Abstract
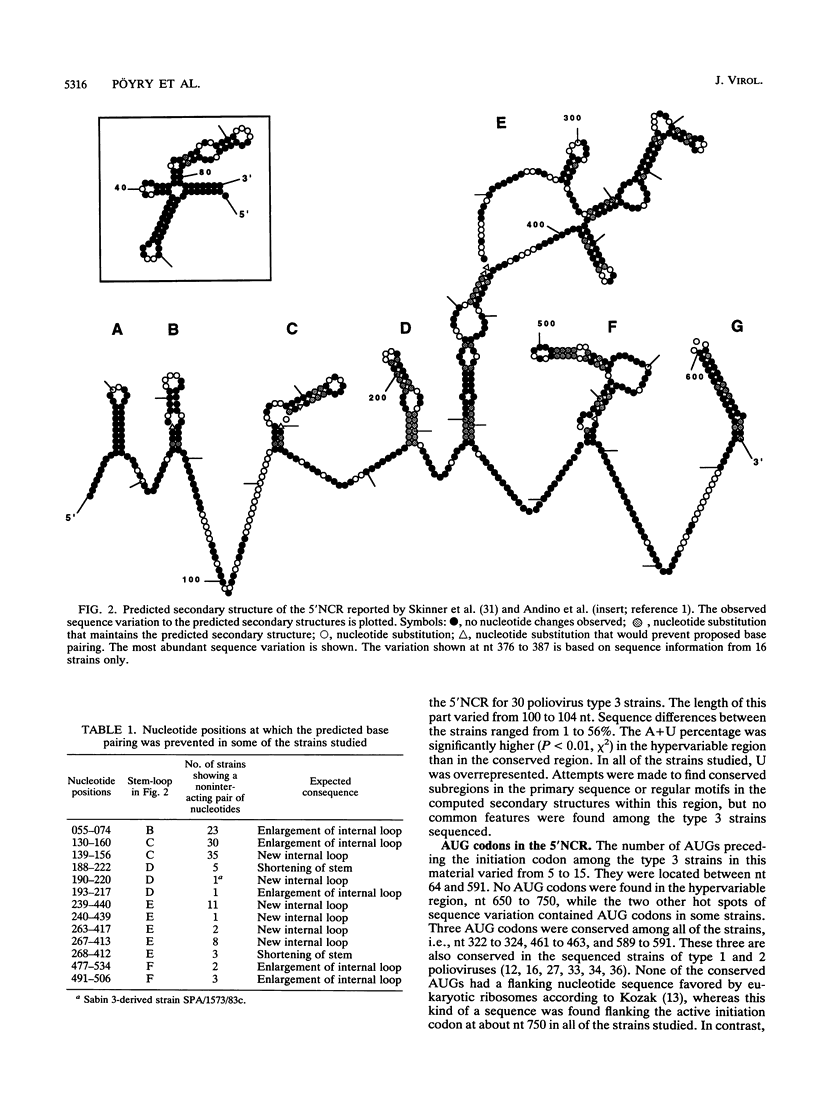
Poliovirus has a single-stranded RNA genome of about 7,440 nucleotides (nt) with an unusually long 750-nt noncoding region in the 5' end (5'NCR). Several regulatory functions have been assigned to the 5'NCR. We sequenced the 5'NCRs of 33 wild-type 3 poliovirus strains to study the range and distribution of naturally occurring sequence variations. In this regard, the 5'NCR can be divided into a conserved part (nt 1 to 650) and a hypervariable part (nt 651 to 750). In the conserved part, altogether 234 unevenly distributed nucleotide positions (36%) showed variation. When these positions were plotted against the predicted secondary-structure models, it was found that the existence of most of the proposed stem-loop structures was supported by extensive structure-conserving substitutions in the stems. Regions with conserved sequences, as well as mutational hot spots, were observed. The hypervariable part of the 5'NCR varied up to 56% between the strains studied. The A + U percentage was significantly higher than in the conserved part. The number of AUG codons varied between 5 and 15 in the conserved part of the 5'NCR, while none was found in the hypervariable part. These results provide information that can be used in site-directed mutagenesis and other approaches targeted to reveal the functional domains of the 5'NCR.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J Virol. 1990 Feb;64(2):607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K., Ehrenfeld E. An internal 5'-noncoding region required for translation of poliovirus RNA in vitro. J Virol. 1988 Aug;62(8):3068–3072. doi: 10.1128/jvi.62.8.3068-3072.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle J. A., Skuce R. A., Fleming C. S., Hoey E. M., Martin S. J. The complete nucleotide sequence of a bovine enterovirus. J Gen Virol. 1988 Feb;69(Pt 2):253–263. doi: 10.1099/0022-1317-69-2-253. [DOI] [PubMed] [Google Scholar]
- Evans D. M., Dunn G., Minor P. D., Schild G. C., Cann A. J., Stanway G., Almond J. W., Currey K., Maizel J. V., Jr Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985 Apr 11;314(6011):548–550. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hughes P. J., Evans D. M., Minor P. D., Schild G. C., Almond J. W., Stanway G. The nucleotide sequence of a type 3 poliovirus isolated during a recent outbreak of poliomyelitis in Finland. J Gen Virol. 1986 Oct;67(Pt 10):2093–2102. doi: 10.1099/0022-1317-67-10-2093. [DOI] [PubMed] [Google Scholar]
- Huovilainen A., Kinnunen L., Ferguson M., Hovi T. Antigenic variation among 173 strains of type 3 poliovirus isolated in Finland during the 1984 to 1985 outbreak. J Gen Virol. 1988 Aug;69(Pt 8):1941–1948. doi: 10.1099/0022-1317-69-8-1941. [DOI] [PubMed] [Google Scholar]
- Kinnunen L., Huovilainen A., Pöyry T., Hovi T. Rapid molecular evolution of wild type 3 poliovirus during infection in individual hosts. J Gen Virol. 1990 Feb;71(Pt 2):317–324. doi: 10.1099/0022-1317-71-2-317. [DOI] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Kawamura N., Nomoto A. Genetic variation occurring on the genome of an in vitro insertion mutant of poliovirus type 1. J Virol. 1989 Mar;63(3):1069–1075. doi: 10.1128/jvi.63.3.1069-1075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5' noncoding sequence in viral replication. J Virol. 1987 May;61(5):1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Monica N., Meriam C., Racaniello V. R. Mapping of sequences required for mouse neurovirulence of poliovirus type 2 Lansing. J Virol. 1986 Feb;57(2):515–525. doi: 10.1128/jvi.57.2.515-525.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Nicholson R., Sonenberg N. In vitro mutational analysis of cis-acting RNA translational elements within the poliovirus type 2 5' untranslated region. J Virol. 1991 Nov;65(11):5895–5901. doi: 10.1128/jvi.65.11.5895-5901.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Pelletier J., Sonenberg N. A cellular protein that binds to the 5'-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989 Jul;3(7):1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- Minor P. D., Dunn G. The effect of sequences in the 5' non-coding region on the replication of polioviruses in the human gut. J Gen Virol. 1988 May;69(Pt 5):1091–1096. doi: 10.1099/0022-1317-69-5-1091. [DOI] [PubMed] [Google Scholar]
- Najita L., Sarnow P. Oxidation-reduction sensitive interaction of a cellular 50-kDa protein with an RNA hairpin in the 5' noncoding region of the poliovirus genome. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5846–5850. doi: 10.1073/pnas.87.15.5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson R., Pelletier J., Le S. Y., Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol. 1991 Nov;65(11):5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Flynn M. E., Kaplan G., Racaniello V., Sonenberg N. Mutational analysis of upstream AUG codons of poliovirus RNA. J Virol. 1988 Dec;62(12):4486–4492. doi: 10.1128/jvi.62.12.4486-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Hellen C. U., Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5' nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991 Nov;65(11):6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E. V., Blinov V. M., Romanova L. I., Sinyakov A. N., Maslova S. V., Agol V. I. Conserved structural domains in the 5'-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989 Feb;168(2):201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- Pöyry T., Kinnunen L., Hovi T. Restricted variability of a 17 nucleotide stretch within the 5'-noncoding region of poliovirus genome. Epidemiol Infect. 1989 Dec;103(3):671–683. doi: 10.1017/s0950268800031071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T., Kinnunen L., Kapsenberg J., Kew O., Hovi T. Type 3 poliovirus/Finland/1984 is genetically related to common Mediterranean strains. J Gen Virol. 1990 Nov;71(Pt 11):2535–2541. doi: 10.1099/0022-1317-71-11-2535. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R., Pallansch M. A., Nottay B. K., Kew O. M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987 Oct;160(2):311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- Rivera V. M., Welsh J. D., Maizel J. V., Jr Comparative sequence analysis of the 5' noncoding region of the enteroviruses and rhinoviruses. Virology. 1988 Jul;165(1):42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- Simoes E. A., Sarnow P. An RNA hairpin at the extreme 5' end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991 Feb;65(2):913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. A., Racaniello V. R., Dunn G., Cooper J., Minor P. D., Almond J. W. New model for the secondary structure of the 5' non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989 May 20;207(2):379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. Poliovirus translation. Curr Top Microbiol Immunol. 1990;161:23–47. doi: 10.1007/978-3-642-75602-3_2. [DOI] [PubMed] [Google Scholar]
- Stanway G., Cann A. J., Hauptmann R., Hughes P., Clarke L. D., Mountford R. C., Minor P. D., Schild G. C., Almond J. W. The nucleotide sequence of poliovirus type 3 leon 12 a1b: comparison with poliovirus type 1. Nucleic Acids Res. 1983 Aug 25;11(16):5629–5643. doi: 10.1093/nar/11.16.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Reeve P., Minor P. D., Schild G. C., Almond J. W. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1539–1543. doi: 10.1073/pnas.81.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y. V., Maslova S. V., Agol V. I. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology. 1985 Dec;147(2):243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Kohara M., Kataoka Y., Suganuma T., Omata T., Imura N., Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984 Apr 25;174(4):561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- Trono D., Andino R., Baltimore D. An RNA sequence of hundreds of nucleotides at the 5' end of poliovirus RNA is involved in allowing viral protein synthesis. J Virol. 1988 Jul;62(7):2291–2299. doi: 10.1128/jvi.62.7.2291-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukkonen P., Huovilainen A., Hovi T. Detection of poliovirus antigen by enzyme immunoassay. J Clin Microbiol. 1986 Dec;24(6):954–958. doi: 10.1128/jcm.24.6.954-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrop G. D., Wareham K. A., Evans D. M., Dunn G., Minor P. D., Magrath D. I., Taffs F., Marsden S., Skinner M. A., Schild G. C. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J Virol. 1989 Mar;63(3):1338–1344. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Angel R. M., Papavassiliou A. G., Fernández-Tomás C., Silverstein S. J., Racaniello V. R. Cell proteins bind to multiple sites within the 5' untranslated region of poliovirus RNA. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8299–8303. doi: 10.1073/pnas.86.21.8299. [DOI] [PMC free article] [PubMed] [Google Scholar]



