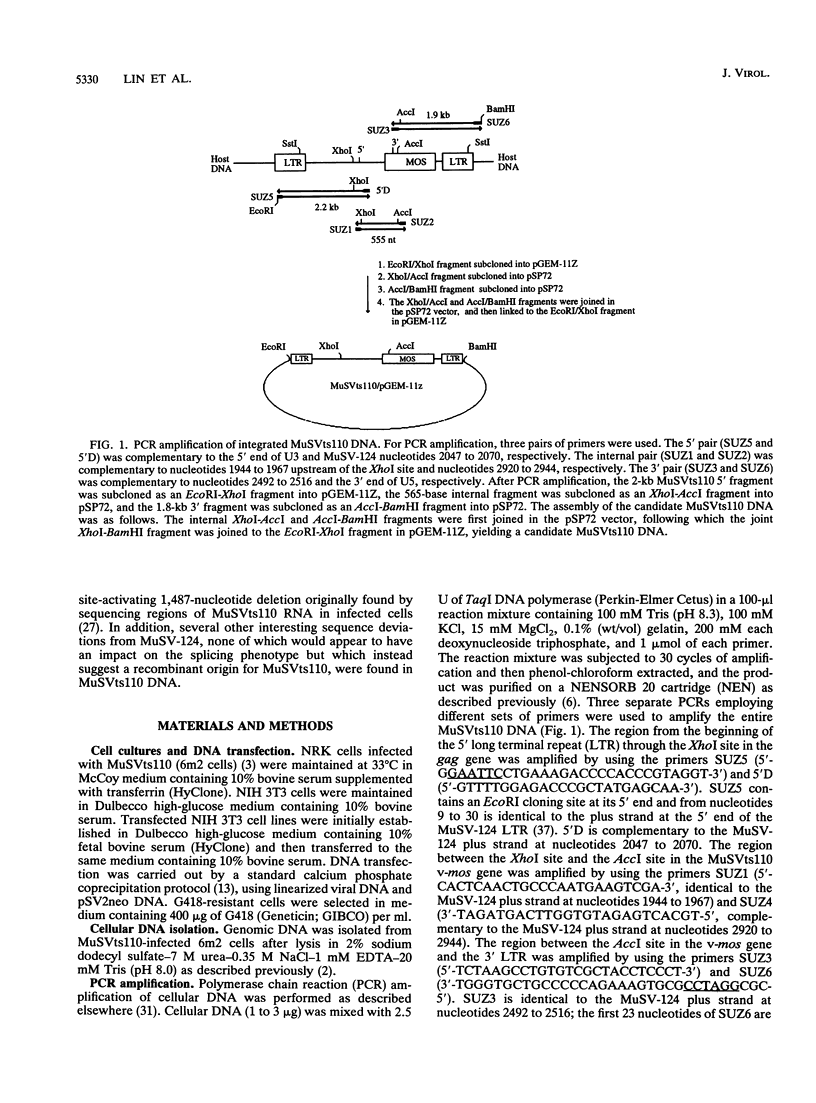
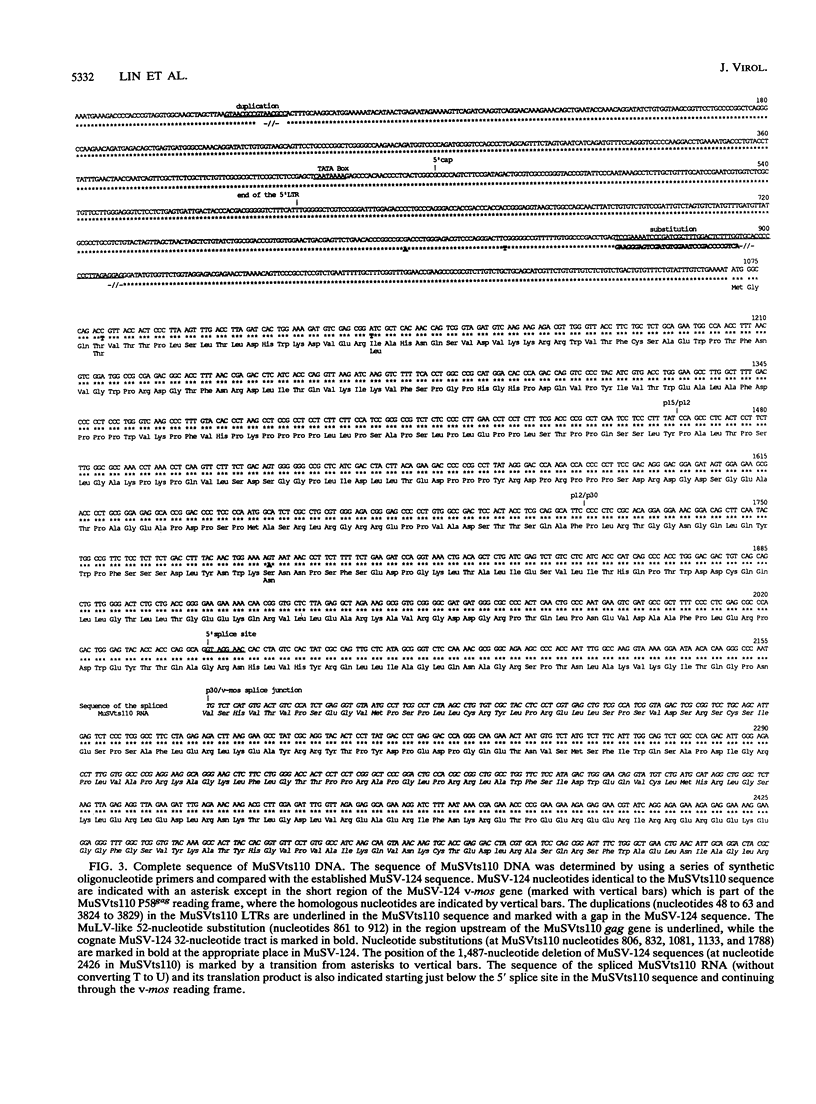
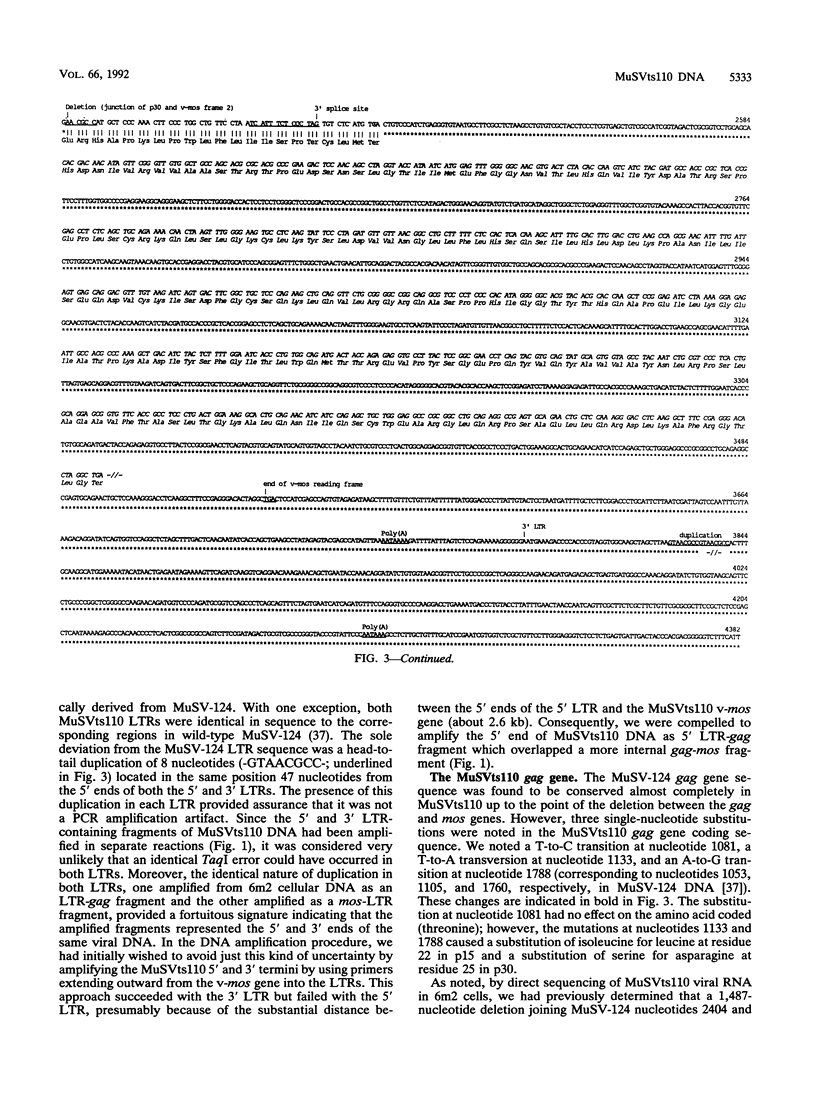
Abstract
We have cloned Moloney murine sarcoma virus (MuSV) MuSVts110 DNA by assembly of polymerase chain reaction (PCR)-amplified segments of integrated viral DNA from infected NRK cells (6m2 cells) and determined its complete sequence. Previously, by direct sequencing of MuSVts110 RNA transcribed in 6m2 cells, we established that the thermosensitive RNA splicing phenotype uniquely characteristic of MuSVts110 results from a deletion of 1,487 nucleotides of progenitor MuSV-124 sequences. As anticipated, the sequence obtained in this study contained precisely this same deletion. In addition, several other unexpected sequence differences were found between MuSVts110 and MuSV-124. For example, in the noncoding region upstream of the gag gene, MuSVts110 DNA contained a 52-nucleotide tract typical of murine leukemia virus rather than MuSV-124, suggesting that MuSVts110 originated as a MuSV-helper murine leukemia virus recombinant during reverse transcription rather than from a straightforward deletion within MuSV-124. In addition, both MuSVts110 long terminal repeats contained head-to-tail duplications of eight nucleotides in the U3 region. Finally, seven single-nucleotide substitutions were found scattered throughout MuSVts110 DNA. Three of the nucleotide substitutions were in the gag gene, resulting in one coding change in p15 and one in p30. All of the remaining nucleotide changes were found in the noncoding region between the 5' long terminal repeat and the gag gene. In NIH 3T3 cells transfected with the cloned MuSVts110 DNA, the pattern of viral RNA expression conformed with that observed in cells infected with authentic MuSVts110 virus in that viral RNA splicing was 30 to 40% efficient at growth temperatures between 28 and 33 degrees C but reduced to trace levels above 37 degrees C.
Full text
PDF



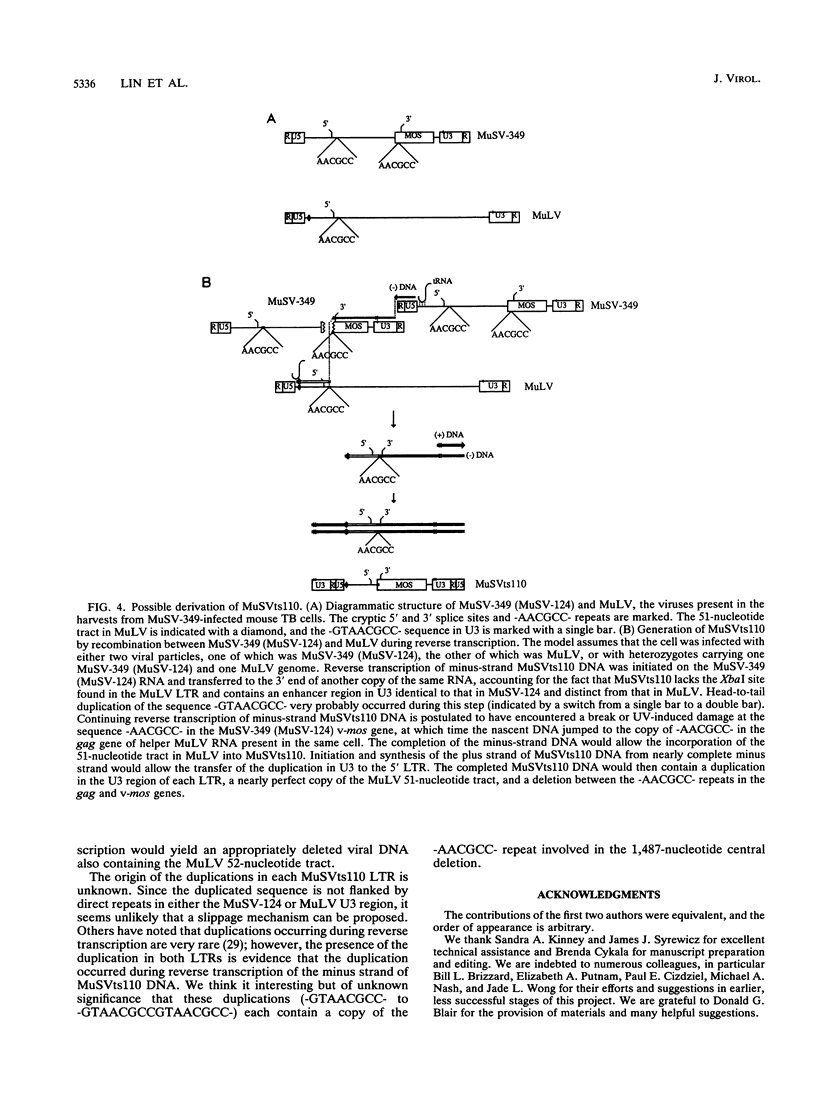

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball J., McCarter J. A., Sunderland S. M. Evidence for helper independent murine sarcoma virus. I. Segregation of replication-defective and transformation-defective viruses. Virology. 1973 Nov;56(1):268–284. doi: 10.1016/0042-6822(73)90305-x. [DOI] [PubMed] [Google Scholar]
- Biggart N. W., Gallick G. E., Murphy E. C., Jr Nickel-induced heritable alterations in retroviral transforming gene expression. J Virol. 1987 Aug;61(8):2378–2388. doi: 10.1128/jvi.61.8.2378-2388.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. G., Hull M. A., Finch E. A. The isolation and preliminary characterization of temperature-sensitive transformation mutants of Moloney sarcoma virus. Virology. 1979 Jun;95(2):303–316. doi: 10.1016/0042-6822(79)90486-0. [DOI] [PubMed] [Google Scholar]
- Budowsky E. I., Abdurashidova G. G. Polynucleotide-protein cross-links induced by ultraviolet light and their use for structural investigation of nucleoproteins. Prog Nucleic Acid Res Mol Biol. 1989;37:1–65. doi: 10.1016/s0079-6603(08)60694-7. [DOI] [PubMed] [Google Scholar]
- Chen-Kiang S., Nevins J. R., Darnell J. E., Jr N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979 Dec 15;135(3):733–752. doi: 10.1016/0022-2836(79)90174-8. [DOI] [PubMed] [Google Scholar]
- Chiocca S. M., Sterner D. A., Biggart N. W., Murphy E. C., Jr Nickel mutagenesis: alteration of the MuSVts110 thermosensitive splicing phenotype by a nickel-induced duplication of the 3' splice site. Mol Carcinog. 1991;4(1):61–71. doi: 10.1002/mc.2940040110. [DOI] [PubMed] [Google Scholar]
- Cizdziel P. E., Nash M. A., Blair D. G., Murphy E. C., Jr Molecular basis underlying phenotypic revertants of Moloney murine sarcoma virus MuSVts110. J Virol. 1986 Jan;57(1):310–317. doi: 10.1128/jvi.57.1.310-317.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizdziel P. E., de Mars M., Murphy E. C., Jr Exploitation of a thermosensitive splicing event to study pre-mRNA splicing in vivo. Mol Cell Biol. 1988 Apr;8(4):1558–1569. doi: 10.1128/mcb.8.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mars M., Sterner D. A., Chiocca S. M., Biggart N. W., Murphy E. C., Jr Regulation of RNA splicing in gag-deficient mutants of Moloney murine sarcoma virus MuSVts110. J Virol. 1990 Apr;64(4):1421–1428. doi: 10.1128/jvi.64.4.1421-1428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis P., Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991 Feb 15;266(5):3172–3177. [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hamelin R., Brizzard B. L., Nash M. A., Murphy E. C., Jr, Arlinghaus R. B. Temperature-sensitive viral RNA expression in Moloney murine sarcoma virus ts110-infected cells. J Virol. 1985 Feb;53(2):616–623. doi: 10.1128/jvi.53.2.616-623.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin R., Kabat K., Blair D., Arlinghaus R. B. Temperature-sensitive splicing defect of ts110 Moloney murine sarcoma virus is virus encoded. J Virol. 1986 Jan;57(1):301–309. doi: 10.1128/jvi.57.1.301-309.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn J. P., Wood T. G., Murphy E. C., Jr, Blair D. G., Arlinghaus R. B. A selective temperature-sensitive defect in viral RNA expression in cells infected with a ts transformation mutant of murine sarcoma virus. Cell. 1981 Jul;25(1):37–46. doi: 10.1016/0092-8674(81)90229-4. [DOI] [PubMed] [Google Scholar]
- Hwang L. H., Gilboa E. Expression of genes introduced into cells by retroviral infection is more efficient than that of genes introduced into cells by DNA transfection. J Virol. 1984 May;50(2):417–424. doi: 10.1128/jvi.50.2.417-424.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R. P., Murphy E. C., Jr, Arlinghaus R. B. Electron microscopic analysis of ts1 10 Moloney mouse sarcoma virus, a variant of wild-type virus with two RNAs containing large deletions. J Mol Biol. 1982 Oct 25;161(2):229–250. doi: 10.1016/0022-2836(82)90150-4. [DOI] [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985 Sep;5(9):2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzer W. S., Maxwell S. A., Arlinghaus R. B. Further characterization of the P85gag-mos -associated protein kinase activity. Virology. 1984 Oct 15;138(1):143–155. doi: 10.1016/0042-6822(84)90154-5. [DOI] [PubMed] [Google Scholar]
- Kloetzer W. S., Maxwell S. A., Arlinghaus R. B. P85gag-mos encoded by ts110 Moloney murine sarcoma virus has an associated protein kinase activity. Proc Natl Acad Sci U S A. 1983 Jan;80(2):412–416. doi: 10.1073/pnas.80.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D., Verma I. M. Two base changes restore infectivity to a noninfectious molecular clone of Moloney murine leukemia virus (pMLV-1). J Virol. 1984 Jan;49(1):214–222. doi: 10.1128/jvi.49.1.214-222.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. D., Bose K. K., Rabkin S. D., Strauss B. S. Sites of termination of in vitro DNA synthesis on ultraviolet- and N-acetylaminofluorene-treated phi X174 templates by prokaryotic and eukaryotic DNA polymerases. Proc Natl Acad Sci U S A. 1981 Jan;78(1):110–114. doi: 10.1073/pnas.78.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P., Strauss B. S. Sites of inhibition of in vitro DNA synthesis in carcinogen- and UV-treated phi X174 DNA. Nature. 1979 Apr 12;278(5705):664–666. doi: 10.1038/278664a0. [DOI] [PubMed] [Google Scholar]
- Nash M. A., Brizzard B. L., Wong J. L., Murphy E. C., Jr Murine sarcoma virus ts110 RNA transcripts: origin from a single proviral DNA and sequence of the gag-mos junctions in both the precursor and spliced viral RNAs. J Virol. 1985 Feb;53(2):624–633. doi: 10.1128/jvi.53.2.624-633.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M., Brown N. V., Wong J. L., Arlinghaus R. B., Murphy E. C., Jr S1 nuclease mapping of viral RNAs from a temperature-sensitive transformation mutant of murine sarcoma virus. J Virol. 1984 May;50(2):478–488. doi: 10.1128/jvi.50.2.478-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991 Sep;65(9):4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Spontaneous variation and synthesis in the U3 region of the long terminal repeat of an avian retrovirus. J Virol. 1982 Jan;41(1):163–171. doi: 10.1128/jvi.41.1.163-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dane R. W. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982 Jun;42(3):918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- de Mars M., Cizdziel P. E., Murphy E. C., Jr Activation of cryptic splice sites in murine sarcoma virus-124 mutants. J Virol. 1990 Nov;64(11):5260–5269. doi: 10.1128/jvi.64.11.5260-5269.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mars M., Cizdziel P. E., Murphy E. C., Jr Activation of thermosensitive RNA splicing and production of a heat-labile P85gag-mos kinase by the introduction of a specific deletion in murine sarcoma virus-124 DNA. J Virol. 1988 Jun;62(6):1907–1916. doi: 10.1128/jvi.62.6.1907-1916.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]