Abstract
An imbalance in photosynthetic electron transfer is thought to be redressed by photosynthetic control of the rate of expression of genes encoding apoproteins of photosystem (PS)-I and PS-II in response to the redox state of plastoquinone (PQ), which is a connecting electron carrier. PS stoichiometry is then adjusted to enhance photosynthetic efficiency. In prokaryotes, sigma factors are well known for their participation in the control of RNA polymerase activity in transcription, whereas there have been no reports concerning their association with redox regulation. We have found that the phosphorylation of SIG1, the major sigma factor (SIG), is regulated by redox signals and selectively inhibits the transcription of the psaA gene, which encodes a PS-I protein. We produced transgenic Arabidopsis plants with or without the putative phosphorylation sites for SIG1 and demonstrated through in vivo labeling that Thr-170 was involved in the phosphorylation. We analyzed the in vivo and in vitro transcriptional responses of the transgenic Arabidopsis plants to the redox status in regard to involvement of the phosphorylation site. We revealed an enhanced phosphorylation of SIG1 under oxidative conditions of PQ in a form associated with the molecular mass of the holoenzyme. Phosphorylation of SIG1 proved crucial through a change in the promoter specificity for sustaining balanced expression of components in PS-I and PS-II and was responsible for harmonious electron flow to maintain photosynthetic efficiency.
Keywords: chloroplast, redox regulation, plastoquinone, RNA polymerase, Arabidopsis
Plants are required for the survival of humans as food supplies and as alternative sources of energy in the form of biofuel, and are important in conservation of the environment. These contributions of plants are based on the function of chloroplasts. Chloroplasts are thought to have originated from ancestral cyanobacteria, which became symbionts in eukaryotic host cells. Plants have evolved as sessile organisms that use their chloroplasts to exploit solar energy. They are unable to escape from undesirable environmental conditions and must adapt to survive. When plants are exposed to sunlight, their chloroplast photosystems (PSs) excite electrons donated by water, which are transferred to NADP+ and other recipients. Altered light conditions influence the efficiency of electron excitation in PS-I and PS-II, leading to an imbalance in photosynthetic electron transfer (1–3) and injury caused by uncoupled excess energy. This imbalance is thought to be redressed by the photosynthetic control of PS stoichiometry (2), where the rate of expression of genes for apoproteins of PS-I and PS-II is regulated in response to the redox state of plastoquinone (PQ) (4, 5). However, it is not known how the regulation is achieved in a signal transduction pathway through redox states of PQ. We have focused on the activity of RNA polymerase as one of the critical steps. The transcription of most photosynthesis genes in the chloroplast is accomplished with bacterial-type plastid-encoded RNA polymerase (PEP), which requires multiple nucleus-encoded sigma factors (SIGs) (6–9) destined for the chloroplast. To reveal the underlying mechanisms we focused on SIGs, which are well characterized in prokaryotes as components of RNA polymerase and engaged in transcription in chloroplasts.
Results and Discussion
Phosphorylation Site of the Sigma Factor.
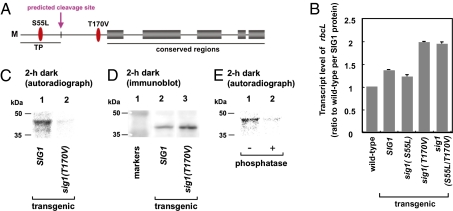
The amino acid sequences of all of the sigma proteins, SIG1 to SIG6, were searched for sites that could be phosphorylated. Among multiple categories of predicted sites, we initiated mutagenesis in those sites that had a minimal number of candidates. SIG1 transcripts and protein are the most abundant of the six kinds of SIGs in leaves grown in light for 2–4 wk, a finding that led us to focus on SIG1 (Fig. S1). SIG1 has sites that can be phosphorylated by Ser/Thr protein kinases, and those immediately ahead of region 1.2 are conserved in Arabidopsis SIG3 and SIG5 (Fig. S2) and in SIG1 orthologs. We produced transgenic Arabidopsis derivatives carrying wild-type SIG1 (referred to as SIG1) and derivatives with individual putative phosphorylation sites that were deleted [sig1(S55L), sig1(T170V) and sig1(S55L/T170V); Fig. 1A]. As SIG genes were expressed in the presence of light (6), 3-wk-old Arabidopsis plants were maintained in the dark for 1 wk to lower endogenous SIG levels before analyzing their chloroplast gene transcripts. In each transgenic line, we investigated the expression of rbcL, which is known to be transcribed by PEP, to determine whether the phosphorylation of sigma factors influences transcription in chloroplasts. The transcript levels of rbcL were normalized with the SIG protein level by immunoblotting with antibodies against recombinant SIG1 (anti-SIG1; Fig. 1B). In lines transformed with sig1(T170V) and sig1(S55L/T170V), transcript levels were higher than in those harboring SIG1 or sig1(S55L) (Fig. 1B). Therefore, Ser-55 in SIG1 is not involved in transcript level regulation. This finding is consistent with it being located in the putative transit peptide that may be eliminated to generate mature SIG1, whereas Thr-170 might play a role in transcription regulation. Transcript levels were normalized with SIG1 protein content, which eliminated the possibility that the amino acid substitution interfered with the SIG1 protein degradation. Hence, we conclude that Thr-170 among the predicted phosphorylation sites is responsible for reducing the levels of transcripts of chloroplast genes in the dark. The phenotypes are reproducible in multiple independent lines [wild-type, SIG1, and sig1(S55L), or sig1(T170V) and sig1(S55L/T170V)] (Fig. 1B), suggesting both that the status of phosphorylation in SIG1 may affect chloroplast gene expression and that potential position effects or dominant-negative effects may not occur. This inference was also supported by the analysis of the number and location of inserted T-DNA sites in the genome (SI Text).
Fig. 1.
Effects of Ser/Thr phosphorylation of SIG1 on the transcript level of a chloroplast gene. (A) Predicted Ser/Thr phosphorylation sites in Arabidopsis SIG1. (B) Levels of rbcL transcripts in wild-type plants and transgenic lines sig1(S55L), sig1(T170V), and sig1(S55L/T170V) when adapted to the dark for 1 wk. Levels determined by quantitative RT-PCR were standardized by using ACT2 transcripts and were further normalized by the content of SIG1 protein determined by immunoblotting. Vertical bars at the top of histograms represent the SDs of three independent experiments. (C–E) Phosphorylation of SIG1 and sig1(T170V). (C) Plants (3-wk-old) were incubated with [32P]orthophosphate in the dark. Immunoprecipitates with anti-SIG1 were fractionated by SDS/PAGE, and radioactivity on the dried gel was detected with Storm 820 (Amersham Biosciences). (D) Immunoblotting of extracts shown in C with anti-SIG1. (E) Extracts from SIG1-transgenic plants labeled with 32P in the dark were prepared without phosphatase inhibitors only for this experiment and were immunoprecipitated with anti-SIG1. The immunoprecipitates were incubated with or without 25 units of calf intestinal alkaline phosphatase for 30 min, followed by SDS/PAGE and autoradiography.
The effect of the amino acid substitution at Thr-170 might have resulted from a change of protein conformation rather than phosphorylation status. To test this hypothesis, we dipped the roots of SIG1-transgenic plants in 32P in the dark and determined the amount of 32P in their SIG proteins. Although the amounts of SIG1 protein expressed were roughly equivalent in Arabidopsis transformed with SIG1 and in sig1(T170V) (Fig. 1D), there was a strong 32P signal in the SIG1-transgenic line and a faint signal in the sig1(T170V) transgenic line (Fig. 1C). Phosphorylated SIG1 detected by autoradiograph (Fig. 1C) was at a higher molecular weight than SIG1 detected by immunoblotting. The faint signal of the upper band in the immunoblot is due to a smaller amount of phosphorylated SIG1 than nonphosphorylated SIG1, which is consistent with the result that most of the SIG1 proteins existed without phosphorylation at Thr-170, free from the core enzyme (see below).
We determined the ratio of transcripts derived from exogenous sig1(T170V) to those from the endogenous wild-type SIG1 by sequencing individual cDNA clones and found that sig1(T170V) showed a 20-fold higher expression than endogenous SIG1. The low level of expression of endogenous SIG1 was consistent with the faint signal obtained from sig1(T170V) plants. The 32P signals diminished when extracts were treated with calf intestinal alkaline phosphatase (Fig. 1E). Hence, the Thr-170 residue of wild-type SIG1 could be phosphorylated. Together with previous experimental data, these findings show that phosphorylation of Thr-170 of SIG1 interferes with chloroplast gene transcription.
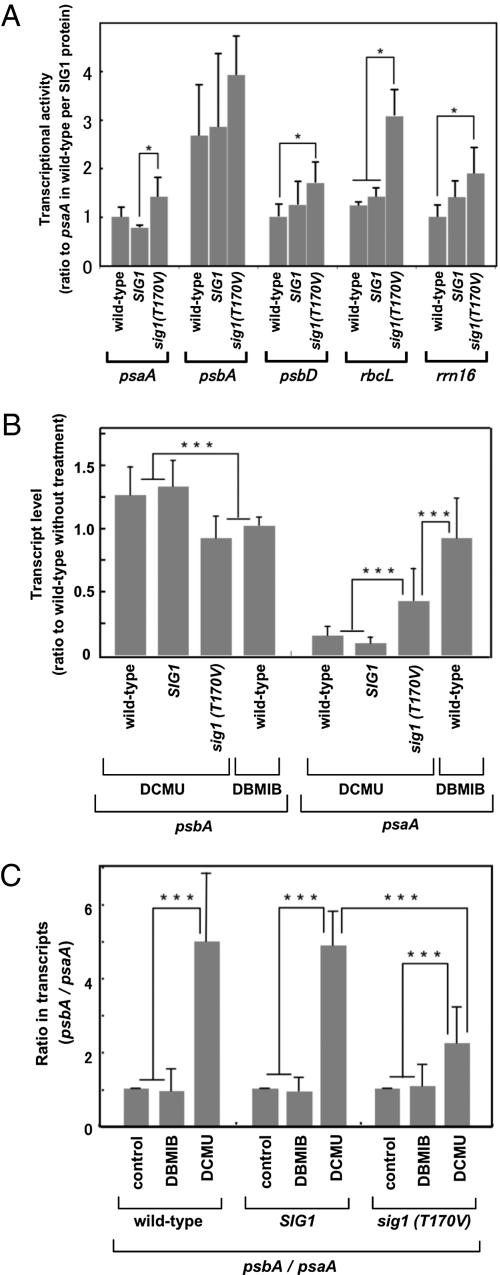
To confirm that SIG1 phosphorylation controls transcriptional activity, we performed chloroplast transcription run-on experiments (Fig. 2A). Transgenic lines for SIG1 and sig1(T170V), and wild-type plants, were grown for 4 wk and transferred to the dark for 2 h. Chloroplasts were isolated and used for run-on transcription. Transcriptional activities were expressed per SIG1 molecule. In the sig1(T170V)-transgenic line, transcription of psbA, psbD, psaA, rbcL, and rrn16 was higher than that for wild-type SIG1. Hence, phosphorylation of SIG1 reduces the transcription of genes encoded by the chloroplast genome to different extents. The transcript levels per SIG1 molecule in the SIG1-transgenic line were not significantly different from those in the wild-type. Transcript levels of psbA and psaA in the SIG1-transgenic line were 0.82-fold and 0.64-fold lower, respectively, than in the sig1(T170V)-transgenic line. Phosphorylation of SIG1 might therefore affect psaA transcription more than psbA transcription. Pfannschmidt et al. (4) showed that the redox state of PQ controls the level of transcription of psbA and psaA. Hence, the phosphorylation of SIG1 might mediate the adjustment of stoichiometry in response to redox signals from PQ.
Fig. 2.
Effects of phosphorylation of SIG1 on the run-on transcription and transcript levels of genes for PS-II (psbA) and PS-I (psaA) at different states of PQ. (A) Run-on transcription of chloroplast genes. Transcriptional activity for each gene is shown as the fraction of that of the wild-type after standardization with the SIG1 content determined by immunoblotting. Error bars represent the variations of three independent experiments. Asterisks indicate statistically significant differences (P < 0.05). (B and C) Transcript levels in plants were determined by real-time RT-PCR. The redox status of PQ was manipulated with the inhibitors DCMU and DBMIB. (B) Ratios of transcript levels of each gene in the treated transgenic lines to those in the untreated wild-type. (C) Ratio of transcripts of psbA to psaA in each plant. “Control” means the plants were not treated. Error bars represent the variation of three independent experiments. Triple asterisks indicate statistically significant differences (P < 0.01).
Redox Regulation of SIG1 Phosphorylation.
We altered the redox state of PQ in plants by using 3-(3′,4′-dichlorophenyl)-1,1′-dimethyl urea (DCMU) for the oxidative state and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) for the reductive state. psaA expression in the wild-type was diminished with DCMU but not with DBMIB, whereas the reverse was observed for psbA expression (Fig. 2B). These agents therefore acted as expected on the basis of a previous report (4). psaA expression was not enhanced, but tended to diminish, under oxidative conditions with DCMU in SIG1-transgenic plants, whereas it was stimulated in sig1(T170V)-transgenic plants (Fig. 2B). Hence, the effect on psaA transcripts in sig1(T170V)-transgenic plants was different from that in SIG1-transgenic and wild-type plants, and the inhibitory effect was stronger to a different degree than in wild-type plants under reductive conditions with DBMIB. By contrast, psbA expression was similar in wild-type and SIG1-transgenic plants with DCMU, whereas it declined in sig1(T170V)-transgenic plants to the level found in wild-type plants under reductive conditions with DBMIB (Fig. 2B). The results for sig1(T170V)-transgenic plants show that SIG1 is phosphorylated by oxidative signals, and the phosphorylation of SIG1 shifts the expression pattern from the reductive type to the oxidative type. Fig. 2C shows the ratios of accumulation of the psbA transcript to the psaA transcript (psbA/psaA), as used to define the stoichiometry of the adjustment between the two PSs. The psbA/psaA ratio in wild-type and SIG1-transgenic plants varied from one to five when changing from reductive conditions to oxidative conditions, respectively. However, the psbA/psaA ratio under oxidative conditions was lower in sig1(T170V)-transgenic plants than in the other plant lines. This result has been reproduced with three additional transgenic lines for both SIG1 and sig1(T170V) (Fig. S3). Overall, these results indicate that the psaA transcript level was elevated in sig1(T170V)-transgenic plants under oxidative conditions, whereas the psbA transcript level was not. Hence, the phosphorylation preferentially reduces the transcription of psaA. We conclude that a change in the promoter specificity of phosphorylated SIG1 leads to a difference between the transcription of psaA and psbA, and the phosphorylation of SIG1 causes the changes of stoichiometry that depend on the redox state of PQ.
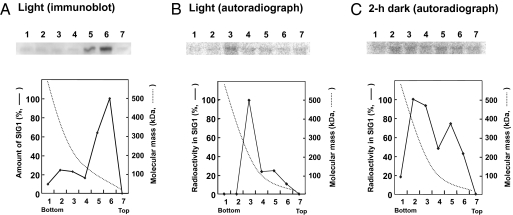
Are phosphorylated SIG1 factors free or associated with the RNA polymerase core enzyme in vivo? Under moderate light intensity, SIG1 exists mainly in the free form in wild-type plants, based on its molecular mass in sucrose density gradients (Fig. 3A). A minor peak of SIG1 was found in fraction 2 at ≈400 kDa (Fig. 3A), where the β-subunit of the core was detected with antibodies against the rpoB product (Fig. S4). Plants transgenic for wild-type SIG1 were labeled with [32P]orthophosphate in the light and dark. After sucrose density-gradient centrifugation, the 32P incorporated into SIG1 was detected by immunoprecipitation with anti-SIG1 (Fig. 3 B and C). The profiles of phosphorylated SIG1 molecules differed in the light where most sedimented in fraction 3 at ≈200 kDa (Fig. 3B), whereas in the dark they formed two peaks at fractions corresponding to the RNA polymerase holoenzyme (fraction 2) and free SIG1 according to molecular mass (Fig. 3C). Hence, SIG1 in the holoenzyme is phosphorylated only in the dark. The preferential inhibition of transcription of psaA relative to psbA can be ascribed to the presence of phosphorylated SIG1 in the holoenzyme. Photosynthetic control of transcription by the redox signal from the reduced and oxidized forms of PQ might be explained as follows. In the reductive state, the holoenzyme contains nonphosphorylated SIG1 and transcribes psaA and psbA equally. In the oxidative state, the holoenzyme contains phosphorylated SIG1 and transcribes psbA efficiently and psaA poorly (Fig. 4).
Fig. 3.
Effect of phosphorylation of SIG1 on association with the RNA polymerase core enzyme in vivo. (A) Leaf extracts of wild-type plants harvested in the daytime were subjected to sucrose density-gradient centrifugation, followed by immunoblotting with anti-SIG1. Molecular masses are based on those of the native proteins in each fraction, as further analyzed by nondenaturing PAGE (4–12%, wt/vol, polyacrylamide gradient). (B and C) SIG1 transgenics (3-wk-old) were incubated with [32P]orthophosphate in light at 75 μmol·m−2·s−1 (B) or in the dark (C). The extracted proteins were fractionated by sucrose density-gradient centrifugation, and each fraction was then immunoprecipitated with anti-SIG1, run on SDS/PAGE, and autoradiographed.
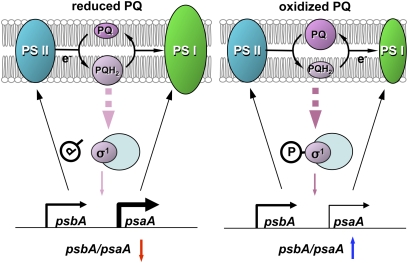
Fig. 4.
Model of the photosynthetic control of chloroplast gene transcription via phosphorylation of a SIG. The redox status of PQ is changed by the intensity and wavelength of light: Irradiance or shorter-wavelength light (680 nm) generates PQH2, and low light or longer-wavelength light (700 nm) at sunrise and sunset oxidizes PQ. The chemically reduced form of PQH2 is produced by electron flow from PS-II and converted to PQ after electron flow to PS-I. Electron flow from PS-II to PQ can be inhibited by DCMU, and electron flow from PQ to PS-I can be inhibited by DBMIB. The expression of psaA, encoding the apoprotein of the PS-I reaction center, is induced by PQH2 and repressed when PQH2 is oxidized. The expression of psbA encoding the D1 protein of the PS-II reaction center is less affected by redox status. The core enzyme associated with dephosphorylated SIG has high transcriptional activity for psaA and psbA, and phosphorylation of SIG causes a greater reduction of the transcription of psaA than psbA.
Pathway of the Redox Signal Started from PQ States.
The study of the phosphorylation of sigma factors was initiated by Tiller et al. (10). Their group showed that sigma-like factors in mustard were phosphorylated and dephosphorylated in etioplasts and chloroplasts, respectively. In addition, they reported that plastid transcription kinase (PTK) affected the phosphorylation of an RNA polymerase moiety in mustard plastids through the reduction of glutathione in response to irradiance (11), and that cpCK2 was an enzyme corresponding to PTK on the criterion of their common properties (12). The group has recently found that cpCK2 phosphorylates multiple sites in SIG6, and that mutations at these sites cause the malfunction of RNA polymerase (13). We show here that the phosphorylation of sigma factors is engaged in the stochiometry adjustment of PSs and involved in a kind of regulation different from that of cpCK2. There are two kinds of redox-signaling pathways originating from PSs, namely the “priming signal” generated from PQ and the “thiol-mediated signal” (14). The former signal transduction pathway may be involved in the phosphorylation of sigma factors observed in this investigation. This phosphorylation is promoted by the priming signal, starting from the oxidized state of PQ that results from low-intensity or higher-wavelength light. Although the components meditating the signal transduction pathway for the stoichiometry adjustment of PSs have not yet been clarified, Puthiyaveetil et al. (15) reported that chloroplast sensor kinase (CSK) is required to control the transcription of chloroplast genes after the redox state. CSK might be involved in an early step in the signal transduction pathway leading to the dephosphorylation of sigma factors under the control of the priming signal.
Various redox effects impact the transcription and transcript stability of photosynthetic genes (16, 17), transcription of nuclear genes (18, 19), and de novo protein synthesis (20, 21). In higher plants, Pfannschmidt et al. (4) demonstrated direct regulatory coupling between the redox state of PQ and the transcription of specific chloroplast genes. We have shown that the stoichiometry of the two PSs is adjusted by modification of a sigma factor in response to a redox signal. Redox regulation typically transduces signals rapidly, directly, and simply (4). Instead of the de novo synthesis of proteins and mRNA or the participation of multiple transcriptional factors, a sigma factor is modified by phosphorylation or dephosphorylation. Each bacterial sigma factor plays a specific role in gene expression under different conditions, and many cooperate to adapt to changed environments (22–24). However, we have shown that modification of a sigma factor causes a change of promoter preference in higher plants.
Nucleic acids and proteins inside prokaryotic endosymbionts or chloroplasts are exposed to oxygen-free radicals, which cause mutations and protein degradation (25). Land plants cannot escape light illumination and have developed systems for preventing damage (1–4, 25–32). To minimize damage caused by the generation of reactive oxygen species (ROS), plants have evolved mechanisms of photoprotection (28–32). PS-I must be induced to accept electrons from PQ to prevent the generation of ROS, when PS-II is strongly active in the daytime by shorter-wavelength light. Our results elucidate a mechanism of induction of PS-I through the dephosphorylation of sigma factors by redox signals of the PQ state (Fig. 4). On the other hand, longer-wavelength light, which is enriched at sunrise and sunset, must be absorbed more efficiently by PS-I than PS-II, resulting in the excitement of electrons in PS-I and, to a lesser degree, in PS-II. Formation of PS-I may then be repressed to coordinate with the activity of PS-II, leading to a reduction of the rate of cyclic electron flow (31, 32) and the detoxification of ROS (30). Sigma factors play a role in this regulation through their phosphorylation in response to the oxidized state of PQ (Fig. 4). Consequently, the phosphorylation of sigma factors under redox control may therefore be involved in maintaining the photosynthetic efficiency.
Materials and Methods
Transformation of Arabidopsis.
Constructs for ectopic expression of SIGs were made with the 35S-sGFP(S65T) vector and pSMAB701. Details of in vitro mutagenesis of SIG1 and plasmid construction can be found in SI Text and Table S1.
32P-labeling and Immunoprecipitation.
Whole plants were placed in Petri dishes containing 10 mL of [32P]orthophosphate solution (H332PO4, ICN; 5.55 MBq) for 2 h. They were rinsed with distilled water to remove excess 32P. Protein extracts were mixed with Protein-G-Sepharose (Amersham Biosciences) at 4 °C for 1 h. The resulting supernatants were reacted with anti-SIG1 and incubated at 4 °C for 1 h, followed by passage through Protein-G-Sepharose and incubation at 4 °C for 1 h. The immunoprecipitated complexes were washed with chilled PBS, and subjected to SDS polyacrylamide gel electrophoresis (SDS/PAGE; 10%, wt/vol, polyacrylamide).
Analysis of Gene Expression and Protein Products.
Real-time PCR was performed with a LightCycler (Roche), and detected with LightCycler DNA Master SYBR Green I (Roche). Transcripts for cytosolic actin (33) (ACT2) were measured as an internal standard.
Chloroplasts prepared from 4-wk-old plants were incubated in the dark for 2 h and subjected to run-on assays. Values were normalized on the basis of SIG1 content determined by immunoblotting, and the means of three independent experiments are shown.
Protein extracts were subjected to SDS/PAGE (10%, wt/vol, polyacrylamide), and SIGs were detected with ECL Plus Western Blotting Detection System (Amersham Biosciences). Details of the protocols are available in SI Text.
Treatment of Plants with Electron-Transport Inhibitors.
Seedlings grown on MS solid medium in the dark for 5 d were cultured hydroponically with MS liquid medium additionally for 5 d. The medium was then replaced with that containing 0.5 μM DCMU or 0.2 μM BDMIB and plants were incubated in the light for 2 d.
Supplementary Material
Acknowledgments
We thank Mamoru Isemura for guidance in the preparation of rabbit antisera, Hiroaki Ichikawa of the National Institute of Agrobiological Resources in Japan for pSMAB701, Takanori Hirano and Yasuo Niwa for intermediate constructs used in cloning in our laboratory, Csaba Koncz at Max Planck Institute for Plant Breeding Research in Cologne in Germany for Agrobacterium tumefaciens GV3101, Akira Ishihama for technical comments and encouragement, Kozi Asada and Kimiyuki Satoh for discussion on photo-oxidative damage, and Philip Hawke for English editing. The research was supported by Grants-in-Aid, the 21st Century COE Program and the Global COE Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Monbukagakusho), and by grants from the Toray Science Foundation and the Salt Science Research Foundation in Japan. This paper is dedicated to the late Lawrence Bogorad, who originally characterized chloroplast RNA polymerase, as well as the psbA gene for triazine receptor protein and the psaA gene for PSI-A1, which he described as “photogenes.” H. Kobayashi and other colleagues had the honor of sharing time and common interests with him at Harvard University until he passed away in 2003.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911692107/-/DCSupplemental.
References
- 1.Fujita Y, Murakami A, Ohki K. Regulation of photosystem composition in the cyanobacterial photosynthetic system: The regulation occurs in response to the redox state of the electron pool located between the two photosystems. Plant Cell Physiol. 1987;28:283–292. [Google Scholar]
- 2.Chow WS, Melis A, Anderson JM. Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA. 1990;87:7502–7506. doi: 10.1073/pnas.87.19.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JH, Glick RE, Melis A. Dynamics of photosystem stoichiometry adjustment by light quality in chloroplasts. Plant Physiol. 1993;102:181–190. doi: 10.1104/pp.102.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- 5.Allen JF, Pfannschmidt T. Balancing the two photosystems: Photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 2000;355:1351–1359. doi: 10.1098/rstb.2000.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isono K, et al. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of σ70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison LA. The role of sigma factors in plastid transcription. Biochimie. 2000;82:537–548. doi: 10.1016/s0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- 8.Tsunoyama Y, et al. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishizaki Y, et al. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- 10.Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Link G. Redox regulation of chloroplast transcription. Antioxid Redox Signal. 2003;5:79–87. doi: 10.1089/152308603321223568. [DOI] [PubMed] [Google Scholar]
- 12.Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem. 2002;269:3329–3337. [PubMed] [Google Scholar]
- 13.Schweer J, Turkeri H, Link B, Link G. AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J. 2010;62:192–202. doi: 10.1111/j.1365-313X.2010.04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 15.Puthiyaveetil S, et al. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci USA. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oswald O, Martin T, Dominy PJ, Graham IA. Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA. 2001;98:2047–2052. doi: 10.1073/pnas.021449998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liere K, Link G. Chloroplast endoribonuclease p54 involved in RNA 3′-end processing is regulated by phosphorylation and redox state. Nucleic Acids Res. 1997;25:2403–2408. doi: 10.1093/nar/25.12.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escoubas JM, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fey V, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- 20.Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- 21.Trebitsh T, Danon A. Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc Natl Acad Sci USA. 2001;98:12289–12294. doi: 10.1073/pnas.211440698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 23.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 24.Brahamsha B, Haselkorn R. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: Cloning, expression, and inactivation of the sigB and sigC genes. J Bacteriol. 1992;174:7273–7282. doi: 10.1128/jb.174.22.7273-7282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 26.Allen JF, Raven JA. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J Mol Evol. 1996;42:482–492. doi: 10.1007/BF02352278. [DOI] [PubMed] [Google Scholar]
- 27.Pfannschmidt T. Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 2003;8:33–41. doi: 10.1016/s1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 28.Müller P, Li XP, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen JF. Cyclic, pseudocyclic and noncyclic photophosphorylation: New links in the chain. Trends Plant Sci. 2003;8:15–19. doi: 10.1016/s1360-1385(02)00006-7. [DOI] [PubMed] [Google Scholar]
- 30.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munekage Y, et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 32.DalCorso G, et al. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132:273–285. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 33.An YQ, et al. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.