Abstract
The Rad6-Rad18 mediated monoubiquitylation of proliferating cell nuclear antigen (PCNA) at lys 164 plays a crucial role in promoting the access of translesion synthesis (TLS) DNA polymerases (Pols) to PCNA in the replication fork stalled at a lesion site. Although a number of genetic and biochemical observations have provided strong evidence that TLS Pols bind PCNA at its interdomain connector loop (IDCL) via their PCNA-interacting protein (PIP) domain, a more recent proposal formulates that TLS Pols bind PCNA at two sites, to the IDCL via their PIP domain and to lys-164 linked ubiquitin (Ub) via their ubiquitin-binding domain. To ascertain the relative contributions of the PIP and Ub-binding zinc finger (UBZ) domains of human Polη in TLS, we have determined whether the C-terminal truncations of hPolη that contain the PIP1 domain but lack the UBZ and PIP2 domains can still function in TLS in human cells. Our observations that such C-terminally truncated proteins promote efficient TLS opposite a cis-syn TT dimer and confer a high degree of UV resistance to XPV cells provide unambiguous evidence that the binding of PCNA via its PIP domain is essential as well as sufficient for providing hPolη the ability to carry out TLS in human cells.
Keywords: human Pol eta, proliferating cell nuclear antigen ubiquitylation, translesion DNA synthesis, ubiquitin-binding zinc finger domain
In humans, a number of translesion synthesis (TLS) DNA polymerases (Pols) promote replication through DNA lesions. Biochemical and structural studies have indicated that they function in TLS in a highly specialized manner, providing an efficient and relatively accurate way to synthesize DNA opposite a diverse array of DNA lesions (1). Among TLS Pols, Polη is highly efficient at replicating through UV-induced cyclobutane pyrimidine dimers (CPDs); and surprisingly, in spite of its rather low intrinsic fidelity of 1 × 10-2 to 3 × 10-2 (2, 3), Polη promotes highly error-free replication through CPDs in human cells (4). Inactivation of Polη in humans causes the cancer-prone syndrome, the variant form of xeroderma pigmentosum (XPV) (5, 6).
The stalling of the replication fork at a DNA lesion induces Rad6-Rad18-dependent ubiquitylation of proliferating cell nuclear antigen (PCNA) at its lys 164 residue (7). Genetic studies in yeast have shown that PCNA ubiquitylation is a necessary prerequisite for TLS (8, 9); however, it still remains unclear how this PCNA modification modulates the access of TLS Pols to PCNA stalled at a lesion site. Because TLS Pols such as η, ι, and κ contain an ubiquitin-binding domain (UBD) in addition to a PCNA-interacting protein (PIP) domain, a view that continues to be generally accepted is that TLS Pols bind to PCNA not only at its interconnector loop domain via their PIP domain, but also to the lys 164-linked ubiquitin moiety via their UBD domain, and that the binding of ubiquitin (Ub) on PCNA is crucial for the TLS Pols to function in lesion bypass (10–12). To determine the relative significance of the PIP and UBD domains in PCNA binding, in a previous study we examined the roles of these domains in human Polη. From biochemical and genetic analyses of the two PIP domains, PIP1 and PIP2, that are present in the C-terminal portion of Polη, we showed that mutational inactivation of both the PIP domains completely inactivated the ability of Polη to bind PCNA; accordingly, whereas the Polη protein mutated in either the PIP1 or PIP2 domain colocalizes with PCNA in replication foci in UV irradiated human cells, Polη protein bearing mutations in both the PIP1 and PIP2 domains showed no evidence of colocalization with PCNA (13). Consequently, mutational inactivation of both the domains completely abrogates the TLS function of Polη in human cells, as was inferred from the lack of complementation of the UV sensitivity of XPV cells by mutant Polη (13).
Human Polη has a ubiquitin-binding zinc finger (UBZ) domain present in its C-terminal portion between residues 631–659. From mutational analyses of several conserved residues in this domain that have been shown to be important for ubiquitin binding, we showed that they varied in their effects that ranged from moderate levels of inactivation of the biological function of Polη to no effect as inferred from the UV sensitivity of these mutations (13). However, because some of the conserved residues in the UBZ domain affected Polη biological function to a significant degree, the possibility that the binding of the UBZ domain to the lys 164-linked Ub moiety on PCNA is important for the ability of Polη to function in TLS could not be entirely discounted from these previous observations. That is because it could still be argued that only the mutations that adversely affected hPolη function were defective in binding to Ub on PCNA, whereas the mutations that retained function have little or no defect in PCNA binding. On the other hand, an alternative explanation for the adverse effects of some of the UBZ mutations is that they do not result from the inability of Polη to bind Ub on PCNA, rather they arise from the dominant negative effects of these mutations on Polη function. Because this idea presumes that the UBZ domain is not required for PCNA binding, we decided to test whether the dominant negative effects of UBZ mutations could be bypassed by deleting the entire UBZ domain, and because hPolη has two PIP domains, either of which could bind to PCNA, we chose to delete the entire C terminus beyond PIP1.
Here we show that such C-terminally truncated hPolη proteins, which lack both the UBZ and PIP2 domains, can promote TLS opposite CPDs in human cells. Hence, we conclude that the ability of hPolη to bind PCNA via its PIP motif is sufficient for it to perform its biological role in lesion bypass, and that the binding of Ub on PCNA via its UBZ motif is not required.
Results
C-Terminal Truncations of hPolη.
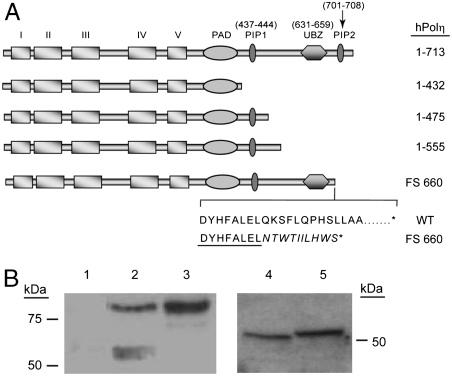
To determine whether the UBZ domain was dispensable for Polη function in human cells, we generated two C-terminal truncations which contained residues 1–475 and 1–555, respectively, and where both the UBZ and PIP2 domains have been deleted. We additionally constructed a C-terminal truncation which contains residues 1–432 from which the PIP1 domain has also been deleted (Fig. 1A). Using these three constructs, we could test whether hPolη that can bind PCNA via its PIP1 motif but lacks both the UBZ and PIP2 motifs could still perform its biological role in TLS. The wild-type and C-terminally truncated Polη proteins were expressed in human cells, and by immunoblotting we verified that the proteins produced were of the expected size (Fig. 1B).
Fig. 1.
Mutations of hPolη in the C terminus. (A) C-terminal truncations of hPolη. The positions of the five conserved Pol domains (I–V), and the polymerase associated domain, PIP1, UBZ, and PIP2 domains are shown. The residues that span the PIP1, UBZ, and PIP2 domains are also indicated above each of them. For the FS660 mutation, the sequence of the last 18 amino acids in the mutant protein is compared to the sequence in the wild-type protein. The FS660 mutation changes the Q residue present in the wild-type protein at position 660 to N and this residue plus the additional nine residues that result from the altered reading frame are shown in italics. The residues D Y H F A L E L form the C-terminal part of the UBZ domain and are underlined in the FS660 protein sequence. Because the frameshift mutation occurs at position 660 right after the last conserved residue of the UBZ domain, the entire UBZ sequence remains unaltered in the mutant protein. An asterisk (*) denotes the termination codon in the wild-type and FS660 proteins. (B) Expression of wild-type and mutant hPolη proteins in XPV cells. XPV cells stably expressing wild-type and mutant forms of hPolη were cultured and lysed with radioimmunoprecipitation assay buffer. The proteins were identified by Western blotting using polyclonal antibodies against hPolη. The expression level of hPolη carried on the CMV vector in XPV cells was similar to the endogenous level of hPolη in normal HF cells. Lane 1, vector control; lane 2, wild-type hPolη; lane 3, FS660 mutant hPolη; lane 4, C-terminally deleted hPolη (1-432); and lane 5, C-terminally deleted hPolη (1–475).
PIP1 Domain is Sufficient for Enhancement of the DNA Synthesis Activity of hPolη upon PCNA Binding.
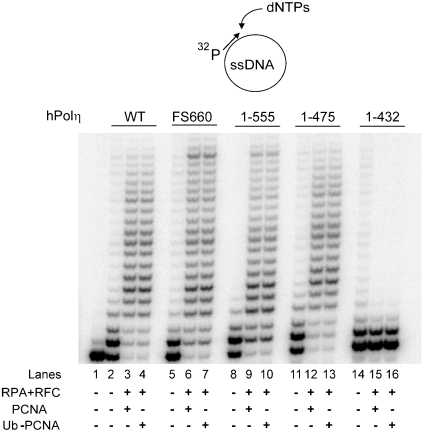
DNA synthesis by hPolη as well as by the other Y-family Pols ι and κ, is stimulated upon PCNA binding (13–16). Previously, we showed that the PIP1 and PIP2 domains of hPolη can substitute for one another in PCNA binding and that mutational inactivation of both PIP domains renders it completely defective for PCNA binding (13). Here, we examine the effects of C-terminal truncations on DNA synthesis by hPolη in the presence of PCNA, replication factor C (RPC), and replication protein A (RPA), using single-stranded circular DNA primed with a 5′ 32P-labeled oligomer at a unique site. As shown in Fig. 2, the DNA synthesis activity of wild-type hPolη is enhanced upon PCNA binding, and the C-terminally truncated hPolη (1–555) and (1–475) proteins exhibit a similar degree of enhancement upon PCNA binding. The hPolη (1–432) protein, however, shows no stimulation of its DNA synthesis activity, because in the absence of the PIP1 domain, the truncated hPolη protein can no longer bind PCNA. These observations with the wild-type and truncated proteins were verified with different concentrations of hPolη and at different time points.
Fig. 2.
Stimulation of DNA synthesis activity of C-terminal truncations and FS660 mutation of hPolη upon PCNA binding. DNA synthesis by 1.0 nM of wild-type hPolη (lanes 2–4), FS660 mutant hPolη (lanes 5–7), or by C-terminally truncated hPolη proteins 1–555 (lanes 8–10), 1–475 (lanes 11–13), and 1–432 (lanes 14–16) was examined on pBluescript circular DNA annealed to radiolabeled primer (10 nM) in the absence or presence of PCNA or Ub-PCNA (100 ng), RFC (50 ng), RPA (200 ng), all four dNTPs (each at 100 μM), and ATP (500 μM).
Absence of the UBZ Domain Has no Adverse Effect on the Ability of hPolη to Replicate Through CPDs in Human Cells.
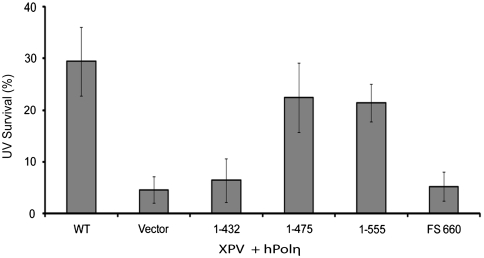
To determine whether the C-terminally truncated proteins lacking both the UBZ and PIP2 domains could carry out the biological role of hPolη in TLS, we transfected the various truncated forms into XPV cells and examined their ability to confer UV resistance (Fig. 3). Interestingly, both the (1–555) and (1–475) truncated proteins increase the UV resistance of XPV cells almost to wild-type levels, indicating that hPolη lacking the entire UBZ domain as well as the PIP2 domain can carry out the biological function to a significant degree. However, the (1–432) truncated protein conferred no increase in UV resistance to XPV cells; thus the ability of hPolη to bind PCNA via one of its two PIP domains is necessary for it to perform its role in TLS in human cells.
Fig. 3.
Complementation of UV sensitivity of XPV cells by C-terminal truncations of hPolη. XPV cells transfected with the wild-type or various mutant forms of hPolη were washed with PBS and irradiated with 5 J/m2 of ultraviolet C light in the presence of PBS. After UV irradiation, fresh growth media with 1 mM caffeine were added to the cells; following incubation for 48 h, UV cytotoxicity was determined by the MTT assay (Promega). The data represent the mean and standard deviations of results of four independent experiments.
Because Polη plays a major role in promoting efficient and error-free replication through CPDs (4), next we determined whether the hPolη proteins lacking the C terminus beyond the PIP1 domain could promote TLS opposite CPDs in human cells. For this purpose, we examined the frequency of TLS opposite a cis-syn TT dimer carried on the leading-strand template in the duplex plasmid where bidirectional replication initiates from an SV40 origin of replication. With this plasmid system, TLS frequencies are determined from the number of blue colonies among the total kan+ colonies (4). As shown in Table 1, TLS opposite a cis-syn TT dimer occurs with a frequency of ∼9% in XPV cells carrying the vector plasmid, and this frequency rises to ∼17% in XPV cells carrying full-length wild-type hPolη which is similar to the TLS levels seen in wild-type cells. Importantly, both the C-terminally truncated hPolη (1–555) and (1–475), restore wild-type levels of TLS to XPV cells, whereas no increase in TLS frequencies occurs in XPV cells carrying (1–432) hPolη.
Table 1.
TLS opposite a cis-syn TT dimer carried on the leading-strand template in normal HF cells or in XPV HF cells that harbor the wild type (WT) or mutant hPolη gene on a plasmid
| HF cells | Polη | Number of kan+ colonies | Number of blue colonies among kan+ | TLS, % | UV sensitivity* |
| Normal | WT | 568 | 101 | 17.8 | + |
| XPV | Vector control | 782 | 72 | 9.2 | +++ |
| XPV | WT | 672 | 116 | 17.3 | + |
| XPV | 1-555 | 686 | 134 | 19.5 | + |
| XPV | 1-475 | 682 | 122 | 17.9 | + |
| XPV | 1-432 | 712 | 70 | 9.8 | +++ |
| XPV | FS660 | 741 | 76 | 10.3 | +++ |
*+, wild-type UV resistance; +++, increased UV sensitivity similar to that of XPV cells.
We have shown previously that Pols η, κ, and ζ provide three alternative pathways for promoting replication through CPDs in human as well as mouse cells, wherein Polη functions in a highly error-free manner and Pols κ and ζ function in a more mutagenic manner (4). Hence the frequency of mutagenic TLS rises in the absence of Polη. To ascertain that the absence of the C terminus beyond PIP1 had no adverse effect on the in vivo fidelity of hPolη in TLS opposite CPDs, we analyzed the frequency of mutagenic TLS in XPV cells carrying wild-type hPolη or the various truncated forms of hPolη. Sequence analyses of ∼200 TLS products from XPV cells carrying the (1–555) or the (1–475) protein showed that the frequency of mutagenic TLS opposite a cis-syn TT dimer in XPV cells was reduced similarly (∼2.5-fold) by the truncated forms as by wild-type hPolη.
We conclude from these observations that hPolη lacking the UBZ as well as the PIP2 domain but containing the PIP1 domain can function in TLS opposite CPDs in human cells with almost the same degree of efficiency and accuracy as the full-length, wild-type protein.
Varying Effects of Conserved Residues in the UBZ Domain on the TLS Function of hPolη in Human Cells.
Previously, we have reported studies on mutations in residues that have been conserved in the UBZ domain of hPolη and which are involved in the binding of ubiquitin (13). These mutations produced varying effects on hPolη TLS function in human cells as judged from their UV sensitivity. For example, the C635A mutation in the highly conserved cysteine required for the coordination of a zinc ion had no significant effect on UV sensitivity, whereas the D652A and F655A mutations conferred a moderate degree of UV sensitivity (13). To verify that the different effects of these UBZ mutations on UV sensitivity reflected their ability to promote lesion bypass, we determined TLS frequency opposite a cis-syn TT dimer carried on the leading-strand template of the duplex plasmid. As shown in Table 2, hPolη harboring the C635A mutation promotes TLS to the same extent as wild-type hPolη and the D652A and F655A mutations support an intermediate level of TLS that lies between that observed in wild-type cells and in XPV cells. Hence the effects of UBZ mutations on TLS parallel their effects on UV sensitivity.
Table 2.
TLS opposite a cis-syn TT dimer carried on the leading-strand template in XPV HF cells that harbor the WT or mutant hPolη gene on a plasmid
| HF cells | Polη | Number of kan+ colonies | Number of blue colonies among kan+ | TLS, % | UV sensitivity* |
| XPV | Vector control | 782 | 72 | 9.2 | +++ |
| XPV | WT | 672 | 116 | 17.3 | + |
| XPV | C635A | 625 | 114 | 18.2 | + |
| XPV | D652A | 652 | 89 | 13.7 | ++ |
| XPV | F655A | 688 | 85 | 12.4 | ++ |
*+, wild-type UV resistance; ++, moderate UV sensitivity intermediate between that of wild-type and XPV cells; +++, increased UV sensitivity similar to that of XPV cells.
Frameshift Mutation in the C Terminus That Lies Just After the UBZ Domain Inhibits hPolη’s Ability to Carry Out Lesion Bypass.
It is quite intriguing that, although hPolη lacking the C terminus beyond PIP1, as in the (1–475) protein, can carry out TLS opposite CPDs in human cells almost as well as the wild-type protein, certain mutations in the UBZ domain, such as D652A and F655A, impair hPolη function in TLS to a significant degree (Tables 1 and 2). These observations could reflect the possibility that certain mutations in the C terminus generate dominant negative effects on hPolη function because they affect protein conformation or other such structure-related aspects; implicit in this idea is the supposition that UBZ mutations affect hPolη function because of their secondary effects on protein structure/conformation and not because of defects in the binding of the Ub moiety on PCNA. To test the plausibility of this idea, we generated a frameshift mutation at amino acid position 660 (FS660) that creates a C-terminally deleted protein in which the last 10 residues from positions 660–669 result from the changed reading frame, and therefore are unrelated to those present in wild-type protein (Fig. 1A). Because the UBZ domain which spans residues 631–659 is left intact in the FS660 mutant, any defects that are observed with this mutation may result from the impairment of hPolη in ways that are unrelated to UBZ.
The FS660 mutant protein was expressed in human cells, and that the protein of nearly the expected size was produced was verified by immunoblotting (Fig. 1B, lane 3). Next we examined whether the DNA synthesis activity of the mutant protein was enhanced upon PCNA binding, and as shown in Fig. 2, PCNA or Ub-PCNA were as stimulatory to synthesis by the FS660 protein as was the wild-type protein. Although the FS660 mutation confers no obvious defects in the DNA synthesis activity of hPolη or on its ability to bind PCNA or Ub-PCNA, the mutation is completely defective in performing its biological function, because the introduction of the plasmid that expresses the FS660 protein in XPV cells caused no increase in UV resistance (Fig. 3). Accordingly, the TLS frequency opposite a cis-syn TT dimer remained about the same in XPV cells carrying the FS660 mutant hPolη as in XPV cells that harbor the vector control and thus lack any hPolη (Table 1).
Discussion
Although genetic studies from yeast have provided strong evidence that PCNA monoubiquitylation at its K164 residue is indispensable for TLS, it has remained unclear how this modification modulates the access of TLS Pols to PCNA in the replication fork stalled at a lesion site. The observation that the various Y-family Pols contain ubiquitin-binding domains has led to the proposal that the TLS Pols bind directly to the Ub moiety on PCNA via the UBD and that this binding mode is indispensable for their ability to function in lesion bypass.
To examine the role of UBDs in PCNA binding, we have focused our studies on the UBZ domain of hPolη because evidence from studies with mutations of conserved residues in this domain have been used extensively in support of the model (10, 12, 17). In a previous study, we showed that hPolη has two PIP motifs in its C terminus, which can substitute for one another and that mutational inactivation of both the PIP motifs renders hPolη nonfunctional in human cells, as the protein can no longer localize onto PCNA in UV irradiated cells and is unable to complement the UV sensitivity of XPV cells (13). Mutational analyses of five of the conserved residues in the UBZ domain showed that the effects of these mutations varied from no defect in Polη function for the C635A and H650A mutations to a moderate defect for the D652A, H654A, and F655A mutations (13). Although the observations that some of the highly conserved residues that are important for the ability of UBZ to bind Ub (18) conferred no significant effect on the biological function of hPolη had indicated that the Ub-binding ability of the UBZ domain was dispensable, we wished to determine whether the UBZ mutations that impair hPolη’s ability to function in TLS do so because they elicit dominant negative effects on some aspects of hPolη structure/function and not because their ability to bind PCNA is affected.
Here, we show that the two C-terminally deleted hPolη proteins (1–475) and (1–555) that contain the PIP1 domain but lack the UBZ and PIP2 domains complement the UV sensitivity of XPV cells and are able to carry out TLS opposite a cis-syn TT dimer to the same extent as the wild-type protein. The (1–432) protein that additionally lacks the PIP1 domain, however, is completely defective in TLS. We conclude from these results that the ability of hPolη to bind PCNA via its PIP domain is sufficient for it to function in TLS. Furthermore, the observation that the (1–475) protein can perform TLS opposite CPDs with the same efficiency and fidelity as the wild-type protein implies that this protein lacking the C-terminal 238 residues can participate in all the protein–protein interactions necessary for hPolη to associate with the components of the replication fork stalled at a lesion site.
To explore the possibility that certain mutations in the C terminus generate dominant negative effects upon Polη structure/function, we constructed a frameshift mutation at position 660 which lies just after the UBZ domain and which contains 10 residues unrelated to the normal sequence. Our observation that upon PCNA binding the DNA synthesis activity of the FS660 protein is stimulated to the same extent as is wild-type protein indicated that the protein is not impaired in its capacity for PCNA binding. However, the mutant protein is completely defective in performing TLS in human cells, as indicated from its inability to complement the UV sensitivity of XPV cells and to carry out TLS opposite a cis-syn TT dimer in human cells. These observations with the FS660 mutation taken together with the ability of the C-terminally truncated (1–475) protein to function in TLS opposite CPDs in human cells would suggest that the FS660 mutation impairs hPolη function indirectly by affecting its structure/conformation such that the ability of the protein to properly associate with components of the replication fork is compromised.
Both of our results that the (1–475) protein functions normally in TLS opposite to a cis-syn TT dimer but the FS660 mutation is highly defective (Table 1) could be explained if hPolη were comprised of two structurally distinct domains in which the N-terminal domain contains all the elements necessary for DNA synthesis, for PCNA binding, and for mediating the various protein–protein interactions needed for the assembly of the protein into the replication complex stalled at the lesion site. The C-terminal domain would contain UBZ and PIP2 and, although this portion of the protein is not required for any of the key aspects involving DNA synthesis, the ability to bind PCNA, and to assemble with the replication machinery, certain mutations in this domain could adversely affect the ability of the N-terminal portion to function in TLS because of the long-range deleterious effects they produce on the globular structure of the protein.
The interpretation of our observations with the C-terminally truncated (1–475) hPolη vs. the FS660 mutant protein can also be extended to explain the deleterious effects of mutations such as D652A and F655A in the UBZ domain of hPolη. Thus, the observation that, although the complete absence of the UBZ domain in the (1–475) protein has no adverse effect on TLS opposite a cis-syn TT dimer, the D652A and F655A mutations impair TLS opposite this lesion to a significant degree (Tables 1 and 2) could be similarly explained from the dominant negative effects of these UBZ mutations on hPolη function.
Even though the C-terminal portion of hPolη can be deleted without any significant effect on its TLS function opposite CPDs, we consider it highly unlikely that this portion including the UBZ domain plays no direct role in some aspects of hPolη function. The major conclusion we can draw from our results is that the PIP motif is required for PCNA binding, and that the UBZ domain’s function in PCNA binding, if any, is dispensable. However, our data provide no clue for the possible role(s) of the UBZ domain and for the C-terminal portion as a whole, as to whether they affect TLS opposite certain specific types of DNA lesions or whether they modulate aspects of hPolη function(s) unrelated to lesion bypass.
Materials and Methods
Generation of hPolη Mutations.
The C-terminal deletion constructs were generated either by PCR or by ligating a linker DNA at a suitable restriction site in the wild-type hPolη construct, pBJ762. The C-terminal truncations that contain residues (1–432) and (1–475) of hPolη were generated by amplifying these segments of the hPolη ORF by PCR. The (1–555) hPolη construct was generated by inserting a SMURFT linker at the unique NcoI site of the hPolη gene. All the constructs were verified by automated DNA sequencing.
Proteins.
Full-length hPolη and C-terminal truncations of hPolη were purified as glutathione-S-transferase fusion proteins from yeast strain BJ5464. The proteins were purified and the glutathione-S-transferase portion of the fusion protein was removed by treatment with PreScission protease (Amersham Pharmacia). Protein concentrations were determined using a Bio-Rad protein assay with bovine serum albumin as a standard.
DNA Pol Assays.
The single-stranded circular DNA (pBluescript, 3KB) was annealed to a 5′-32P-labeled primer which is complementary to the F1 origin of plasmid (5′-CCC CCG ATT TAG AGC TTG ACG GGG AAA CCG GCG AAC GTG GC-3′) and used as a substrate for the DNA polymerase assay. The standard DNA Pol reaction mixture contained 10 nM DNA substrate, 40 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg of BSA/mL, 500 μM ATP, and 100 μM each dGTP, dATP, dTTP, and dCTP. The reaction was carried out in the absence or presence of PCNA or Ub-PCNA (100 ng), RFC (50 ng), and RPA (200 ng) at 37 °C for 10 min after the addition of WT or mutant hPolη (1 nM) protein to the reaction mixture. The reaction was stopped by the addition of loading buffer (40 μL) containing EDTA (20 nM), 95% formamide, 0.3% bromophenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. The reaction products were visualized with a Molecular Dynamics STORM PhosphorImager
Stable Expression of Wild-Type and Mutant hPolη in XPV Cells.
Wild-type and mutant hPolη genes were subcloned into pSilencer 4.1-CMV puro vector (Ambion). The vectors were transfected into XPV (XP30RO) human fibroblast (HF) cells by Lipofectamine 2000 reagent (Invitrogen). After 24 h incubation, 0.2 μg of puromycin was added to the culture media. After 3 days of incubation, cells were washed with PBS buffer and were continuously cultured with the media containing 50 ng of puromycin for ∼2 weeks. For checking protein expression by Western blot analysis, cells were washed with PBS buffer and lysed with radioimmunoprecipitation assay buffer (1x PBS, 1% IP-40, 0.5% sodium deoxycholate, 0.1% SDS). After 1 h incubation on ice, the cellular mixture was centrifuged and the supernatant was collected. Equivalent amounts (approximately 30 μg) of prepared cellular extracts were separated on a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane (Bio-rad). The membranes were probed with affinity purified rabbit polyclonal antibodies against hPolη followed by secondary antibodies conjugated with horse radish peroxidase. The signals were detected using ECL-Plus (Amersham).
UV Cytotoxicity Assays.
Stable transfected XPV cells were plated in six-well plates with 50% confluence. After 24 h of incubation, cells were treated with UV. For UV irradiation, cells were washed with PBS buffer and irradiated with 5 J/m2 of ultraviolet C light in the presence of PBS buffer. After irradiation, fresh growth medium containing 1 mM caffeine was added to the cells. Cells were incubated for additional 48 h after UV irradiation. UV cytotoxicity was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Promega) following the manufacture’s manual. Briefly, 200 μL of MTT assay solutions were added to each well and incubated for 30 min. Cell viability was determined by measuring the OD at 490 nM.
In Vivo Translesion Synthesis Assays in XPV Cells.
The duplex plasmid that carries a cis-syn TT dimer on the leading-strand or the lagging-strand template has been described previously (4). Stably transfected XPV cells were plated in a six-well plate with 80% confluence. After 24 h incubation, the duplex plasmid DNA (2 μg) carrying the cis-syn TT dimer on the leading-strand template was transfected with Lipofectamine 2000 (Invitrogen). After 30 h incubation, plasmid DNA was rescued from cells by the alkaline lysis method and digested with DpnI to remove unreplicated plasmid DNA. The plasmid DNA was then transformed into Escherichia coli XL1-Blue super competent cells (Stratagene). Transformed bacterial cells were diluted in 1 ml superoptimal broth with catabolite repression media and plated on both LB/amp (50 μg/mL ampicillin, Sigma) and LB/kan (25 μg/mL kanamycin, Sigma) plates containing 1 μM isopropyl-1-thio-β-D-galactopyranoside (Roche) and 100 μg/mL of X-Gal (Roche). After 16 h incubation at 37 °C, blue and white colonies were counted from kanamycin plates. The actual TLS frequency was determined from the number of blue colonies out of total colonies growing on LB/kan plates. Plasmid DNA obtained from blue colonies was analyzed to determine the mutation frequency and the mutational changes incorporated during TLS. For details of these methods, see Yoon et al. (4).
Acknowledgments.
We are grateful to Ildiko Unk and Lajos Haracska for Ub-PCNA. This work was supported by National Institutes of Health Grant ES012411.
Footnotes
The authors declare no conflict of interest.
References
- 1.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 4.Yoon J-H, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci USA. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 6.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 7.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 8.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 10.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 11.Guo C, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plosky BS, et al. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acharya N, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haracska L, et al. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haracska L, et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haracska L, et al. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bienko M, et al. Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 18.Bomar MG, Pai M-T, Tzeng S-R, Li SS-C, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]