ubiquitinated inclusion bodies in human disease. J Pathol. 1992;166(1):61–68. doi: 10.1002/path.1711660110. [DOI] [PubMed] [Google Scholar]
- 8.Renkawek K, et al. alpha B-crystallin is present in reactive glia in Creutzfeldt-Jakob disease. Acta Neuropathol. 1992;83(3):324–327. doi: 10.1007/BF00296796. [DOI] [PubMed] [Google Scholar]
- 9.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3(10):1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelmus MM, et al. Small heat shock proteins inhibit amyloid-beta protein aggregation and cerebrovascular amyloid-beta protein toxicity. Brain Res. 2006;1089(1):67–78. doi: 10.1016/j.brainres.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 11.Hatters DM, Lindner RA, Carver JA, Howlett GJ. The molecular chaperone, alpha-crystallin, inhibits amyloid formation by apolipoprotein C-II. J Biol Chem. 2001;276(36):33755–33761. doi: 10.1074/jbc.M105285200. [DOI] [PubMed] [Google Scholar]
- 12.Carver JA, et al. The interaction of the molecular chaperone alpha-crystallin with unfolding alpha-lactalbumin: A structural and kinetic spectroscopic study. J Mol Biol. 2002;318(3):815–827. doi: 10.1016/S0022-2836(02)00144-4. [DOI] [PubMed] [Google Scholar]
- 13.Devlin GL, Carver JA, Bottomley SP. The selective inhibition of serpin aggregation by the molecular chaperone, alpha-crystallin, indicates a nucleation-dependent specificity. J Biol Chem. 2003;278(49):48644–48650. doi: 10.1074/jbc.M308376200. [DOI] [PubMed] [Google Scholar]
- 14.Raman B, et al. AlphaB-crystallin, a small heat-shock protein, prevents the amyloid fibril growth of an amyloid beta-peptide and beta2-microglobulin. Biochem J. 2005;392(Pt 3):573–581. doi: 10.1042/BJ20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ecroyd H, et al. Mimicking phosphorylation of alphaB-crystallin affects its chaperone activity. Biochem J. 2007;401(1):129–141. doi: 10.1042/BJ20060981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Ferrone FA, Wetzel R. Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci USA. 2002;99(18):11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ignatova Z, Gierasch LM. Extended polyglutamine tracts cause aggregation and structural perturbation of an adjacent beta barrel protein. J Biol Chem. 2006;281(18):12959–12967. doi: 10.1074/jbc.M511523200. [DOI] [PubMed] [Google Scholar]
- 18.Ellisdon AM, Pearce MC, Bottomley SP. Mechanisms of ataxin-3 misfolding and fibril formation: Kinetic analysis of a disease-associated polyglutamine protein. J Mol Biol. 2007;368(2):595–605. doi: 10.1016/j.jmb.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 19.Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem. 2006;281(25):16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 20.Thakur AK, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16(4):380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulone D, Masino L, Thomas DJ, San Biagio PL, Pastore A. The interplay between PolyQ and protein context delays aggregation by forming a reservoir of protofibrils. PLoS ONE. 2006;1:e111. doi: 10.1371/journal.pone.0000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Chiara C, et al. The AXH domain adopts alternative folds the solution structure of HBP1 AXH. Structure. 2005;13(5):743–753. doi: 10.1016/j.str.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Wyttenbach A, et al. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11(9):1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- 24.Carra S, Brunsting JF, Lambert H, Landry J, Kampinga HH. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2{alpha} phosphorylation. J Biol Chem. 2009;284(9):5523–5532. doi: 10.1074/jbc.M807440200. [DOI] [PubMed] [Google Scholar]
- 25.Carra S, Sivilotti M, Chavez Zobel AT, Lambert H, Landry J. HspB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum Mol Genet. 2005;14(12):1659–1669. doi: 10.1093/hmg/ddi174. [DOI] [PubMed] [Google Scholar]
- 26.Robertson AL, et al. The structural impact of a polyglutamine tract is location-dependent. Biophys J. 2008;95(12):5922–5930. doi: 10.1529/biophysj.108.138487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rekas A, et al. Interaction of the molecular chaperone alphaB-crystallin with alpha-synuclein: Effects on amyloid fibril formation and chaperone activity. J Mol Biol. 2004;340(5):1167–1183. doi: 10.1016/j.jmb.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 28.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97(14):7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 30.Masino L, et al. Characterization of the structure and the amyloidogenic properties of the Josephin domain of the polyglutamine-containing protein ataxin-3. J Mol Biol. 2004;344(4):1021–1035. doi: 10.1016/j.jmb.2004.09.065. [DOI] [PubMed] [Google Scholar]
- 31.Liepinsh E, et al. Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer. Structure. 2001;9(6):457–471. doi: 10.1016/s0969-2126(01)00607-4. [DOI] [PubMed] [Google Scholar]
- 32.Pintacuda G, Otting G. Identification of protein surfaces by NMR measurements with a pramagnetic Gd(III) chelate. J Am Chem Soc. 2002;124(3):372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 33.Dougan L, Li J, Badilla CL, Berne BJ, Fernandez JM. Single homopolypeptide chains collapse into mechanically rigid conformations. Proc Natl Acad Sci USA. 2009;106(31):12605–12610. doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci USA. 2006;103(45):16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein FA, et al. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: Towards a length-dependent toxicity gradient. J Mol Biol. 2007;371(1):235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 36.Behrends C, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23(6):887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Nozaki K, Onodera O, Takano H, Tsuji S. Amino acid sequences flanking polyglutamine stretches influence their potential for aggregate formation. Neuroreport. 2001;12(15):3357–3364. doi: 10.1097/00001756-200110290-00042. [DOI] [PubMed] [Google Scholar]
- 38.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103(29):11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branco J, et al. Comparative analysis of genetic modifiers in Drosophila points to common and distinct mechanisms of pathogenesis among polyglutamine diseases. Hum Mol Genet. 2008;17(3):376–390. doi: 10.1093/hmg/ddm315. [DOI] [PubMed] [Google Scholar]
- 40.Tam S, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16(12):1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chow MK, Mackay JP, Whisstock JC, Scanlon MJ, Bottomley SP. Structural and functional analysis of the Josephin domain of the polyglutamine protein ataxin-3. Biochem Biophys Res Commun. 2004;322(2):387–394. doi: 10.1016/j.bbrc.2004.07.131. [DOI] [PubMed] [Google Scholar]
- 42.Lindner RA, Kapur A, Mariani M, Titmuss SJ, Carver JA. Structural alterations of alpha-crystallin during its chaperone action. Eur J Biochem. 1998;258(1):170–183. doi: 10.1046/j.1432-1327.1998.2580170.x. [DOI] [PubMed] [Google Scholar]
- 43.Lindner RA, Kapur A, Carver JA. The interaction of the molecular chaperone, alpha-crystallin, with molten globule states of bovine alpha-lactalbumin. J Biol Chem. 1997;272(44):27722–27729. doi: 10.1074/jbc.272.44.27722. [DOI] [PubMed] [Google Scholar]
- 44.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: Selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9(19):2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 45.Nicastro G, Habeck M, Masino L, Svergun DI, Pastore A. Structure validation of the Josephin domain of ataxin-3: Conclusive evidence for an open conformation. J Biomol NMR. 2006;36(4):267–277. doi: 10.1007/s10858-006-9092-z. [DOI] [PubMed] [Google Scholar]
- 46.Nicastro G, et al. Josephin domain of ataxin-3 contains two distinct ubiquitin binding sites. Biopolymers. 2009;91(12):1204–1214. doi: 10.1002/bip.21210. [DOI] [PubMed] [Google Scholar]
- 47.Nicastro G, et al. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci USA. 2005;102(30):10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plater ML, Goode D, Crabbe MJ. Effects of site-directed mutations on the chaperone-like activity of alphaB-crystallin. J Biol Chem. 1996;271(45):28558–28566. doi: 10.1074/jbc.271.45.28558. [DOI] [PubMed] [Google Scholar]
- 49.Aquilina JA, Watt SJ. The N-terminal domain of alphaB-crystallin is protected from proteolysis by bound substrate. Biochem Biophys Res Commun. 2007;353(4):1115–1120. doi: 10.1016/j.bbrc.2006.12.176. [DOI] [PubMed] [Google Scholar]
- 50.Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci USA. 2009;106(37):15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow MK, Ellisdon AM, Cabrita LD, Bottomley SP. Polyglutamine expansion in ataxin-3 does not affect protein stability: Implications for misfolding and disease. J Biol Chem. 2004;279(46):47643–47651. doi: 10.1074/jbc.M405799200. [DOI] [PubMed] [Google Scholar]
- 52.Horwitz J, Huang QL, Ding L, Bova MP. Lens alpha-crystallin: Chaperone-like properties. Methods Enzymol. 1998;290:365–383. doi: 10.1016/s0076-6879(98)90032-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.