Abstract
Hox genes play evolutionarily conserved roles in specifying axial position during embryogenesis. A prevailing paradigm is that changes in Hox gene expression drive evolution of metazoan body plans. Conservation of Hox function across species, and among paralogous Hox genes within a species, supports a model of functional equivalence. In this report, we demonstrate that zebrafish hoxa3a (zfhoxa3a) expressed from the mouse Hoxa3 locus can substitute for mouse Hoxa3 in some tissues, but has distinct or null phenotypes in others. We further show, by using an allele encoding a chimeric protein, that this difference maps primarily to the zfhoxa3a C-terminal domain. Our data imply that the mouse and zebrafish proteins have diverged considerably since their last common ancestor, and that the major difference between them resides in the C-terminal domain. Our data further show that Hox protein function can evolve independently in different cell types or for specific functions. The inability of zfhoxa3a to perform all of the normal roles of mouse Hoxa3 illustrates that Hox orthologues are not always functionally interchangeable.
Keywords: evolution, Hox, vertebrate, thymus, parathyroid
Hox genes encode a family of transcription factors with conserved roles in patterning the anterior–posterior axis during embryogenesis in all bilaterian animals (1). Mice and other mammals have 39 Hox genes arranged in four clusters located on four different chromosomes, whereas zebrafish and other teleosts have 48 Hox genes in seven clusters resulting from a genome-wide duplication (2). Hox genes from the same group (transparalogous genes or paralogues) arose from duplication, and share more similarity in protein sequence and expression pattern than genes within a cluster. Paralogous Hox genes often play diverse biological roles, as evidenced by their mutant phenotypes, but also show extensive redundancy and functional overlap. When vertebrate Hox genes have been expressed in Drosophila, they often provide similar functions as their Drosophila orthologues, supporting a model of functional equivalence of Hox genes cross phyla. In contrast to this striking functional conservation during evolution, many studies have shown a correlation between changes in Hox expression pattern and variation in morphological pattern (3, 4). These results have lead to a widely accepted model that cis-element evolution is the main driving force of morphological evolution, and is a major mechanism whereby Hox genes participate in this process (5). Although specific instances of protein functional divergence have been correlated with morphological evolution in arthropods (6), the degree to which cis-regulatory versus protein function changes influence morphological evolution remains controversial (5, 7). Furthermore, the largely nonsegmented body plan in vertebrates and increased potential for redundancy as a result of extra genome duplications raises the question of which mechanism(s) are operational in vertebrates.
The group 3 Hox genes are required for patterning part of the anterior body plan during embryogenesis. Extensive genetic studies of mouse Hoxa3 have demonstrated roles in patterning and development of endodermal, mesodermal, and ectodermal derivatives, and in cell migration, proliferation, apoptosis, and differentiation. Hoxa3 is expressed in the third and fourth pharyngeal pouch endoderm and in pharyngeal arch mesenchyme, and has similar anterior boundaries in multiple tissues (8). Null mutation of mouse Hoxa3 causes neonatal lethality, pharyngeal organ defects or aplasia, and defects in the tracheal epithelium, soft palate, pharyngeal skeleton, the IX cranial nerve, and the carotid body (8–12). Hoxa3 mutation also exacerbates the defects of single or compound mutants of Group 3 Hox paralogues in the axial skeleton and neural tube (10, 13, 14).
The striking functional equivalence of Hox3 paralogues was most dramatically demonstrated by swapping the Hoxa3 and Hoxd3 protein coding sequences, leaving the regulatory regions intact (15). Expressing either protein under the control of the other’s regulatory sequences resulted in a WT phenotype, providing strong evidence that these two proteins are functionally equivalent despite their different single mutant phenotypes and diverged protein sequences. Hoxa1 and Hoxb1 were also largely interchangeable by a similar approach (16). These and other studies support the functional equivalence of paralogous Hox proteins, and suggest that the overall quantity of Hox proteins may be more important than the specific proteins present (17, 18).
In this study we tested the conservation of Hoxa3 orthologues between mouse and zebrafish, the two major vertebrate model organisms. To test whether the zebrafish hoxa3a protein can substitute for mouse Hoxa3, we generated an allele in which mouse Hoxa3 coding sequence was precisely replaced with that of zfhoxa3a, the only Hoxa3 orthologue in zebrafish (19). The zebrafish gene expressed from mouse locus complemented some defects seen in the mouse Hoxa3 null mutant, consistent with the equivalence of mouse Hoxa3 and Hoxd3, showing that all three proteins share conserved biological functions. However, the Hoxa3zf allele was equivalent to the null in the development of the IXth cranial nerve, thymus, and parathyroid, and had a neomorphic pharyngeal skeleton phenotype. Using a second strain in which only the C-terminal half of the protein is from zebrafish, we show that these functional differences primarily map to the domain downstream of the homeodomain. Although protein sequence alignment showed that overall, the zebrafish hoxa3a and mouse Hoxa3 and Hoxd3 proteins showed similar degrees of conservation, zebrafish hoxa3a appears to have undergone extremely rapid molecular evolution relative to other vertebrate Hoxa3 orthologues. These data provide evidence that the zebrafish hoxa3a and mouse Hoxa3 proteins have functionally diverged since their respective taxa last shared a common ancestor, and suggest that these differences are to the results of changes outside the homeodomain.
Results
Expression of Zebrafish hoxa3a Protein from Mouse Hoxa3 Locus.
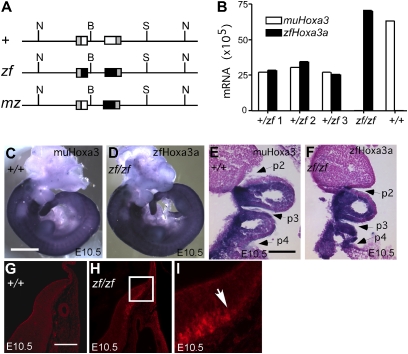
To generate a new Hoxa3 allele (Hoxa3zf) that expressed the zfhoxa3a protein from the endogenous mouse Hoxa3 locus, the mouse Hoxa3 protein coding sequences were replaced with those of zebrafish hoxa3a by gene targeting, and a C-terminal HA tag was added, similar to the strategy used previously for the mouse Hoxa3-Hoxd3 swap (Fig. 1A and Fig. S1) (15). All sequences outside the protein coding domains, including the intron between the two coding exons, were from the mouse locus. Another allele, Hoxa3mz, was produced as a consequence of recombination occurring within the mouse intron and in the homologous sequences 3′ of the neoR cassette (Fig. 1A and Fig. S1). Hoxa3mz encodes a protein with mouse N-terminal domain (NTD) and hexapeptide sequences and zebrafish homeodomain and C-terminal domain (CTD).
Fig. 1.
Structure and expression of Hoxa3 alleles. (A) Scheme of Hoxa3 WT (+), Hoxa3zf (zf), and Hoxa3mz (mz) alleles. Horizontal thin lines represent noncoding genomic DNA at the mouse Hoxa3 locus. Boxes represent exons as follows: gray, 5′ or 3′UTR of mouse Hoxa3; white, mouse Hoxa3 coding sequences; black, zebrafish hoxa3a coding sequences. N, NotI; B, BamHI; S, SpeI. (B) Quantitative RT-PCR shows equivalent mRNA levels for the zfhoxa3a (zf) and mouse Hoxa3 (WT) transcripts in individual whole heterozygous embryos (+/zf 1–3), or in homozygotes (zf/zf, +/+). Whole mount (C and D) and coronal paraffin section (E and F) in situ hybridization of embryonic d 10.5 embryos using allele-specific probes shows identical spatial expression patterns for the WT murine and zf alleles. (G–I) Immunofluorescence detection of the HA tag in the zfHoxa3 protein in the hindbrain at embryonic d 10.5. Box in H corresponds to panel in I. Arrow in I shows anterior limit of protein detection. (Scale bars: 1 mm in C and D; 200 μm in G–I.)
As recent data have identified transcription factor binding sites within the coding sequence of the Hoxa2 gene (20), we tested whether the zf and mz alleles had the same mRNA expression patterns and levels as the WT mouse allele. At embryonic d 10.5, the Hoxa3zf allele was expressed correctly, with the same spatial and temporal pattern and at the same level as the WT Hoxa3 mRNA (Fig. 1 B–F). Analysis of the zebrafish hoxa3a protein using the HA tag showed that the protein was present and had the correct anterior limit in the hindbrain (Fig. 1 G–I).
Conserved Hoxa3 Protein Functions.
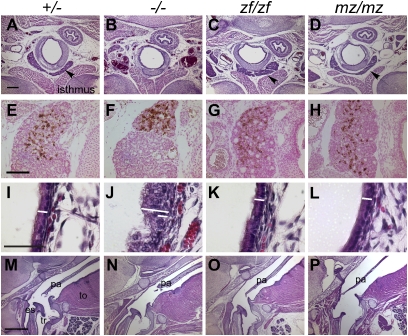
We tested whether zebrafish hoxa3a protein was able to substitute for mouse Hoxa3 in mice that expressed only the zf or mz alleles (zf/zf or mz/mz; Table 1), or in compound heterozygotes with the null allele (zf/null or mz/null). In the Hoxa3-null mutant mouse, the ventral thyroid isthmus is absent or ectopic (Fig. 2 A and B), and the ultimobranchial body–derived C-cells fail to mix with the thyroid proper (Fig. 2 E and F) (8). Neither of these phenotypes were present in Hoxa3zf/zf and Hoxa3mz/mz mice (Fig. 2 C, D, G, and H). The tracheal epithelium in Hoxa3-null mutants has a thicker epithelial layer and a convoluted surface (8) (Fig. 2 I and J). In all the Hoxa3zf/zf and Hoxa3mz/mz animals examined, this epithelium was normal (Fig. 2 K and L). These phenotypes were also rescued by only one copy of the Hoxa3zf or Hoxa3mz allele (zf/null and mz/null; Fig. S2 E–J).
Table 1.
Summary of the phenotypes, showing that Hoxa3mz allele functions virtually the same as Hoxa3zf allele
| Location | Hoxa3null/null | Hoxa3zf/zf | Hoxa3mz/mz |
| Thyroid isthmus | Deleted or ectopic | WT | WT |
| Ultimobranchial body | Separated from thyroid | WT | WT |
| Tracheal epithelium | Disorganized | WT | WT |
| Soft palate | Truncated | WT | WT |
| IX cranial nerve | Disconnected or fused to X | Null | Null |
| Thymus | Athymia | Null | Null |
| Parathyroid | Aparathyroidism | Null | Null |
| Throat cartilage | Malformed | Null | Null |
| Hyoid lesser horn | Deleted | Neomorphic* | Neomorphic† |
*The hyoid lesser horn is different in morphology from WT.
†The lesser horn of Hoxa3mz/mz appears different from either WT or Hoxa3zf/zf.
Fig. 2.
Zebrafish hoxa3a can substitute for mouse Hoxa3 in thyroid, ultimobranchial body, tracheal epithelium, and soft palate development. Transverse (A–L; dorsal is up) or sagittal (M–P; anterior is up, dorsal is to the left.) paraffin sections of newborn animals; genotypes apply to each column. Scale bars apply to each row. (A–D) The thyroid isthmus (arrow) is deleted in Hoxa3null/null (−/−) mice (B), but restored in Hoxa3zf/zf (zf/zf) (C) and Hoxa3mz/mz (mz/mz) (D). (E–H) Transverse sections of newborn mice stained with anticalcitonin antibody (brown). Integration of ultimobranchial body–derived C cells is restored in zf/zf (G) and mz/mz (H) mice. (I–L) The disorganized tracheal epithelium in the Hoxa3-null mutant (J) was not seen in zf/zf (K) or mz/mz (L) animals. The white bar in each panel shows the thickness of the WT epithelium, contrasted with the null mutant (long bar in J). (M–P) The posterior palate (velum) is shortened in Hoxa3-null mutants (N), but is normal in zf/zf and mz/mz mice (M, O, and P). tr, trachea; es, esophagus; pa, palate; to, tongue. (Scale bars: 200 μm in A–D; 100 μm in E–H; 50 μm in I–L; 800 μm in M–P.)
Hoxa3null/null animals have a truncated secondary palate (9) that can result in a bloated abdomen caused by breathing air into the esophagus (Fig. 2 M and N and Fig. S2 A and B). In all Hoxa3zf/zf, Hoxa3mz/mz, and Hoxa3mz/null newborns analyzed, the secondary palate was normal, and the bloated abdomen phenotype was never present (Fig. 2 O and P, Table S1, and Fig. S2L). The exception was in zf/null mice, which had only a partial rescue of palate length that was unable to prevent bloating (Fig. S2 D and K). Thus, the zf and mz alleles were able to completely substitute for mouse Hoxa3 in the thyroid/ultimobranchial body and tracheal epithelium, and in most cases in the soft palate. Interestingly, mutants of all genotypes (zf/zf, mz/mz, zf/null, mz/null) died within hours after birth, similar to the null mutant (9), indicting that other phenotypes contribute to the neonatal lethality of Hoxa3 null mutants.
Diverged Hoxa3 Protein Functions.
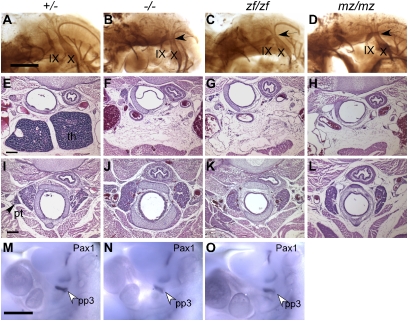
Although zebrafish hoxa3a was sufficient for the normal development of some tissues, it was equivalent to a null allele in other aspects of the phenotype. In the majority of Hoxa3-null mutants, the IXth cranial nerve is either fused to the vagus nerve (X) th or disconnected from the hindbrain (10, 12). Similar defects at similar frequencies were observed in Hoxa3zf/zf and Hoxa3mz/mz embryos (Fig. 3 A–D, Table 1 and Table S2).
Fig. 3.
Cranial nerve, thymus, and parathyroid defects are not rescued by zfhoxa3a. (A–D) Whole-mount antineurofilament staining of embryonic d 10.5 control, Hoxa3null/null (−/−), Hoxa3zf/zf (zf/zf), and Hoxa3mz/mz (mz/mz) embryos. In the control, the IX cranial nerve is connected to hindbrain. (B) In −/− embryos, the IX cranial nerve is often fused (arrow) to the X cranial ganglia. (C and D) The same fusion is observed in zf/zf and mz/mz mutants. (E–L) Transverse paraffin sections of newborn animals stained with hematoxylin and eosin (dorsal is up). (Scale bar: 200 μm.) The thymus (F–H) and parathyroids (J–L) are absent in −/−, zf/zf, and mz/mz mutant mice. th, thymus; pt, parathyroid. (M–O) Whole-mount in situ hybridization for Pax1 at embryonic d 10.5. Pax1 expression in the third pouch (pp3) is reduced in −/− embryo, but expression in the other pouches is unchanged. zf/zf shows a similar pattern as −/− (cranial is up). (Scale bar: 500 μm.)
The most consistent phenotype of the Hoxa3-null mutant is the absence of thymus and parathyroids. Neither organ was detected at the normal or any ectopic location in Hoxa3zf/zf and Hoxa3mz/mz animals (Fig. 3 E–L). These morphological defects were supported by earlier changes in marker expression associated with thymus and parathyroid organogenesis. The thymus-specific marker, Foxn1 (21), was absent from the third pharyngeal pouch in both Hoxa3null/null and Hoxa3zf/zf E11.5 embryos (Fig. S2 O–Q). Expression of Gcm2, which is required for parathyroid organogenesis (22, 23), was also greatly reduced in both mutants (Fig. S2 R–T). Like the null mutant, the expression of Pax1, a potential downstream target of Hoxa3, was also reduced specifically in the third pouch of the Hoxa3zf/zf embryos at embryonic d 10.5 (8) (Fig. 3 M–O).
These results show that both Hoxa3zf and Hoxa3mz act as null alleles in cranial nerve IX, thymus, and parathyroid development. As the homeodomain is identical between the mouse and zebrafish proteins, these functional differences map to the downstream CTD.
Morphological Differences in Hyoid Development from zf and mz Alleles.
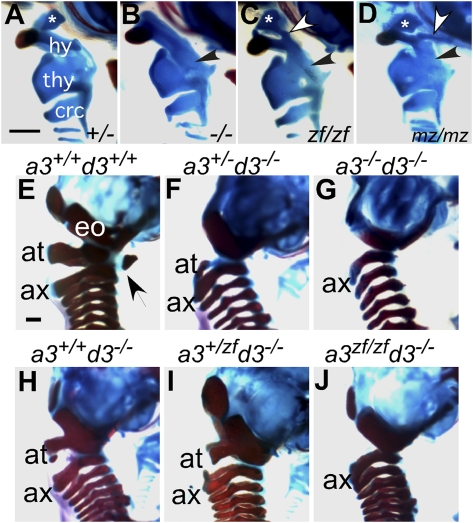
The lesser and greater horns of the hyoid bone are derived from the second and third pharyngeal arches, respectively. In the Hoxa3null/null mouse, the lesser horn is absent or greatly reduced and the greater horn is malformed and fused to the thyroid cartilage (10, 14) (Fig. 4B). The Hoxa3zf/zf and Hoxa3mz/mz mutants had similar greater horn phenotypes as the null mutant, with fusions to the thyroid cartilage, which was also malformed (Fig. 4 C and D, black arrows). Although both these mutants did rescue the presence of a lesser horn, in both cases it had morphologies different from WT. The zf/zf lesser horn had a teardrop shape that was fused to the middorsal cranial aspect of the greater horn (Fig. 4C, white arrow). The lesser horn in the mz/mz mutant was more square than WT or zf/zf, and there was an extra cartilage extension from the greater horn back to the base of the skull that was not seen in the WT or zf/zf genotypes (Fig. 4D, white arrow). Neither phenotype was affected by reducing the dose in zf/null or mz/null mice (Fig. S2 M and N). These data suggest that hyoid lesser horn patterning is sensitive to a species difference in Hoxa3 protein function, and that both the NTD and CTD may contribute. These phenotypes were always recessive to the WT morphology, as heterozygotes for either the zf or mz alleles had normal morphologies, showing that the mouse protein was dominant to the zebrafish protein in establishing pharyngeal skeletal morphology.
Fig. 4.
Novel pharyngeal skeleton morphologies in Hoxa3zf/zf and Hoxa3mz/mz mice, and skeletal phenotype of compound mutants with Hoxd3. (A–D) Lateral views of the throat cartilages in cleared newborn skeletal preparations. Anterior is up, dorsal is to the right. Asterisk indicates the lesser horn of hyoid; hy, greater horn of hyoid; thy, thyroid cartilage; crc, cricoid cartilage. (Scale bar: 500 μm.) In Hoxa3null/null (−/−), Hoxa3zf/zf (zf/zf), and Hoxa3mz/mz (mz/mz), the greater horn is malformed and fused to the thyroid cartilage (black arrows in B–D). (B) In the null, the lesser horn of the hyoid is greatly reduced or deleted. (C and D) The zf/zf and mz/mz mutants have distinct hyoid morphologies, and are different from WT or null. White arrows show extra cartilage structures in these mutants. (E–J) Lateral views of the cervical region in cleared skeleton preparations of the indicated genotypes for Hoxa3 (a3) or Hoxd3 (d3). Anterior is up, dorsal is to the left. Exoccipital (eo) bone, atlas (at), axis (ax), and anterior arch of atlas (arrowhead) are indicated. Note that G and J are similar, whereas I is more similar to H than to F. (Scale bar: 1 mm.)
Quantitative Analysis of Zebrafish-Derived Proteins in Mice.
Although the zf and mz alleles were correctly expressed at the mRNA level, it was possible that their failure to fully substitute for the mouse protein was a result of reduced translation or protein stability. We tested whether the mouse and zebrafish proteins had similar steady-state protein levels in vivo using a proteomics approach to quantify protein levels using MS. We used the mz allele for this test, as it shared the NTD with the mouse WT protein and is functionally similar to the zf allele, and measured the amount of each protein in whole mz/mz homozygous or WT E10.5 embryos. Using a tryptic peptide from the mouse Hoxa3 NTD that was not found in other Hox proteins, the normalized area under the peak from a reconstructed ion chromatogram was compared between the two samples. The ratio of Hoxa3 between mz/mz and WT mice was calculated as 1.2:1; this slight difference was not significant. The similar levels and localization of mRNA and protein for the mouse and zebrafish proteins show that any functional differences between these alleles are a result of functional differences between these proteins.
Interaction Between zfHoxa3 and Hoxd3 in the Axial Skeleton.
Although Hoxa3null/null mutants have a normal axial skeleton, reducing Hoxa3 dosage in a Hoxd3 mutant background reveals a dosage-dependent functional redundancy for these genes (10, 14). Reducing the dose of Hoxa3 in Hoxd3 homozygotes leads to progressively more severe defects of the atlas and basioccipital bone, with the entire atlas deleted in Hoxa3−/−;Hoxd3−/− mice (Fig. 4 E–H) (14). Hoxa3zf/zf;Hoxd3−/− mice had the same phenotype as Hoxa3−/−;Hoxd3−/− (Fig. 4 G and J), suggesting that Hoxa3zf allele might function as a null allele in the cervical vertebrae. However the zf allele did not show the same dose dependency as the mouse null, as Hoxa3+/zf;Hoxd3−/− mice had a phenotype more similar to Hoxa3+/+;Hoxd3−/− than Hoxa3+/−;Hoxd3−/− (Fig. 4I). These results suggest that zebrafish hoxa3a has a function intermediate to the null and WT alleles of mouse Hoxa3 in patterning the cervical vertebrae.
zfhoxa3a Failed to Substitute for Mouse Hoxa3 in Neural Crest.
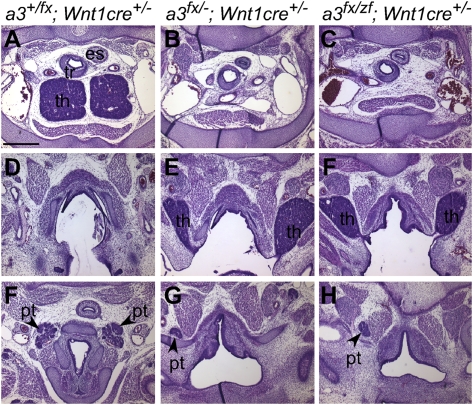
To test the tissue specificity of zfhoxa3-associated phenotypes, we generated Wnt1cre;Hoxa3fx/zf animals, in which NCCs expressed only zfhoxa3a, but all other tissues were heterozygous (+/zf). These mice had phenotypes essentially identical to a NCC-specific KO of Hoxa3 (Fig. 5). Deletion of a conditional allele of Hoxa3 in neural crest cells (NCCs) using Wnt1cre caused defects in thymus and parathyroid morphogenesis, including failure to separate from the pharynx and delayed detachment of the parathyroids in Wnt1cretg/0;Hoxa3fx/null mice (Fig. 5 A, B, D, and E). Thymic lobes were ectopic and still connected to the pharynx (Fig. 5C), with overall size and organization of these lobes similar to the WT thymus. The parathyroid was ectopic but normal in morphology (Fig. 5 F–H). Thus, zebrafish hoxa3a failed to substitute for mouse Hoxa3 for its function in NCCs to support later stages of thymus and parathyroid morphogenesis.
Fig. 5.
Hoxa3zf allele has null function in NCCs. Hematoxylin and eosin staining of transverse paraffin sections from embryonic d 15.5 embryos (dorsal is up). Genotypes apply to each column; panels in each row are from a comparable anterior–posterior location. In the control embryo, the thymus (th) is located in the chest (A), and the parathyroids (pt) are embedded in the thyroid (F). In embryos with a NCC-specific deletion of mouse Hoxa3 (Hoxa3fx/-;Wnt1cre+/−) the thymic lobes are absent from the normal position (B), and are instead still attached to the pharynx and are ectopic (E). Parathyroids are also ectopic and anterior to the thymus (G). (C, F, and H) Embryos in which only the zf allele is expressed in the NCC (Hoxa3fx/zf;Wnt1cre+/−) have a phenotype identical to NCC-specific Hoxa3 deletion. (Scale bar: 400 μm.)
Accelerated Evolution of the zfhoxa3a Protein.
To further map the sequences responsible for these phenotypic differences, we compared the protein sequences of mouse Hoxa3, Hoxd3, and zebrafish hoxa3a genes. As Hoxa3 and Hoxd3 are functionally similar, their comparison acts as a baseline for estimating similarity based on primary protein sequence. The overall identity between zfhoxa3a and muHoxa3 (59%) was slightly higher than that between muHoxa3 and muHoxd3 (52%; Fig. S3A). Murine Hoxa3 and zfhoxa3a have identical hexapeptide and homeodomain sequences, and the region between these domains was also highly conserved; in these DNA binding and cofactor interaction domains, the mouse and zebrafish Hoxa3 proteins were more similar than were the mouse Hoxa3 and Hoxd3 proteins. The region N-terminal to the hexapeptide contained a proline-rich domain in mouse Hoxa3 that was absent in mouse Hoxd3 and zfhoxa3a. The CTDs of the three proteins shared higher homology than the N-terminal regions, and had several short regions that were highly conserved in all three proteins. Overall, there were no obvious regions of high homology between the two mouse proteins that were absent or diverged in the zebrafish protein.
As a more sensitive test of sequence divergence, we performed molecular evolutionary analyses of select Hoxa3 proteins (without homeodomain) from cartilaginous fishes, bony fishes, and various tetrapods (Figs. S3B, S4, and S5 and Table S3). A neighbor-joining tree rooted to the cartilaginous fish lineage showed that the zfhoxa3a sequence is considerably distant from other vertebrate Hoxa3 sequences, including those of other teleost fishes (Fig. S3B). This suggests that its rate of molecular evolution may be accelerated relative to other vertebrates. Relative rate tests (RRTs) using the Tajima method (24) confirmed that the zebrafish gene has undergone a significantly faster rate of evolution than orthologues from all other vertebrate taxa, including bony fishes (Table S3). Interestingly, although functional differences primarily mapped to the CTD, we did not observe overt differences in tree topology or relative rates of evolution between the NTD versus CTD of Hoxa3 when the respective sequences were stratified.
Discussion
Cross-species functional tests have provided valuable information in understanding the evolution of gene function, including Hox genes. To our knowledge, the present study is the first cross-species study to use precise gene replacement in mice as previously used to show functional homology of paralogous mouse Hox proteins. We have used this approach to test the functional equivalence of the Hoxa3 gene from two distantly related vertebrate species. Mouse Hoxa3 is required for the development of a diverse range of structures and tissues, providing a unique opportunity to test Hox gene function. The application of quantitative proteomics technology to measure steady-state protein levels provides a high degree of confidence that the zebrafish-derived mRNA sequences are correctly transcribed and translated, and that the resulting fish-derived proteins have similar steady-state levels as the mouse protein. The combination of this genetic approach with the ability to assay a wide range of phenotypic parameters allowed us to test with great sensitivity whether zebrafish hoxa3a is equivalent to mouse Hoxa3 when they are expressed in the same biological context.
Nonequivalence of Mouse and Zebrafish Hoxa3 Proteins.
Our data show that the mouse and zebrafish Hoxa3 proteins have nonequivalent in vivo functions. This result is not predicted by the paradigm of functional equivalence, which holds that conserved transcriptional regulators, such as Hox proteins, are generally equivalent between paralogues and orthologues. The use of genetic complementarity studies across species was an early aspect of vertebrate Hox gene studies, and the ability of human, mouse, and chicken Hox genes to rescue Drosophila Hox mutations or to mimic overexpression phenotypes revealed a high degree of functional equivalence between distant Hox orthologues (25–27). The ability of vertebrate Hox genes to function in an insect indicates that vertebrate Hox proteins have retained many ancestral functions across long evolutionary distances and dramatic morphological changes. However, it is possible that Hox proteins may have also evolved novel functions during vertebrate evolution, in addition to retaining ancient functions.
Our data suggest that, unlike the mouse paralogous Hox3 genes, Hoxa3 protein function has evolved considerably since the divergence of the ancestral species that gave rise to mammals and teleosts. This result is surprising, as the genome-wide duplication that generated the Hoxa3 and Hoxd3 paralogs occurred at least 450 Mya, and before the divergence of teleost fishes from the vertebrate lineage leading to mammals (2). As a result, zebrafish hoxa3a diverged from mouse Hoxa3 more recently than mouse Hoxd3, but shows clear evidence of rapid molecular evolution in both the NTD and CTD (Fig. S4B and Table S3). However, only the CTD showed evidence of functional divergence. The combination of molecular evolution analysis with the genetic test of functional equivalence provides strong evidence that the zebrafish hoxa3a is functionally distinct. The inability of zfhoxa3a to perform all of the functions of Hoxa3 shows that orthologues are not always functionally equivalent, and shows that Hox protein divergence appears to occur in vertebrates, as has also been shown in arthropods (6). Analysis of the coelacanth genome compared with mammalian and teleost genomes, and particularly of Hox gene clusters, indicates that teleosts have considerable genetic plasticity relative to the lineage leading to mammals, with an extra genome duplication and rapid genetic and phenotypic radiation (28–30). Our data suggest that the zebrafish hoxa3a gene has evolved at both the gene and protein function level compared with its mammalian orthologs. Although the additional genome duplication in zebrafish and subsequent subfunctionalization could account for individual gene differences in some cases, in this case the duplicated gene (i.e., Hoxa3b) has not been retained and therefore cannot be providing the missing functions, further supporting our conclusion of protein functional divergence.
Several molecular mechanisms could explain the failure of zfhoxa3a to substitute fully for mouse Hoxa3. Hoxa3 and hoxa3a may have independently evolved novel functions in addition to ancestral protein functions and/or lost ancestral functions since the divergence of these two vertebrate lineages. Our evidence for rapid molecular evolution of the zfhoxa3a gene suggests that the zebrafish protein may have acquired or lost functions compared with the mouse. It is also possible that within each species, each protein is required for the same functions, but that the genetic networks within which they function have coevolved such that zfhoxa3a cannot interact properly with components of the mouse network in some cell types. It is intriguing in this respect that only some functions are lost whereas others are retained, suggesting that Hoxa3 may interact with independent gene networks in different cell types or at different times during development.
CTD of the Hoxa3 Protein.
Our data also show that the majority of the functional differences between mouse and zebrafish Hoxa3 proteins maps to the second coding exon, which includes the homeodomain and CTD. Although differences in homeodomain sequence in different mouse paralogous Hox groups can be associated with functional differences (31, 32), zebrafish hoxa3a has an identical homeodomain to mouse Hoxa3. Thus, the functional differences that we have identified between these two Hox proteins must reside downstream of the homeodomain. Our data may provide the first in vivo evidence for specific required functions for vertebrate Hox proteins outside of the conserved hexapeptide and homeodomain regions. Interestingly, only group 2 and group 3 Hox proteins have a long CTD, which is present in group 3 Hox proteins at least as far back as tunicates (33). Therefore, this domain could be a critical target for the functional evolution of Hox3 proteins. CTD sequences that are conserved between mouse Hoxa3 and Hoxd3 proteins, but diverged in zebrafish hoxa3a, are thus candidates for mediating this functional divergence.
Hox Protein Function and Morphological Evolution.
Hoxa3 was the first Hox gene to be mutated in mice (9), and the diversity of phenotypes in Hoxa3-null mutants has allowed us to analyze the functional equivalence of the mouse and zebrafish orthologues in much greater detail than has been possible in prior studies of cross-species functional conservation (8–10, 13, 14). Given the different morphologies in the pharyngeal regions of fish and mammals and the hypothesized role of Hox genes in morphological evolution, it is tempting to speculate whether the differences that we see between the mouse and zebrafish Hoxa3 orthologues are caused in part by differing demands of mouse and fish anatomy. Such questions are significantly complicated by the difficulty of unambiguously assigning structural homology in organisms as diverged as mice and fish; however, for some phenotypes such as the lack of a thymus, it is clear that the zebrafish gene is failing to function in the generation of a structure that is present in both species. As a hoxa3a-null zebrafish mutant is not available, we do not know what functions zfhoxa3a performs in zebrafish. There are a total of four Hox3 genes in teleosts, one each for the a3, b3, c3, and d3 groups (19). Unlike in mouse, at least two and possibly three group 3 Hox genes are expressed in the pouch endoderm in zebrafish (Fig. S6). A previous report using morpholino (MO) knockdown suggested that loss of hoxa3a alone had little phenotypic effect, but showed redundant function with Hoxb3a in the development of gill-related structures (34). However, the phenotypic effects occur at relatively late stages, and it was not clear whether the MOs remained effective. We performed a similar analysis using splice-blocking MOs (Fig. S6). Although the MOs were effective at 24 h, by 52 h, spliced mRNAs were readily apparent, indicating that the lack of phenotypes at later stages may not reflect the full range of gene function. Thus, it remains possible that zfhoxa3a has some similar functions to the mouse gene. Alternatively, some aspects of Hoxa3 protein function could be performed by other Hox3 genes in zebrafish. If so, this would represent a novel combination of gene expression and protein function-based subfunctionalization during the evolution of Hox3 genes in vertebrates.
The intriguing result that some functions seem to be shared between the mouse and zebrafish Hoxa3 proteins whereas others are not may have important implications for the ability of “toolkit” transcription factors to evolve at the protein level to effect morphological change (5). Hoxa3 is an excellent example of “mosaic pleiotropy,” in which a single transcription factor has diverse functions in different structures and at different times. This concept has been proposed as a principal reason why any change in function of toolkit transcription factors would not likely be tolerated (5). That the ability of the zebrafish protein to substitute for mouse Hoxa3 is not universal, and ranges between completely WT to completely null phenotypes, indicates that Hoxa3 may perform different roles or interact with different partners in different cell types, and that these functions may evolve independently to some extent. Furthermore, these differences map outside of the conserved homeodomain, indicating that subtle changes in less well conserved domains may have major and specific effects on protein function without affecting DNA binding, and thus may serve as important sites of Hox protein evolution.
Materials and Methods
Gene Targeting.
The mouse Hoxa3zf and Hoxa3mz alleles were generated by homologous recombination. The Hoxa3 locus was targeted with a vector based on a 12-kb Not I fragment of C57Bl6 genomic DNA that was linearized and electroporated into LK-1 C57Bl6 ES cells. ES cell line derivation, electroporation, and injection were performed in the Mouse ES Cell and Transgenic Core Facility at the Medical College of Georgia. Clones were screened by Southern blot with 5′ and 3′ franking probes and an internal probe. Details on targeting vector construction, Southern blot screening, mouse strain generation, and genotyping with PCR can be found in SI Materials and Methods.
Hoxa3 mRNA and Protein Quantification.
First-strand cDNA was reverse transcribed from total RNA from embryonic d 10.5 embryos. Quantitative PCR was performed on an ABI 7500 real-time PCR system with SYBR green PCR master mix (Applied Biosystems). For proteomics analysis, the target peptide (SPLLNSPTVGK) was chosen from the tryptic peptides of the mouse Hoxa3 NTD. Proteins extracted from mouse embryos were digested with trypsin (Promega) following reduction and alkylation. Resulting peptides were separated with an offline strong cation exchange chromatography. Seven fractions were collected and analyzed in parent ion monitoring mode via LC-MS/MS (LTQ-Orbitrap XL; ThermoFisher). Acquired spectra were searched against a mouse protein database (Swissprot, updated March 24, 2009) using Bioworks (version 3.3.1, SP1; ThermoFisher). The calculation of the ratio was based on the peak area of the reconstructed ion chromatogram of respective peptides following normalization by a high-scoring tryptic peptide that coeluted from titin. Details are provided in SI Materials and Methods.
Phenotyping.
In situ hybridization, histology, immunohistochemistry, skeleton preparation, and imaging are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
L.C. thanks Z. Liu for help with in situ probes, J. Gordon for help with histology, and K. Masuda for Hoxa3−/− embryos and for sharing unpublished data. This work was supported by National Science Foundation Grants 0131314 (to N.R.M.) and 0719558 (to C.T.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005129107/-/DCSupplemental.
References
- 1.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Amores A, et al. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 4.Gellon G, McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays. 1998;20:116–125. doi: 10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 6.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 7.Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- 8.Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121:1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- 9.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350:473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 10.Manley NR, Capecchi MR. Hox group 3 paralogous genes act synergistically in the formation of somitic and neural crest-derived structures. Dev Biol. 1997;192:274–288. doi: 10.1006/dbio.1997.8765. [DOI] [PubMed] [Google Scholar]
- 11.Kameda Y, Nishimaki T, Takeichi M, Chisaka O. Homeobox gene hoxa3 is essential for the formation of the carotid body in the mouse embryos. Dev Biol. 2002;247:197–209. doi: 10.1006/dbio.2002.0689. [DOI] [PubMed] [Google Scholar]
- 12.Watari N, Kameda Y, Takeichi M, Chisaka O. Hoxa3 regulates integration of glossopharyngeal nerve precursor cells. Dev Biol. 2001;240:15–31. doi: 10.1006/dbio.2001.0447. [DOI] [PubMed] [Google Scholar]
- 13.Manley NR, Capecchi MR. Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Dev Biol. 1998;195:1–15. doi: 10.1006/dbio.1997.8827. [DOI] [PubMed] [Google Scholar]
- 14.Condie BG, Capecchi MR. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature. 1994;370:304–307. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 15.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 16.Tvrdik P, Capecchi MR. Reversal of Hox1 gene subfunctionalization in the mouse. Dev Cell. 2006;11:239–250. doi: 10.1016/j.devcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Duboule D. Vertebrate Hox genes and proliferation: An alternative pathway to homeosis? Curr Opin Genet Dev. 1995;5:525–528. doi: 10.1016/0959-437x(95)90058-o. [DOI] [PubMed] [Google Scholar]
- 18.Duboule D. Developmental genetics. A Hox by any other name. Nature. 2000;403:607–610. doi: 10.1038/35001179. [DOI] [PubMed] [Google Scholar]
- 19.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 20.Tümpel S, Cambronero F, Sims C, Krumlauf R, Wiedemann LM. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc Natl Acad Sci USA. 2008;105:20077–20082. doi: 10.1073/pnas.0806360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon J, Bennett AR, Blackburn CC, Manley NR. Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech Dev. 2001;103:141–143. doi: 10.1016/s0925-4773(01)00333-1. [DOI] [PubMed] [Google Scholar]
- 22.Günther T, et al. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature. 2000;406:199–203. doi: 10.1038/35018111. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Yu S, Manley NR. Gcm2 is required for the differentiation and survival of parathyroid precursor cells in the parathyroid/thymus primordia. Dev Biol. 2007;305:333–346. doi: 10.1016/j.ydbio.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuzinger S, et al. Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development. 1998;125:1703–1710. doi: 10.1242/dev.125.9.1703. [DOI] [PubMed] [Google Scholar]
- 26.Acampora D, et al. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- 27.Lutz B, Lu HC, Eichele G, Miller D, Kaufman TC. Rescue of Drosophila labial null mutant by the chicken ortholog Hoxb-1 demonstrates that the function of Hox genes is phylogenetically conserved. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- 28.Koh EG, et al. Hox gene clusters in the Indonesian coelacanth, Latimeria menadoensis. Proc Natl Acad Sci USA. 2003;100:1084–1088. doi: 10.1073/pnas.0237317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crow KD, Stadler PF, Lynch VJ, Amemiya C, Wagner GP. The “fish-specific” Hox cluster duplication is coincident with the origin of teleosts. Mol Biol Evol. 2006;23:121–136. doi: 10.1093/molbev/msj020. [DOI] [PubMed] [Google Scholar]
- 30.Noonan JP, et al. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Potter SS. Functional specificity of the Hoxa13 homeobox. Development. 2001;128:3197–3207. doi: 10.1242/dev.128.16.3197. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Potter SS. Functional comparison of the Hoxa 4, Hoxa 10, and Hoxa 11 homeoboxes. Dev Biol. 2002;244:21–36. doi: 10.1006/dbio.2002.0595. [DOI] [PubMed] [Google Scholar]
- 33.Pierce RJ, et al. Evidence for a dispersed Hox gene cluster in the platyhelminth parasite Schistosoma mansoni. Mol Biol Evol. 2005;22:2491–2503. doi: 10.1093/molbev/msi239. [DOI] [PubMed] [Google Scholar]
- 34.Hogan BM, et al. Zebrafish gcm2 is required for gill filament budding from pharyngeal ectoderm. Dev Biol. 2004;276:508–522. doi: 10.1016/j.ydbio.2004.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.