Abstract
AIM: To assess the relationship between vitamin D receptor (VDR) gene polymorphisms and the presence of hepatocellular carcinoma (HCC).
METHODS: Two-hundred forty patients who underwent liver transplantation were studied. The etiologies of liver disease were hepatitis C (100 patients), hepatitis B (37) and alcoholic liver disease (103). A group of 236 healthy subjects served as controls. HCC in the explanted liver was detected in 80 patients. The following single nucleotide gene polymorphisms of the VDR were investigated by polymerase chain reaction and restriction fragment length polymorphism: FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) (BAT).
RESULTS: The frequencies of genotypes in patients without and with HCC were for FokI F/F = 69, F/f = 73, f/f = 18 and F/F = 36, F/f = 36, f/f = 8; BsmI b/b = 45, B/b = 87, B/B = 28 and b/b = 33, B/b = 35, B/B = 12; for ApaI A/A = 53, A/a = 85, a/a = 22 and A/A = 27, A/a = 38, a/a = 15; for TaqI T/T = 44, T/t = 88, t/t = 28 and T/T = 32, T/t = 38, t/t = 10. Carriage of the b/b genotype of BsmI and the T/T genotype of TaqI was significantly associated with HCC (45/160 vs 33/80, P < 0.05 and 44/160 vs 32/80, P < 0.05, respectively). The absence of the A-T-C protective allele of BAT was significantly associated with the presence of HCC (46/80 vs 68/160, P < 0.05). A strong association was observed between carriage of the BAT A-T-C and G-T-T haplotypes and HCC only in alcoholic liver disease (7/46 vs 12/36 vs 11/21, P < 0.002, respectively).
CONCLUSION: VDR genetic polymorphisms are significantly associated with the occurrence of HCC in patients with liver cirrhosis. This relationship is more specific for patients with an alcoholic etiology.
Keywords: Alcohol, Hepatocellular carcinoma, Liver cirrhosis, Vitamin D receptor polymorphisms
INTRODUCTION
Hepatocellular carcinoma (HCC), the fifth most common cause of cancer and the third leading cause of cancer-related death worldwide[1], accounts for 85% to 90% of primary liver cancers. It is characterized by dynamic temporal trends and marked demographic, geographic and ethnic variations[1]. In western countries, HCC is most often superimposed on cirrhosis, its major risk factor[2,3]. Alcohol, hepatitis B virus (HBV) and hepatitis C virus (HCV) are the main etiologic agents of cirrhosis and HCC worldwide. HBV and HCV supposedly exert a direct carcinogenic effect, accounting for the high prevalence of liver cancer among infected patients[2,3]; the same might not be true for alcohol[1]. Overall, it is prudent to affirm that differences in the incidence rates and the strong gender distribution in HCC are not entirely due to differences in the exposure to the three causative agents mentioned above. Genetic factors could also contribute, particularly gene polymorphisms of inflammatory cytokines and growth factor ligands and receptors[4].
The vitamin D receptor (VDR) is a member of the nuclear receptor super-family of ligand-inducible transcription factors which are involved in many physiological processes, including cell growth and differentiation, embryonic development and metabolic homeostasis. The transcriptional activity of this receptor is modulated by ligands, such as steroids, retinoids and other lipid-soluble compounds, and by nuclear proteins acting as co-activators and co-repressors[5,6]. The liganded VDR heterodimerizes with the retinoid X receptor and binds to vitamin D response elements in the promoter of target genes, thereby affecting their transcription. The genomic organization of the VDR at locus 12q13.1 shows that the VDR gene itself is quite large (over 100 kb) and has an extensive promoter region capable of generating multiple tissue-specific transcripts[7].
Several single nucleotide restriction fragment length polymorphisms have been described in the VDR gene in association with neoplastic and non-neoplastic diseases. In particular, VDR polymorphisms have been related to, although with conflicting observations, cancers of the breast, prostate, skin, colon-rectum, bladder and kidney[8-10]. Furthermore, VDR polymorphisms were found to influence the prognosis of prostate and breast cancer, renal cell carcinoma and malignant melanoma[11]. VDR polymorphisms have been investigated in the context of some chronic liver diseases, such as primary biliary cirrhosis and auto-immune hepatitis[12-16]. Surprisingly, however, there are no data in the literature on the possible association between VDR polymorphisms and the occurrence of HCC.
The aims of the present paper were (a) to investigate the possible relationship between VDR polymorphisms and the occurrence of HCC in patients with liver cirrhosis and (b) to assess whether the etiology of their liver disease exerts a role in modulating such a relationship.
MATERIALS AND METHODS
Patients
The study included 240 consecutive patients who underwent liver transplantation (LT) for end-stage liver disease due to hepatitis B (n = 37, 15.4%), hepatitis C (n = 100, 41.7%) and alcoholic liver disease (n = 103, 42.9%). The main demographic and clinical characteristics are reported in Table 1. Two-hundred thirty-six healthy community blood donors served as controls. They were 164 males (69.5%) and 72 females (30.5%); the median age was 48 years with a range of 18-77 years. The male gender distribution in the patients and controls (74% vs 70%) was similar, whereas these two groups differed in age: 54 ± 8 years vs 46 ± 13 years (mean ± SD, P < 0.0001). Control subjects did not have any clinical and/or laboratory evidence of liver disease or other major pathological conditions, such as diabetes mellitus. All patients and controls were Caucasian. Informed consent to participate in the study was obtained from each subject in accordance with the Declaration of Helsinki and following the guidelines of our ethical committee. All study participants approved the storage of their frozen DNA specimens for research purposes in our laboratory.
Table 1.
Main demographic and clinical characteristics of liver transplant recipients (n = 240) according to the presence (n = 80, 33.3%) or absence (n = 160, 66.7%) of HCC
| HCC present (n = 80) | HCC absent (n = 160) | P value | ||
| Male gender | 178 (74.2) | 70 | 108 | < 0.0010 |
| Age1 (yr) | 55 (22-68) | 57.2 ± 5.9 | 52.0 ± 8.6 | < 0.0001 |
| Body mass index1 (kg/m2) | 25.2 (14.8-48.5) | 26.4 ± 4.2 | 24.7 ± 3.3 | < 0.0010 |
| Etiology | ||||
| Viral (n = 137) | 50 | 87 | NS | |
| HBV | 37 (15.4) | |||
| HCV | 100 (41.7) | |||
| Alcoholic | 103 (42.9) | |||
| Child-Pugh score1 | 8 (5-14) | 7 (5-14) | 8 (5-13) | < 0.0001 |
| Child-Pugh score1 > 8 (n = 87) | 22 | 65 | < 0.0500 | |
| Diabetes mellitus | 67 (27.9) | 29 | 38 | < 0.0500 |
The continuous variables are reported as means (standard deviation), whereas the categorical variables are reported as frequencies (%). Statistical analysis was performed using the Student’s t-test for continuous variables and the Pearson χ2 test for categorical variables.
At transplant operation. HCC: Hepatocellular carcinoma; HBV: Hepatitis B virus; HCV: Hepatitis C virus; NS: Not significant.
Histology
All total hepatectomy specimens were sectioned at intervals of approximately one cm to search for suspicious focal hepatic lesions. Standard histological staining techniques were applied to confirm the presence of HCC and to evaluate the characteristics of the identified tumors, such as pathologic tumor grade (Edmondson grade) and macro- or micro-vascular invasion.
Vitamin D assay
In 113 patients (47.1%), serum samples, which were collected the day before the transplant operation and were separated and stored at -80°C until analysis, were available to assay the pre-LT serum vitamin D concentration. Circulating 25-hydroxyvitamin D levels were measured using a chemo-luminescent immunoassay implemented on a Liaison automatic analyzer (DiaSorin Inc, Stillwater, MN, USA). Data were expressed in ng/mL. Reference values of serum vitamin D adopted in this study were in accordance with those proposed by the Scientific Advisory Committee of Nutrition[17], which considers serum vitamin D levels < 10-15 ng/mL to be inadequate for bone and overall health in healthy individuals.
Molecular biology
Genomic DNA was isolated from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Milan, Italy) according to the manufacturer’s instructions. Four diallelic polymorphisms of the VDR were genotyped: FokI C>T (rs10735810) and TaqI T>C (rs10735810) polymorphic sites on the coding sequence, BsmI A>G (rs1544410) and ApaI G>T (rs7975232) on the last intron. For the detection of the VDR polymorphisms, the polymerase chain reaction (PCR) technique was applied and followed by restriction fragment length polymorphism assays. The PCR amplifications were carried out in a total volume of 10 μL containing 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 0.01% Tween-20, 0.2 mmol/L deoxyribonucleotides, 2-4 pmol of each primer, 2.0 mmol/L MgCl2 and 0.5 U hot-start Taq DNA polymerase (RighTaq, Euroclone, Milan, Italy). The sequences of primers used for FokI were (f) 5'-TGCAGCCTTCACAGGTCATA-3', (r) 5'-GGCCTGCTTGCTGTTCTTAC-3'; for TaqI and ApaI were (f) 5'-ACGTCTGCAGTGTGTTGGAC-3', (r) 5'-TCACCGGTCAGCAGTCATAG-3'; for BsmI were (f) 5'-CAGTTCACGCAAGAGCAGAG-3', (r) 5'-ACCTGAAGGGAGACGTAGCA-3'. All the primers were newly designed with the aid of the NCBI Primer-Blast Tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The cycling conditions for all the VDR polymorphisms were set as 40 cycles at 95°C for 30 s, 61°C for 30 s and 72°C for 1 min. In a total volume of 20 μL, amplified DNA (10 μL) was digested overnight with 2 U of restriction endonucleases using the buffers and temperatures recommended by the manufacturers. The presence of restriction sites for the FokI, TaqI, BsmI and ApaI enzymes were coded as ‘f’, ‘t’, ‘b’ and ‘a’ and the absence of restriction sites as ‘F’, ‘T’, ‘B’ and ‘A’, respectively. The FokI C>T (F/f) polymorphism was analyzed by digestion of a 157-bp PCR product with FokI (New England Biolabs, Hitchin, UK), which resulted in two fragments of 121 and 36 bp in the presence of the ‘f’ allele and in an uncut fragment in the presence of the ‘F’ allele. The TaqI T>C (T/t) and ApaI T>G (A/a) polymorphisms were analyzed by digestion of a 211-bp PCR product with TaqI (New England Biolabs, Hitchin, UK), which resulted in two fragments of 172 and 39 bp in the presence of the ‘t’ allele. These same polymorphisms were also analyzed by digestion with ApaI (New England Biolabs, Hitchin, UK), which resulted in two fragments of 121 and 90 bp for the ‘a’ allele. The BsmI A>G (B/b) polymorphism was analyzed by digestion of a 236-bp PCR product with BsmI (New England Biolabs, Hitchin, UK), which resulted in two fragments of 197 and 39 bp in the presence of the ‘b’ allele. All PCR reactions were carried out in a Techne TC-412 thermal cycle, and PCR products were sized by electrophoresis on a 3% agarose gel stained with ethidium bromide.
Statistical analysis
The statistical analysis of data was performed using the BMDP Dynamic Statistical Software Package 7.0 (Statistical Solutions, Cork, Ireland). Continuous variables were presented as median (range) or mean ± SD, whereas categorical variables were expressed as frequencies (%). Differences between continuous variables were assessed by the Student’s t-test, whereas differences between categorical variables were evaluated using the Pearson χ2 test. The χ2 G test “Goodness of Fit” was employed to verify whether the proportions of the four polymorphisms were distributed in controls and in patients in accordance with the Hardy-Weinberg equation. Differences in the allelic and genotypic frequencies between different groups were assessed by means of the Pearson χ2 test and calculation of odds ratios with 95% confidence intervals (CI). Haplotype reconstruction from population genotype data and inferred phased diplotype calculation for each control subject or patient with liver cirrhosis were performed by means of the ARLEQUIN integrated software package for population genetics, version 3.1[18]. Analysis of molecular variance (AMOVA) with a global and a pair-wise approach was performed to assess whether haplotype allelic content differed among groups. Locus-by-locus AMOVA was utilized to assess the statistical contribution of each polymorphism. Pair-wise differences in haplotype frequencies were assessed using the exact test for sample differentiation. Linkage disequilibrium between the four analyzed VDR polymorphisms in the studied population was determined by means of the Haploview software[19]. Stepwise logistic regression analysis with a forward approach was used to verify whether the absence or presence of specific VDR haplotypes was an independent predictor of HCC.
RESULTS
Liver histology
HCC foci were detected in the native livers from 80 (33.3%) of the patients with end-stage liver disease; 33 patients (41.2%) were HCV positive, 17 (21.2%) HBV positive, and 30 (29.1%) had alcoholic liver disease. Table 1 illustrates the distribution of the values of the main demographic and clinical variables of cirrhotic patients according to the presence of HCC. Sixteen patients (20.0%) had an Edmondson grade of one, forty-eight patients (60.0%) had a grade of two, fifteen patients (18.8%) had a grade of three and one patient (1.2%) had a grade of four. Macro- and micro-vascular invasion was only observed in five cases (6.2%).
VDR polymorphisms in liver cirrhosis and controls
FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) allele and genotype frequencies are reported in Table 2. No departure from the Hardy-Weinberg equilibrium equation was observed for each polymorphism in patients or controls. No significant difference was detected between patients and controls in allele or genotype frequencies. A strong linkage disequilibrium was detected between BsmI and TaqI (R2 = 0.92); linkage disequilibrium was also detected between ApaI and TaqI (R2 = 0.46) and between BsmI and ApaI (R2 = 0.48) (Figure 1).
Table 2.
Allelic and genotypic frequencies of the VDR FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms in controls and liver cirrhosis patients n (%)
| VDR | Control subjects (n = 236) | Liver cirrhosis (n = 240) | OR | 95% CI | P1 |
| F = 0.646 | F = 0.665 | 1 | Ref. | ||
| FokI | f = 0.354 | f = 0.335 | 1.085 | 0.831-1.417 | NS |
| B = 0.392 | B = 0.421 | 1 | Ref. | ||
| BsmI | b = 0.608 | b = 0.579 | 1.127 | 0.870-1.460 | NS |
| A = 0.570 | A = 0.590 | 1 | Ref. | ||
| ApaI | a = 0.430 | a = 0.410 | 1.084 | 0.838-1.402 | NS |
| T = 0.608 | T = 0.579 | 1 | Ref. | ||
| TaqI | t = 0.392 | t = 0.421 | 0.887 | 0.685-1.149 | NS |
| F/F = 104 (44.1) | F/F = 105 (43.8) | 1 | Ref. | ||
| FokI | F/f = 97 (41.1) | F/f = 109 (45.4) | 1.113 | 0.758-1.635 | NS |
| f/f = 35 (14.8) | f/f = 26 (10.8) | 0.736 | 0.416-1.303 | NS | |
| B/B = 41 (17.4) | B/B = 40 (16.7) | 1 | Ref. | ||
| BsmI | B/b = 103 (43.6) | B/b = 122 (50.8) | 1.214 | 0.732-2.014 | NS |
| b/b = 92 (39.0) | b/b = 78 (32.5) | 0.869 | 0.513-1.473 | NS | |
| A/A = 76 (32.2) | A/A = 80 (33.3) | 1 | Ref. | ||
| ApaI | A/a = 117 (49.6) | A/a = 123 (51.3) | 0.999 | 0.668-1.494 | NS |
| a/a = 43 (18.2) | a/a = 37 (15.4) | 0.817 | 0.477-1.400 | NS | |
| T/T = 89 (37.7) | T/T = 76 (31.7) | 1 | Ref. | ||
| TaqI | T/t = 109 (46.2) | T/t = 126 (52.5) | 1.354 | 0.909-2.017 | NS |
| t/t = 38 (16.1) | t/t = 38 (15.8) | 1.171 | 0.681-2.013 | NS |
The odds ratios were constructed with the wild type for each polymorphism as the reference. The statistical analysis was carried out using the Pearson χ2 test.
Pearson χ2 test; OR: Odds ratio; CI: Confidence interval; VDR: Vitamin D receptor.
Figure 1.
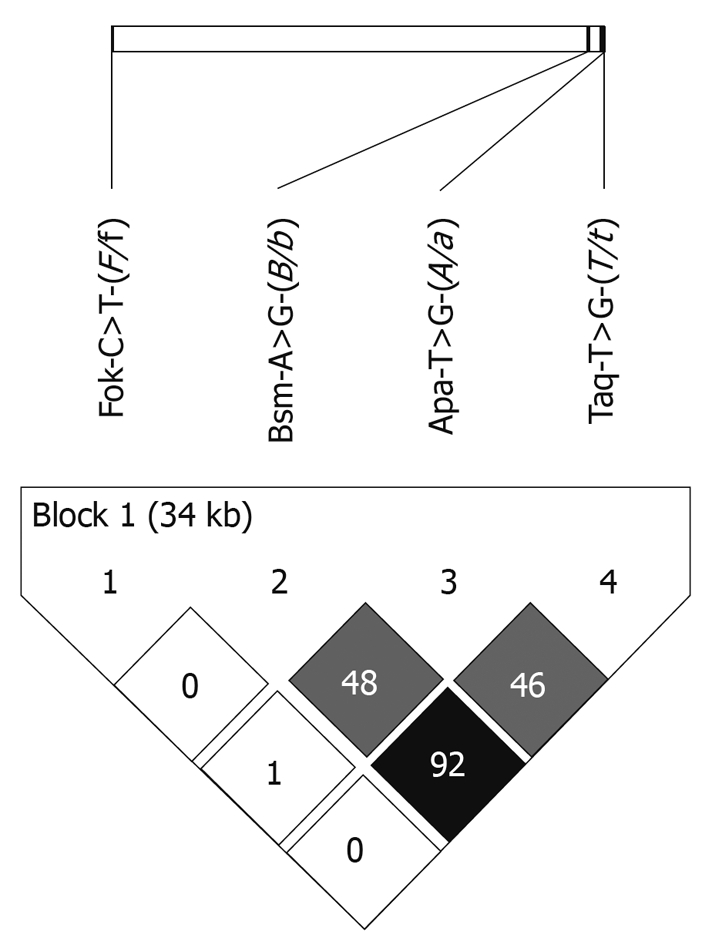
Schematic representation of linkage disequilibrium in the studied population (n = 476) between the four VDR polymorphisms: FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t). R2 for linkage disequilibrium between each marker is reported. Shades of gray are proportional to the R2 value, expressing the strength of the linkage disequilibrium.
VDR polymorphisms in patients and controls in relationship to the etiology of liver disease
AMOVA was performed by grouping the liver cirrhosis patients according to the etiology of their liver disease: patients with cirrhosis of viral origin (n = 137) and patients with cirrhosis of alcoholic origin (n = 103); a third group consisted of control subjects (n = 236). A significant difference was detected among these three populations (P < 0.05) by global AMOVA; the comparison of pairs of population samples demonstrated a significant difference between patients with cirrhosis of viral and alcoholic origin (P < 0.01). Locus-by-locus AMOVA showed the following significant differences for the single nucleotide gene polymorphisms tested: FokI C>T (F/f) P = 0.720, BsmI A>G (B/b) P = 0.047, ApaI T>G (A/a) P = 0.052 and TaqI T>C (T/t) P = 0.065. Estimated VDR haplotype frequencies of FokI C>T, BsmI A>G, ApaI T>G and TaqI T>C polymorphisms are reported in Table 3. A significant difference in haplotype frequencies was detected between patients with cirrhosis of viral and alcoholic origin.
Table 3.
Estimated VDR haplotype frequencies [FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms] in control subjects and in patients with liver cirrhosis of viral and alcoholic origin
|
Control subjects (n = 236) |
Alcoholic cirrhosis (n = 103) |
Viral cirrhosis (n = 137) |
||||||
| Haplotypes | n | Frequencies | Haplotypes | n | Frequencies | Haplotypes | n | Frequencies |
| C-A-T-C | 139 | 0.295 | C-A-T-C | 59 | 0.286 | C-A-T-C | 103 | 0.376 |
| T-G-G-T | 102 | 0.216 | T-G-G-T | 46 | 0.223 | T-G-G-T | 56 | 0.204 |
| C-G-G-T | 99 | 0.210 | C-G-G-T | 50 | 0.243 | C-G-G-T | 43 | 0.158 |
| C-G-T-T | 58 | 0.123 | C-G-T-T | 29 | 0.141 | C-G-T-T | 25 | 0.091 |
| T-A-T-C | 41 | 0.087 | T-A-T-C | 11 | 0.053 | T-A-T-C | 23 | 0.084 |
| T-G-T-T | 23 | 0.049 | T-G-T-T | 6 | 0.029 | T-G-T-T | 19 | 0.069 |
| C-A-T-T | 4 | 0.008 | C-A-T-T | 1 | 0.005 | C-A-T-T | 3 | 0.011 |
| C-G-T-C | 3 | 0.006 | C-G-T-C | 2 | 0.010 | C-G-T-C | 2 | 0.007 |
| C-G-G-C | 2 | 0.004 | C-A-G-C | 2 | 0.010 | |||
| T-A-T-T | 1 | 0.002 | ||||||
The statistical analysis was performed by means of the exact test for sample differentiation based on haplotype frequencies. Alcoholic vs viral liver cirrhosis P < 0.05.
VDR polymorphisms in liver cirrhosis with and without HCC
Table 4 illustrates the genotype frequencies of FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms in patients with liver cirrhosis grouped according to the presence (n = 80) or absence (n = 160) of HCC. Patients with HCC were more likely to carry the b/b genotype compared with the B/B + B/b genotypes of the BsmI A>G (B/b) polymorphism and the T/T genotype compared with the T/t + t/t genotypes of the TaqI T>C (T/t) polymorphism. Considering the BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) (BAT) polymorphisms, carriage of the A-T-C haplotype was associated with the absence of HCC and carriage of the G-T-T haplotype with the presence of HCC. We then grouped the patients as follows: group (a) carriers of the BAT A-T-C haplotype (n = 126), group (b) carriers of both BAT A-T-C and G-T-T haplotypes or of none of the two haplotypes (n = 75) and group (c) carriers of the BAT G-T-T haplotype (n = 39). A significantly linear trend for increasing frequencies of HCC was detected starting with group (a) (34/126, 27.0%) to group (b) (29/75, 38.7%) to group (c) (17/39, 43.6%, P < 0.05). Stepwise logistic regression analysis was performed to verify whether carriage of the BAT A-T-C and G-T-T haplotypes was a predictor of HCC independent of gender, age (≤/> 50 years), body mass index (</≥ 25 kg/m2), Child-Pugh score (≤/> 8) at the transplant operation, viral etiology of liver disease and presence of diabetes mellitus. The analysis confirmed that carriage of the BAT A-T-C and G-T-T haplotypes was a predictor of HCC occurrence (improvement of χ2 P < 0.05, OR, 1.95, 95% CI: 1.06-3.57) independent of age > 50 years (improvement of χ2 P < 0.001, OR 3.81, 95% CI: 1.75-8.29), male gender (improvement of χ2 P = 0.001, OR 4.01, 95% CI: 1.80-8.96), viral etiology of liver disease (improvement of χ2 P < 0.05, OR 2.19, 95% CI: 1.18-4.05) and body mass index ≥ 25 kg/m2 (improvement of χ2 P < 0.05, OR 1.82, 95% CI: 0.99-3.35).
Table 4.
Genotypic frequencies of the VDR FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms in liver cirrhosis patients grouped according to the presence or absence of HCC
| VDR | HCC absent (n = 160) | HCC present (n = 80) | OR | 95% CI | P1 |
| F/F = 69 (43.1) | F/F = 36 (45.0) | 1 | Ref. | ||
| FokI | F/f = 73 (45.6) | F/f = 36 (45.0) | 0.945 | 0.537-1.663 | NS |
| f/f = 18 (11.3) | f/f = 8 (10.0) | 0.852 | 0.345-2.113 | NS | |
| B/B = 28 (17.5) | B/B = 12 (15.0) | 1 | Ref. | 2 | |
| BsmI | B/b = 87 (54.4) | B/b = 35 (43.8) | 0.939 | 0.433-2.028 | NS |
| b/b = 45 (28.1) | b/b = 33 (41.2) | 1.711 | 0.766-3.813 | NS | |
| A/A = 53 (33.1) | A/A = 27 (33.8) | 1 | Ref. | ||
| ApaI | A/a = 85 (53.1) | A/a = 38 (47.5) | 0.878 | 0.483-1.595 | NS |
| a/a = 22 (13.8) | a/a = 15 (18.7) | 1.338 | 0.605-2.968 | NS | |
| T/T = 44 (27.5) | T/T = 32 (40.0) | 1 | Ref. | 3 | |
| TaqI | T/t = 88 (55.0) | T/t = 38 (47.5) | 0.594 | 0.329-1.072 | 0.08 |
| t/t = 28 (17.5) | t/t = 10 (12.5) | 0.491 | 0.212-1.141 | 0.09 |
The odds ratios were constructed with the wild type for each polymorphism as the reference. The statistical analysis was carried out using the Pearson χ2 test.
Pearson χ2 test;
B/B + B/b genotypes vs b/b genotype: P < 0.05;
T/t + t/t genotypes vs T/T genotype: P < 0.05.
VDR polymorphisms in liver cirrhosis with and without HCC in relationship to the etiology of liver disease
AMOVA was performed by grouping the patients with liver cirrhosis according to the etiology of their liver disease (viral origin n = 137 and alcoholic origin n = 103) and the presence (n = 80) or absence (n = 160) of HCC. A significant difference was detected among these four populations (P < 0.002); the comparison of pairs of population samples demonstrated significant differences between patients with liver cirrhosis of alcoholic origin with HCC vs (a) patients with liver cirrhosis of viral origin with HCC (P < 0.0001), (b) patients with liver cirrhosis of viral origin without HCC (P < 0.0001) and (c) patients with liver cirrhosis of alcoholic origin without HCC (P < 0.01). Locus-by-locus AMOVA showed the following significant differences for the single polymorphisms: FokI C>T (F/f) P = 0.923, BsmI A>G (B/b) P = 0.000, ApaI T>G (A/a) P = 0.033 and TaqI T>C (T/t) P = 0.000. Estimated VDR haplotype frequencies of FokI C>T, BsmI A>G, ApaI T>G and TaqI T>C polymorphisms are reported in Table 5. A significant difference in haplotype frequencies was detected between patients with alcoholic liver cirrhosis with HCC vs patients with viral liver cirrhosis without HCC (P < 0.01) and patients with viral liver cirrhosis with HCC (P < 0.05). Although no relationship was detected between carriage of the BAT A-T-C and G-T-T haplotypes and the presence of HCC in patients with viral liver disease, a strong association was observed between carriage of the BAT A-T-C and G-T-T haplotypes and HCC in alcoholic liver disease (27/80 vs 17/39 vs 6/18, P = NS) (7/46 vs 12/36 vs 11/21, P < 0.002).
Table 5.
Estimated VDR haplotype frequencies [FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms] in patients with liver cirrhosis of viral and alcoholic origin according to the presence of HCC
|
Alcoholic cirrhosis (n = 103) |
Viral cirrhosis (n = 137) |
||||||||||
|
HCC present (n = 30) |
HCC absent (n = 73) |
HCC present (n = 50) |
HCC absent (n = 87) |
||||||||
| Haplotypes | n | Frequencies | Haplotypes | n | Frequencies | Haplotypes | n | Frequencies | Haplotypes | n | Frequencies |
| C-A-T-C | 9 | 0.150 | C-A-T-C | 50 | 0.343 | C-A-T-C | 40 | 0.40 | C-A-T-C | 64 | 0.368 |
| T-G-G-T | 16 | 0.267 | T-G-G-T | 30 | 0.205 | T-G-G-T | 15 | 0.15 | T-G-G-T | 38 | 0.218 |
| C-G-G-T | 18 | 0.300 | C-G-G-T | 32 | 0.219 | C-G-G-T | 19 | 0.19 | C-G-G-T | 27 | 0.155 |
| C-G-T-T | 14 | 0.233 | C-G-T-T | 15 | 0.103 | C-G-T-T | 5 | 0.05 | C-G-T-T | 17 | 0.098 |
| T-A-T-C | 1 | 0.017 | T-A-T-C | 10 | 0.068 | T-A-T-C | 7 | 0.07 | T-A-T-C | 15 | 0.086 |
| T-G-T-T | 2 | 0.033 | T-G-T-T | 4 | 0.027 | T-G-T-T | 11 | 0.11 | T-G-T-T | 11 | 0.063 |
| C-A-T-T | 1 | 0.007 | C-A-T-T | 2 | 0.02 | C-G-T-C | 1 | 0.006 | |||
| C-G-T-C | 2 | 0.014 | C-G-T-C | 1 | 0.01 | T-A-T-T | 1 | 0.006 | |||
| C-G-G-C | 2 | 0.014 | |||||||||
The statistical analysis was performed by means of the exact test for sample differentiation based on haplotype frequencies. Non-viral liver cirrhosis with HCC vs (a) viral liver cirrhosis without HCC, P < 0.01, (b) viral liver cirrhosis with HCC, P < 0.05.
Vitamin D levels and VDR haplotypes in relationship to HCC occurrence
A significant linear trend was detected when patients were grouped according to gender and vitamin D serum levels (cut-off level 15 ng/mL) in relation to HCC occurrence. Group A and B comprised female patients with vitamin D serum levels ≤ 15 ng/mL and > 15 ng/mL, respectively; group C and D comprised male patients with serum levels of vitamin D ≤ 15 ng/mL and > 15 ng/mL, respectively. HCC was detected with increasing frequency starting with group A (0/12, 0.0%) to group B (2/12, 16.7%) to group C (16/38, 42.1%) to group D (26/51, 51.0%, P < 0.0005). A synergistic effect was found between vitamin D serum levels > 15 ng/mL and carriage of the BAT ATC haplotype; in these patients, HCC occurred in 5/26 (19.2%) cases compared with 39/87 (44.8%) of the remaining cases (P < 0.02).
DISCUSSION
In the present study, patients with liver cirrhosis were found to present allele and genotype frequencies of FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) polymorphisms close to those observed in control subjects. Associations between specific polymorphisms and auto-immune hepatitis and primary biliary cirrhosis have been observed in several studies, although with conflicting results. In fact, the presence of the B allele of the BsmI A>G (B/b) polymorphism was found to be associated with the occurrence of primary biliary cirrhosis in three studies involving Caucasian and Japanese populations[12-14], whereas in two other studies performed in Caucasian and Chinese populations[15,16], primary biliary cirrhosis was related to the presence of the b allele. Similarly, Vogel et al[15] found an association between chronic auto-immune hepatitis and the F allele of the FokI C>T (F/f) polymorphism, whereas the opposite results were observed by Fan et al[16]. Our study of patients affected by liver cirrhosis of viral (HBV or HCV) and alcoholic origin did not demonstrate any differences in the allele and genotype frequencies of the VDR polymorphisms between the patients and controls. The data agree with the only report in the literature on VDR polymorphisms in patients with chronic liver disease of a non-auto-immune origin. In fact, Suneetha et al[20] did not demonstrate differences in VDR polymorphism allele frequencies between controls and patients with chronic viral hepatitis due to HBV.
Some novel findings were provided by the present investigation. The first is represented by the observation of differences in the VDR polymorphisms in patients with liver cirrhosis in relationship to the etiology of their liver disease. The analysis of molecular variance found a significant difference in allele frequencies between patients with viral (HBV and HCV) and alcoholic liver cirrhosis; this was accounted for by the significant differences of the BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) (BAT) polymorphisms for which a strong linkage disequilibrium was detected. On the contrary, the FokI C>T (F/f) polymorphism was non-significantly distributed between these two groups. Consequently, haplotype analysis highlighted a different distribution between the two groups with liver cirrhosis; the BAT A-T-C haplotype was more represented in patients with viral cirrhosis (0.460%) than in those with alcoholic cirrhosis (0.339%). We are unable to provide a clear explanation of this previously unreported finding especially because the BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) (BAT) polymorphisms are located in a VDR gene region of unknown function. Two possible explanations are hypothesized: (a) the real genetic risk for liver cirrhosis might be in linkage disequilibrium with the observed haplotypes of these polymorphisms and/or (b) gene-environment interactions have a causative role.
The second novel finding of this paper is the significant association between VDR polymorphisms and the presence of HCC in patients with liver cirrhosis. HCC was found to be associated with the b allele of the BsmI A>G (B/b) polymorphism and with the T allele of the TaqI T>C (T/t) polymorphism. The BAT A-T-C haplotype was inversely related to the occurrence of HCC, whereas the BAT G-T-T haplotype was directly associated with this cancer; these associations were independent of the main demographic and clinical variables known to be strong predictors of the occurrence of HCC. VDR polymorphisms have been explored in cancers of epithelial origin, such as breast, ovarian, prostate, lung and skin cancers[20,21]. Even though some studies did not detect associations between the VDR polymorphisms and these diseases, e.g. Gsur et al[23] in prostate cancer and Dunning et al[24] in breast cancer, the majority of the authors found a significant association between the VDR polymorphisms and cancer[8-10,20]. In particular, carriage of the B BsmI A>G (B/b) allele and of the t TaqI T>C (T/t) allele has been described to exert a protective effect in prostate cancer[24], malignant melanoma[25,26] and breast cancer[27]; these results support our findings in the present series on HCC in liver cirrhosis.
The third observation that may be derived from these data concerns the strong interaction between the presence of HCC and the etiology of liver cirrhosis in relationship to VDR polymorphisms. Both the analysis of molecular variance and the estimated haplotype frequencies highlighted the different behaviors that were detected in the VDR polymorphisms in alcoholic patients with HCC, patients with alcoholic cirrhosis without HCC and patients with liver cirrhosis of viral origin complicated or uncomplicated by the presence of HCC. Locus-by-locus analysis demonstrated that the major contribution was provided by the BsmI A>G (B/b) and TaqI T>C (T/t) polymorphisms; the BAT A-T-C haplotype was strongly protective, whereas the G-T-T haplotype was associated with HCC in patients with alcoholic cirrhosis but not in patients with liver cirrhosis of viral origin.
Besides the classic action involving calcium and phosphate homeostasis, vitamin D possesses non-classic actions also known to be mediated through the VDR[28]. First, vitamin D exerts immune modulation activity either by stimulating innate immune function[29] and/or by inhibiting hyper-activity of adaptive immunity[30]. More closely related to the subject of the present study, vitamin D has been extensively evaluated for its potential anticancer activity in animal and cell studies. It has been suggested that the anticancer activity of vitamin D is due to its anti-proliferative and pro-differentiating action in most cell types[31]. From an epidemiological point of view, there is some evidence that low vitamin levels are associated with a higher incidence of cancer[32] even though there is no known beneficial effect of treating cancers with vitamin D[33].
Patients with liver cirrhosis are known to be at high risk for vitamin D deficiency in direct proportion to the severity of their chronic liver disease[34,35]. This observation applies particularly to cirrhotic patients with end-stage liver cirrhosis who are subjected to liver transplantation[36], such as those investigated in the present study. In contrast, chronic alcohol abuse is a factor that can interfere in multiple ways with vitamin D metabolism either through malnutrition with an assumption of reduced nutrient intake and/or reduced exposure to sunlight. Reduced vitamin D levels have been detected in the majority of alcoholic patients without chronic liver disease[37]. It is conceivable, therefore, that in patients with liver cirrhosis, the synergistic action of severely reduced serum vitamin D levels and of a specific VDR haplotype facilitates HCC development. In fact, supporting this hypothesis, we found vitamin D insufficiency (≤ 15 ng/mL) in a large proportion (55.8%) of our patients with end-stage chronic liver disease; moreover, a gender-adjusted association between vitamin D insufficiency and the occurrence of HCC was also detected. Finally, HCC foci were observed less frequently in the native livers of patients carrying the protective BAT A-T-C haplotype and simultaneously possessing serum vitamin D levels > 15 ng/mL.
In conclusion, VDR genetic polymorphisms are significantly associated with the occurrence of HCC in patients with liver cirrhosis. This relationship is more specific for patients with an alcoholic etiology.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer-related death worldwide. Liver cirrhosis due to alcohol consumption or to chronic infection by hepatitis C virus (HCV) and hepatitis B virus (HBV) are considered the main etiologic agents for the development of HCC. The differences in the incidence rates and the strong gender distribution of HCC are probably not entirely due to differences in exposure to the three causative agents mentioned above. Recently, several genetic factors regarding gene polymorphisms of inflammatory cytokines and growth factors have been considered.
Research frontiers
The vitamin D receptor (VDR) is a member of the nuclear receptor superfamily of ligand- inducible transcription factors which are involved in cell growth and differentiation. Several single nucleotide restriction length polymorphisms have been described in the VDR gene in association with cancers of the breast, prostate, colon, bladder and kidney. There are no data in the literature concerning the prevalence of VDR polymorphisms in HCC of different etiologies.
Innovations and breakthroughs
This is the first study to examine the potential role of VDR genetic polymorphisms in the occurrence of HCC in humans. In this study, the authors investigate the VDR single nucleotide gene polymorphisms FokI C>T (F/f), BsmI A>G (B/b), ApaI T>G (A/a) and TaqI T>C (T/t) (BAT) in a large series of patients who underwent liver transplantation due to liver cirrhosis with or without HCC. They demonstrate that carriage of the b/b genotype of BsmI and the T/T genotype of TaqI was significantly associated with HCC, whereas carriage of the BAT A-T-C and G-T-T haplotypes was significantly more prevalent in patients with alcoholic liver disease and HCC.
Applications
The characterization of VDR genetic polymorphisms in patients with liver cirrhosis could help to identify those who are at high risk of developing HCC. This observation could be used to modify the strategy of periodical ultrasound surveillance in this category of patients.
Peer review
In this paper, the relationships between VDR gene polymorphisms and the presence of HCC have been evaluated in patients with viral or alcoholic cirrhosis. The most important result of this study is the observation that some VDR genetic polymorphisms are associated with the occurrence of HCC in patients with liver cirrhosis. The results are convincing, well presented and of some interest.
Footnotes
Supported by Grants from the Ricerca Sanitaria Finalizzata Program, Regione Piemonte, Italy
Peer reviewer: Francesco Feo, Professor, Department of Biomedical Sciences, Section of Experimental Pathology and Oncology, University of Sassari, Via P, Manzella 4, 07100 Sassari, Italy
S- Editor Wang JL L- Editor Webster JR E- Editor Ma WH
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- 5.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez-Martínez R, Zambrano A, Castillo AI, Aranda A. Vitamin D-dependent recruitment of corepressors to vitamin D/retinoid X receptor heterodimers. Mol Cell Biol. 2008;28:3817–3829. doi: 10.1128/MCB.01909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Valdivielso JM, Fernandez E. Vitamin D receptor polymorphisms and diseases. Clin Chim Acta. 2006;371:1–12. doi: 10.1016/j.cca.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Tang C, Chen N, Wu M, Yuan H, Du Y. Fok1 polymorphism of vitamin D receptor gene contributes to breast cancer susceptibility: a meta-analysis. Breast Cancer Res Treat. 2009;117:391–399. doi: 10.1007/s10549-008-0262-4. [DOI] [PubMed] [Google Scholar]
- 10.Raimondi S, Johansson H, Maisonneuve P, Gandini S. Review and meta-analysis on vitamin D receptor polymorphisms and cancer risk. Carcinogenesis. 2009;30:1170–1180. doi: 10.1093/carcin/bgp103. [DOI] [PubMed] [Google Scholar]
- 11.Köstner K, Denzer N, Müller CS, Klein R, Tilgen W, Reichrath J. The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: a review of the literature. Anticancer Res. 2009;29:3511–3536. [PubMed] [Google Scholar]
- 12.Halmos B, Szalay F, Cserniczky T, Nemesanszky E, Lakatos P, Barlage S, Schmitz G, Romics L, Csaszar A. Association of primary biliary cirrhosis with vitamin D receptor BsmI genotype polymorphism in a Hungarian population. Dig Dis Sci. 2000;45:1091–1095. doi: 10.1023/a:1005581414918. [DOI] [PubMed] [Google Scholar]
- 13.Lakatos LP, Bajnok E, Hegedus D, Tóth T, Lakatos P, Szalay F. Vitamin D receptor, oestrogen receptor-alpha gene and interleukin-1 receptor antagonist gene polymorphisms in Hungarian patients with primary biliary cirrhosis. Eur J Gastroenterol Hepatol. 2002;14:733–740. doi: 10.1097/00042737-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka A, Nezu S, Uegaki S, Kikuchi K, Shibuya A, Miyakawa H, Takahashi S, Bianchi I, Zermiani P, Podda M, et al. Vitamin D receptor polymorphisms are associated with increased susceptibility to primary biliary cirrhosis in Japanese and Italian populations. J Hepatol. 2009;50:1202–1209. doi: 10.1016/j.jhep.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Vogel A, Strassburg CP, Manns MP. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35:126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 16.Fan L, Tu X, Zhu Y, Zhou L, Pfeiffer T, Feltens R, Stoecker W, Zhong R. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J Gastroenterol Hepatol. 2005;20:249–255. doi: 10.1111/j.1440-1746.2005.03532.x. [DOI] [PubMed] [Google Scholar]
- 17.Scientific Advisory Committee on Nutrition. Update on vitamin D. Position Statement by the Scientific Advisory Committee on Nutrition. London: The Stationery Office; 2007. pp. 7–9. [Google Scholar]
- 18.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856–863. doi: 10.1016/j.jhep.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Yin M, Wei S, Wei Q. Vitamin D Receptor Genetic Polymorphisms and Prostate Cancer Risk: A Meta-analysis of 36 Published Studies. Int J Clin Exp Med. 2009;2:159–175. [PMC free article] [PubMed] [Google Scholar]
- 22.Zmuda JM, Cauley JA, Ferrell RE. Molecular epidemiology of vitamin D receptor gene variants. Epidemiol Rev. 2000;22:203–217. doi: 10.1093/oxfordjournals.epirev.a018033. [DOI] [PubMed] [Google Scholar]
- 23.Gsur A, Feik E, Madersbacher S. Genetic polymorphisms and prostate cancer risk. World J Urol. 2004;21:414–423. doi: 10.1007/s00345-003-0378-4. [DOI] [PubMed] [Google Scholar]
- 24.Dunning AM, McBride S, Gregory J, Durocher F, Foster NA, Healey CS, Smith N, Pharoah PD, Luben RN, Easton DF, et al. No association between androgen or vitamin D receptor gene polymorphisms and risk of breast cancer. Carcinogenesis. 1999;20:2131–2135. doi: 10.1093/carcin/20.11.2131. [DOI] [PubMed] [Google Scholar]
- 25.Mocellin S, Nitti D. Vitamin D receptor polymorphisms and the risk of cutaneous melanoma: a systematic review and meta-analysis. Cancer. 2008;113:2398–2407. doi: 10.1002/cncr.23867. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Liu Z, Wang LE, Gershenwald JE, Lee JE, Prieto VG, Duvic M, Grimm EA, Wei Q. Haplotype and genotypes of the VDR gene and cutaneous melanoma risk in non-Hispanic whites in Texas: a case-control study. Int J Cancer. 2008;122:2077–2084. doi: 10.1002/ijc.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barroso E, Fernandez LP, Milne RL, Pita G, Sendagorta E, Floristan U, Feito M, Aviles JA, Martin-Gonzalez M, Arias JI, et al. Genetic analysis of the vitamin D receptor gene in two epithelial cancers: melanoma and breast cancer case-control studies. BMC Cancer. 2008;8:385. doi: 10.1186/1471-2407-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikle D. Nonclassic actions of vitamin D. J Clin Endocrinol Metab. 2009;94:26–34. doi: 10.1210/jc.2008-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ustianowski A, Shaffer R, Collin S, Wilkinson RJ, Davidson RN. Prevalence and associations of vitamin D deficiency in foreign-born persons with tuberculosis in London. J Infect. 2005;50:432–437. doi: 10.1016/j.jinf.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Penna G, Adorini L. 1 Alpha,25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J Immunol. 2000;164:2405–2411. doi: 10.4049/jimmunol.164.5.2405. [DOI] [PubMed] [Google Scholar]
- 31.Fleet JC. Molecular actions of vitamin D contributing to cancer prevention. Mol Aspects Med. 2008;29:388–396. doi: 10.1016/j.mam.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 33.Beer TM, Ryan CW, Venner PM, Petrylak DP, Chatta GS, Ruether JD, Redfern CH, Fehrenbacher L, Saleh MN, Waterhouse DM, et al. Double-blinded randomized study of high-dose calcitriol plus docetaxel compared with placebo plus docetaxel in androgen-independent prostate cancer: a report from the ASCENT Investigators. J Clin Oncol. 2007;25:669–674. doi: 10.1200/JCO.2006.06.8197. [DOI] [PubMed] [Google Scholar]
- 34.Monegal A, Navasa M, Guañabens N, Peris P, Pons F, Martinez de Osaba MJ, Rimola A, Rodés J, Muñoz-Gómez J. Osteoporosis and bone mineral metabolism disorders in cirrhotic patients referred for orthotopic liver transplantation. Calcif Tissue Int. 1997;60:148–154. doi: 10.1007/s002239900205. [DOI] [PubMed] [Google Scholar]
- 35.Fisher L, Fisher A. Vitamin D and parathyroid hormone in outpatients with noncholestatic chronic liver disease. Clin Gastroenterol Hepatol. 2007;5:513–520. doi: 10.1016/j.cgh.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Stein EM, Cohen A, Freeby M, Rogers H, Kokolus S, Scott V, Mancini D, Restaino S, Brown R, McMahon DJ, et al. Severe vitamin D deficiency among heart and liver transplant recipients. Clin Transplant. 2009;23:861–865. doi: 10.1111/j.1399-0012.2009.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik P, Gasser RW, Kemmler G, Moncayo R, Finkenstedt G, Kurz M, Fleischhacker WW. Low bone mineral density and impaired bone metabolism in young alcoholic patients without liver cirrhosis: a cross-sectional study. Alcohol Clin Exp Res. 2009;33:375–381. doi: 10.1111/j.1530-0277.2008.00847.x. [DOI] [PubMed] [Google Scholar]


