Abstract
Hypertension is a complex trait that has been studied extensively for genetic contributions of the nuclear genome. We examined mitochondrial genomes of the hypertensive strains: the Dahl Salt-Sensitive (S) rat, the Spontaneously Hypertensive Rat (SHR), and the Albino Surgery (AS) rat, and the relatively normotensive strains: the Dahl Salt-Resistant (R) rat, the Milan Normotensive Strain (MNS), and the Lewis rat (LEW). These strains were used previously for linkage analysis for blood pressure (BP) in our laboratory. The results provide evidence to suggest that variations in the mitochondrial genome do not account for observed differences in blood pressure between the S and R rats. However, variants were detected among the mitochondrial genomes of the various hypertensive strains, S, SHR, and AS, and also among the normotensive strains R, MNS, and LEW. A total of 115, 114, 106, 106, and 16 variations in mtDNA were observed between the comparisons S versus LEW, S versus MNS, S versus SHR, S versus AS, and SHR versus AS, respectively. Among the 13 genes coding for proteins of the electron transport chain, 8 genes had nonsynonymous variations between S, LEW, MNS, SHR, and AS. The lack of any sequence variants between the mitochondrial genomes of S and R rats provides conclusive evidence that divergence in blood pressure between these two inbred strains is exclusively programmed through their nuclear genomes. The variations detected among the various hypertensive strains provides the basis to construct conplastic strains and further evaluate the effects of these variants on hypertension and associated phenotypes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-010-9259-5) contains supplementary material, which is available to authorized users.
Introduction
Hypertension is a complex trait and an important risk factor for common cardiovascular diseases such as myocardial infarction, stroke, renal failure, and congestive heart failure. Changes in mitochondrial functionality are observed in parallel with progression of hypertension (Chan et al. 2009; Thomas et al. 2008; Zimmerman and Zucker 2009), which suggests that variations in the mitochondrial genome could account for differences in blood pressure. Studies on the genetic contribution to hypertension in both humans and animal models are, however, largely reported on the nuclear genome (Aneas et al. 2009; Cicila et al. 2009; Cowley 2006; Cowley et al. 2004; Deng 2007; Gilibert et al. 2009; Graham et al. 2007; Joe and Garrett 2005, 2006; Joe et al. 2009; Lee et al. 2007; Mattson et al. 2007, 2008; Moreno et al. 2003a, 2007; Saad et al. 2007a, b, 2008; Stoll et al. 2001; Toland et al. 2008a, b) with some exceptions such as recent reports of mitochondrial tRNA mutations observed in a Chinese population with essential hypertension (Li et al. 2009; Liu et al. 2009). Confirming these association studies in humans poses a significant problem that could be readily resolved using homozygous genomes of mammalian models of hypertension. Inbred rat and mouse strains with variations in their mitochondrial genomic sequences serve as good substrates for construction of conplastic strains as models for examining genetic contributions of the mitochondrial genome. Such limited experiments in mice and a single study in rats have reported that several complex traits are controlled by genetic elements of the mitochondrial genome (Chen et al. 2008b, c; Gregorova et al. 2008; Gusdon et al. 2007; Pravenec et al. 2007; Yu et al. 2009a, b). However, there are no reports of any studies on the mitochondrial genomic effects on blood pressure.
The goal of this report was to identify inbred rat models widely used in hypertension research as strains that can also be exploited to understand the contributions of the mitochondrial genome to the onset and progression of hypertension. Specifically, the entire mitochondrial genomes of the two selectively bred divergent models of blood pressure, the Dahl Salt-Sensitive (S) rats (also referred to as S/Jr) and the Dahl Salt-Resistant (R) rats (also referred to as R/Jr), the definitive colonies of which are maintained at the University of Toledo College of Medicine (formerly the Medical College of Ohio), two other genetically hypertensive strains, i.e., the Spontaneously Hypertensive Rat (SHR) and the Albino Surgery (AS) rat, and two normotensive strains Milan normotensive strain (MNS) and Lewis (LEW) which were previously used as parental strains for BP linkage analysis (Deng and Rapp 1995; Dukhanina et al. 1997; Garrett et al. 1998) were determined. The results obtained point to mitochondrial genomes of the S or R rat as not being contrasting, but point to comparisons between S and the hypertensive strains SHR or AS or to comparisons between S and the normotensive strains MNS and LEW as appropriate contrasting strains to study the genetic contributions of the mitochondrial genome.
In addition, during the course of our study, errors in public databases of the rat mitochondrial genome are identified and documented as a valuable reference to future investigators of the rat mitochondrial genome.
Materials and methods
Animals
All animal experiments were conducted as per preapproved protocols by the Institutional Animal Care and Use Committee (IACUC) of the University of Toledo College Of Medicine (UTCOM). All rats were between 2 and 3 months of age and maintained on a low-salt (0.4% NaCl) diet. Dahl Salt-Sensitive (S) rats and Dahl Salt-Resistant (R) rats were from our colony maintained at UTCOM. The Spontaneously Hypertensive Rat strain (SHR/Hsd/Mco), referred to as SHR, was originally obtained from Harlan Sprague-Dawley (Indianapolis, IN) and maintained in our colony. Albino Surgery (AS) rats were obtained previously (Garrett et al. 2002) from the National Institute for Medical Research (Mill Hill, UK). The Lewis (LEW/NCrlBR) rats referred to as LEW, were originally obtained from Charles River Laboratories (Wilmington, MA). The Milan Normotensive Strain (MNS) was originally obtained from Veterinary Resources Branch, National Institutes of Health (Bethesda, MD). Both LEW and MNS were maintained in our animal facility.
Isolation of DNA and sequencing
DNA was extracted from tail biopsies using the Promega Wizard kit (Promega, Madison, WI). Polymerase chain reaction (PCR) was carried out using 23 pairs of primers to amplify the entire mitochondrial genome. Each set of primers was designed using the primer 3 program (http://frodo.wi.mit.edu/primer3/) and attached with M13 tags to amplify PCR products that were approximately 500-1000 bp. A list of primer sequences is provided in Supplementary Table 1. These custom primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Amplified products were subsequently purified using the Qiagen PCR purification kit (Qiagen, Valencia, CA). The purified products were sequenced by MWG DNA Sequencing Services (High Point, NC) and analyzed using the software Sequencher v4.9 (Gene Codes Corp., Ann Arbor, MI). All mitochondrial DNA sequences of the S, R, SHR, and AS rats from the current study were deposited in GenBank (Accession Nos. GU997608—SS/Jr, GU997611—SR/Jr, GU997609—AS, GU997610—SHR, HM152027—LEW, HM152028—MNS).
Results
Comparisons of mitochondrial genome sizes
Mitochondrial DNA of the three hypertensive strains studied differed in their overall sizes (Table 1). The mitochondrial DNA of S and R rats, which were both derived from Sprague-Dawley rats, were the shortest (among the strains tested), with 16,309 bp. The strains derived from Wistar rats, i.e., SHR, MNS, LEW, and AS, had mitochondrial genomes longer than that of the strains derived from the Sprague-Dawley rats and ranged from 16,312 to 16,316 bp (Table 1).
Table 1.
Comparisons of total mitochondrial genome sizes
| Strain | mtDNA size (bp) |
|---|---|
| Brown Norway (BN) | 16,313 |
| Dahl Salt-Sensitive (SS/Jr) | 16,309 |
| Dahl Salt-Resistant (SR/Jr) | 16,309 |
| Spontaneously Hypertensive Rat (SHR) | 16,312 |
| Albino Surgery (AS) | 16,314 |
| Lewis (LEW) | 16,316 |
| Milan Normotensive Strain (MNS) | 16,316 |
Strainwise comparisons of mitochondrial variations
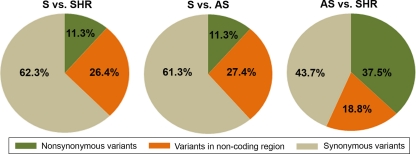
All sequence data obtained from the current study were compared with the reference BN sequence (AC_000022.1). Note that the sequence data obtained from the mtDNA of both the hypertensive Dahl S and the relatively normotensive R rats were identical. Both MNS and LEW mtDNA sequences were identical except for a synonymous variation at position 8844 (position number compared to the BN as reference sequence) where a guanine in LEW was substituted by an adenosine in MNS. Variations were observed among all the hypertensive strains compared. Specifically, 106 variations in mtDNA were observed between two of the most commonly used hypertensive rat models, S and SHR. Of these, 11.3% were nonsynonymous variants, 26.4% were within noncoding RNA gene sequences, and 62.3% were synonymous variants (Fig. 1). Likewise, the mtDNA sequence of the hypertensive AS rat was compared with that of the S rat. A total of 106 variants were detected between S and AS. Among these, 11.3% were in the gene-coding regions, 27.4% were within noncoding RNA gene sequences, and 61.3% were synonymous variants (Fig. 1). There were only 16 variants between SHR and AS mtDNA sequences. Of these, 37.5% were within gene-coding sequences, 18.8% were within noncoding RNA sequences, and 43.7% were synonymous variants (Fig. 1).
Fig. 1.
Overall strainwise comparisons of mitochondrial genomic variations. Sequences obtained from all strains were compared with S as the reference sequence
Nonsynonymous variants
Among the 13 protein-coding genes of the mtDNA, nonsynonymous variations were observed between S, SHR, and AS in eight genes (Table 2). The SHR and AS forms of NADH-coenzyme Q oxidoreductase gene subunit 2 (mt-Nd2) were conserved except at nucleotide position 3956, resulting in substitution of Ala/Val18 in SHR/AS. The mt-Nd2 sequence of the S rat differs from both the SHR and AS rats resulting in five and four amino acid substitutions, respectively. Prominent among these substitutions is the deletion of a histidine residue (His316) in the S rat compared to SHR and AS. The amino acid sequences of the other subunits, mt-Nd3 and mt-Nd5, are identical between S and SHR, which differs from that of the AS rat. Isoleucine9 of mt-Nd3 in S and SHR is replaced by Thr9 in AS rats. Threonine37 of mt-Nd5 in S and SHR is replaced by Ala37 in AS rats.
Table 2.
Mitochondrial DNA variants predicted to cause amino acid substitutions in rat models used in genetic analysis of hypertension
| Gene | mtDNA location (bp) | Triplet codon | Amino acid | Amino acid No. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BNa | S/R | SHR | AS | MNS | LEW | S/R | SHR | AS | MNS | LEW | ||
| mt-Nd2 | 3956 | GTA (3956) | GCA (3956) | GTA (3958) | GTA (3960) | GTA (3960) | Val | Ala | Val | Val | Val | 18 |
| mt-Nd2 | 4352 | AAC (4352) | AGC (4352) | AGC (4354) | AGC (4356) | AGC (4356) | Asn | Ser | Ser | Ser | Ser | 150 |
| mt-Nd2 | 4696 | GCA (4696) | ACA (4696) | ACA (4698) | ACA (4700) | ACA (4700) | Ala | Thr | Thr | Thr | Thr | 265 |
| mt-Nd2 | 4814 | ACA (4814) | ATA (4814) | ATA (4816) | ATA (4818) | ATA (4818) | Thr | Met | Met | Met | Met | 304 |
| mt-Nd2 | 4855 | : (4854.1) | CAC (4855) | CAC (4857) | CAC (4859) | CAC (4859) | : | His | His | His | His | 316 |
| mt-Cox2 | 7498 | ATC (7494) | GTC (7497) | GTC (7499) | GTC (7501) | GTC (7501) | Ile | Val | Val | Val | Val | 165 |
| mt-Atp6 | 8021 | GAA (8017) | AAA (8020) | AAA (8022) | AAA (8024) | AAA (8024) | Glu | Lys | Lys | Lys | Lys | 35 |
| mt-Nd3 | 9476 | ATT (9472) | ATT (9475) | ACT (9477) | ATT (9479) | ATT (9479) | Ile | Ile | Thr | Ile | Ile | 9 |
| mt-Nd4 | 10227 | ATC (10223) | ACC (10226) | ACC (10228) | ACC (10230) | ACC (10230) | Ile | Thr | Thr | Thr | Thr | 23 |
| mt-Nd4 | 11225 | GCA (11221) | ACA (11224) | GCA (11226) | GCA (11228) | GCA (11228) | Ala | Thr | Ala | Ala | Ala | 356 |
| mt-Nd4 | 11360 | GTC (11356) | ATC (11359) | GTC (11361) | GTC (11363) | GTC (11363) | Val | Ile | Val | Val | Val | 401 |
| mt-Nd4 | 11415 | CTA (11411) | CTA (11414) | CCA (11416) | CCA (11418) | CCA (11418) | Leu | Leu | Pro | Pro | Pro | 419 |
| mt-Nd5 | 11844 | ACT (11840) | ACT (11843) | GCT (11845) | GCT (11847) | GCT (11847) | Thr | Thr | Ala | Ala | Ala | 37 |
| mt-Nd6 | 13647 | ATT (13643) | GTT (13646) | GTT (13648) | GTT (13650) | GTT (13650) | Ile | Val | Val | Val | Val | 139 |
| mt-Cytb | 14775 | GAC (14771) | AAC (14774) | AAC (14776) | AAC (14778) | AAC (14778) | Asp | Asn | Asn | Asn | Asn | 214 |
amtDNA sequence in NCBI (GenBank accession No. AC-000022.1or AY172581; http://www.ncbi.nlm.nih.gov/nuccore/26983975?report=genbank). Data presented was collected by setting BN as the reference sequence and aligning all strain sequences in a single contig using the Sequencher software v4.9
Four nonsynonymous variants were detected within the mt-Nd4 gene. Three of these variants resulted in Ile/Thr23, Ala/Thr356, and Val/Ile401 substitutions between S and SHR (Table 2). A significant substitution at amino acid 419 from leucine in both S and SHR to proline in the AS was noted (Table 2). Gene sequence for mt-Nd6 was conserved between SHR and AS, with only one amino acid substitution from Val/Ile139 observed in S rats. All the other genes were also conserved between SHR and AS. The S, however, differed from both the SHR and AS with three nonsynonymous substitutions in each of the genes coding for cytochrome c-oxidase subunit-2 (mt-Cox-2), F0-F1-ATP synthase (mt-Atp-6), and the subunit of ubiquinol cytochrome-c-reductase (mt-Cyt b) (Table 2).
The protein-coding sequences of the two normotensive strains MNS and LEW are similar to that of the AS except for one substitution within the mt-Nd3 gene (Table 2).
Variations in genes for ribosomal and transfer RNA
When compared with the other genetically hypertensive and normotensive strains, S and R rat mitochondrial DNA coding for 12s rRNA differs from that of SHR, AS, MNS, and LEW at two positions, 935 and 942 bp (Table 3). The S rat 16s rRNA sequence is also considerably different from that of the SHR (13 variants) and AS (15 variants) (Table 3). Both of the normotensive strains MNS and LEW were identical in their 16s rRNA sequences but were different from that of the S (17 variants) (Table 3).
Table 3.
Variations in mitochondrial genes for ribosomal and transfer RNA in inbred rat strains
| Gene | Nucleotide location | Single nucleotide polymorphism | ||||
|---|---|---|---|---|---|---|
| BNa | S/Rb | SHR | AS | MNS | LEW | |
| rRNA (12-s) | 935 | A (935) | G (935) | G (935) | G (935) | G (935) |
| rRNA (12-s) | 942 | C (942) | T (942) | T (942) | T 942) | T (942) |
| rRNA (16-s) | 1130 | A (1130) | C (1130) | C (1130) | C (1130) | C (1130) |
| rRNA (16-s) | 1136 | : (1135.1) | C (1136) | C (1136) | C (1136) | C (1136) |
| rRNA (16-s) | 1136.1 | : (1135.2) | C (1137) | C (1137) | C (1137) | C (1137) |
| rRNA (16-s) | 1136.2 | : (1135.3) | : (1137.1) | C (1138) | C (1138) | C (1138) |
| rRNA (16-s) | 1136.3 | : (1135.4) | : (1137.2) | C (1139) | C (1139) | C (1139) |
| rRNA (16-s) | 1136.4 | : (1135.5) | : (1137.3) | : (1139.1) | C (1140) | C (1140) |
| rRNA (16-s) | 1136.5 | : (1135.6) | : (1137.4) | : (1139.2) | C (1141) | C (1141) |
| rRNA (16-s) | 1137 | A (1136) | C (1138) | C (1140) | C (1142) | C (1142) |
| rRNA (16-s) | 1223 | A (1222) | G (1224) | G (1226) | G (1228) | G (1228) |
| rRNA (16-s) | 1248 | C (1247) | T (1249) | T (1251) | T (1253) | T (1253) |
| rRNA (16-s) | 1521 | G (1520) | A (1522) | A (1524) | A (1526) | A (1526) |
| rRNA (16-s) | 1585 | T (1584) | C (1586) | C (1588) | C (1590) | C (1590) |
| rRNA (16-s) | 1653.1 | A (1653) | : (1654.1) | : (1656.1) | : (1658.1) | : (1658.1) |
| rRNA (16-s) | 1653.2 | C (1654) | : (1654.2) | : (1656.2) | : (1658.2) | : (1658.2) |
| rRNA (16-s) | 1716 | T (1717) | C (1717) | C (1719) | C (1721) | C (1721) |
| rRNA (16-s) | 1832 | G (1833) | A (1833) | A (1835) | A (1837) | A (1837) |
| rRNA (16-s) | 2170 | T (2171) | C (2171) | C (2173) | C (2175) | C (2175) |
| tRNA (Cys) | 5200 | A (5196) | G (5199) | G (5201) | G (5203) | G (5203) |
| tRNA (Cys) | 5202 | A (5198) | G (5201) | G (5203) | G (5205) | G (5205) |
| tRNA (Cys) | 5237 | T (5233) | A (5236) | A (5238) | A (5240) | A (5240) |
| tRNA (Tyr) | 5269 | G (5265) | C (5268) | C (5270) | C (5272) | C (5272) |
| tRNA (Asp) | 6978 | G (6974) | A (6977) | A (6979) | A (6981) | A (6981) |
| tRNA (Thr) | 15333 | G (15329) | A (15332) | A (15334) | A (15336) | A (15336) |
| D-Loop region | 15460 | C (15456) | T (15459) | C (15461) | C (15463) | C (15463) |
| D-Loop region | 15549 | C (15545) | T (15548) | T (15550) | T (15552) | T (15552) |
| D-Loop region | 15589 | A (15585) | G (15588) | G (15590) | G (15592) | G (15592) |
| D-Loop region | 16313 | A (16309) | G (16312) | G (16314) | G (16316) | G (16316) |
amtDNA sequence in NCBI (GenBank accession No. AC-000022.1or AY172581; http://www.ncbi.nlm.nih.gov/nuccore/26983975?report=genbank)
bS and R rat mtDNA sequences are identical
Genes for tRNA are conserved among S, SHR, and AS, except for tRNA for four amino acids that were polymorphic in the S compared with SHR and AS. Three variations were detected within the tRNA for cysteine and one each for tRNA sequences of tyrosine, aspartic acid, and threonine (Table 3). Within the functionally important D-loop region of mtDNA, four variants were noted between S and SHR at positions 15,460, 15,549, 15,589 and 16,313 (Table 3). The AS sequence was identical to the S at 15,460 but identical to the SHR at 15,549, 15,589, and 16,313 (Table 3). MNS and LEW sequences for all tRNAs are identical to that of the SHR and AS.
Comparisons with published hypertensive rat mitochondrial genomic sequences
There are two other published mtDNA sequences of different stocks of S rats used in hypertensive research, i.e., that of SSMCW (Schlick et al. 2006) and that of DS (Chen et al. 2008a). A single variant was detected between S and the reported mtDNA sequence of SSMCW (C to A, respectively, DQ673914) at 11,334 bp of the S mitochondrial genome [the location corresponds to 11,338 bp in the reference BN mtDNA (GenBank accession No. AC_000022.1)]. However, there were seven variants between the mtDNA sequences of S and the published DS (Table 4). Besides the S rat, the sequence of the SHR obtained was compared with the available mtDNA sequence of SHR/OlaIpcv (Pravenec et al. 2007). There are no variants between SHR sequenced in our laboratory and the published SHR mtDNA (Pravenec et al. 2007).
Table 4.
Mitochondrial DNA variations between different stocks of Dahl salt-sensitive rats
| S (S/Jr) | SSMCW | DS(Iwai) | BNa mtDNA location (bp) |
|---|---|---|---|
| A | A | DEL | 1,130 |
| DEL | DEL | A | 1,131 |
| A | A | T | 1,209 |
| C | C | DEL | 4,848 |
| C | C | DEL | 4,849 |
| A | A | DEL | 4,850 |
| T | T | C | 11,799 |
| C | A | Unknown | 11,334 |
S = sequence data presented in the current report from the original stock of Dahl salt-sensitive (S) rat at the University of Toledo (formerly Medical College of Ohio); SSMCW = sequence data from the Dahl salt-sensitive rat at the Medical College of Wisconsin (Schlick et al. 2006); DS(Iwai) = Dahl salt-sensitive rat data from Chen et al. (2008a); DEL = deletion
aPositions obtained from the mtDNA sequence in NCBI (GenBank accession No. AC-000022.1 or AY172581, http://www.ncbi.nlm.nih.gov/nuccore/26983975?report=genbank)
Finally, a short note on an inconsistency that we have resolved for future reference: The GenBank accession number of the BN (BN/SsNHsdMCW) strain is cited in previous references (Chen et al. 2008a; Pravenec et al. 2007; Schlick et al. 2006) as AC_000022. However, the current record (November 2009) of AC_000022 in GenBank is that of the Wistar rat, which when compared to the BN rat mtDNA (BN/SsNHsdMCW) harbors 330 variants (data not shown). The correct accession numbers and their historical derivatives of these accession numbers is presented clearly in Table 5 to serve as a future reference for investigators.
Table 5.
Clarification of GenBank accession numbers of rat mtDNA
| Strain | GenBank accession no. | mtDNA length (bp) | ||
|---|---|---|---|---|
| Current | Alternative | Origin of sequence | ||
| BN (BN/SsNHsdMCW) | AC_000022.2 | AY172581 | GI:110189662 | 16,313 |
| Wistar | AC_000022.1 | AC_000022 | X14848 | 16,300 |
Discussion
Inbred rat strains previously have been used extensively in our laboratory to study the contributions of their nuclear genomes to hypertension (Joe and Garrett 2005, 2006; Joe et al. 2009; Saad et al. 2007a, b, 2008; Toland et al. 2008b). The purpose of this study was to define combinations of these rats that can be exploited to study the genetic contributions of the mitochondrial genome to hypertension. The lack of any sequence variants between the mitochondrial genomes of S and R rats provides conclusive evidence that divergence in blood pressure between these two inbred strains is programmed exclusively through their nuclear genomes. The S and R rats originally were bred selectively from an outbred stock of Sprague-Dawley rats for differences in blood pressure in response to a salt diet. Because the mitochondrial genomes are maternally inherited, the identical mitochondrial genomic sequence between S and R indicates that they may have been derived from a single female rat. This inference is consistent with the documentation in the rat genome database (http://rgd.mcw.edu) that the outbred stock of Sprague-Dawley rats was indeed originally initiated in 1925 by R. Dawley at the Sprague-Dawley Company in Madison, WI, by breeding a hybrid hooded male of unknown origin to a white female and subsequently to his white female offspring for seven generations. The data also suggest that either during the selection process or during inbreeding and thereafter, no polymorphisms/mutations have accumulated to date in the mitochondrial genomes of either S or R rats.
Rat mitochondrial genomic polymorphisms have been reported for two other S rat stocks that are used extensively in research on the genetics of hypertension (Chen et al. 2008a; Schlick et al. 2006). These are the Dahl S rats maintained at the Medical College of Wisconsin (SSMcw) and in Japan (Dahl-Iwai S or DS). Unfortunately, the inbred S rats commercially available from Harlan Sprague-Dawley were genetically contaminated in 1992-1993 (Lewis et al. 1994; St Lezin et al. 1994). Although a test of commercial stocks indicated that this problem was apparently corrected (Walder et al. 1996), recent genome-wide single nucleotide polymorphism data indicate that the nuclear genome of at least one of the S rat subsets, the SSMcw, is 2.52% polymorphic compared to the original Dahl S rat inbred at our institution (S compared to SSMcw at http://rgd.mcw.edu). Contrary to these polymorphisms reported on their nuclear genomes, data from the current study suggests that mitochondrial genomes of S rats inbred and maintained in our laboratory and SSMcw are nearly identical with only one variation, which may perhaps be attributed to an error in sequencing. The mitochondrial genome of the Dahl-Iwai S rat having more variations compared to either S or SSMcw remains puzzling. Unlike the S or SSMcw, which represent the lineage of inbred rats by Rapp and Dene (1985), the Dahl-Iwai S was inbred by Iwai and Heine (1986). Because all of the S and R rats originated from Sprague-Dawley rats, which in turn originated from a single female rat, it is likely that any variants observed between the mtDNA of stocks of S rats either represent errors in sequencing or mutations that have accumulated since their inbreeding.
For the purpose of assessing the contributions of the mitochondrial genomes to hypertension, the variations observed between the genetically hypertensive strains are more relevant than the variations observed between hypertensive and relatively normotensive strains. Some of these variations were nonsynonymous and within genes coding proteins of the electron transport chain. These variants may or may not contribute to the status of hypertension but are nevertheless useful for dissecting key cellular mechanisms. For instance, molecules of the TCA cycle, including fumarate and succinate, are reported as being linked to the genetics of hypertension (Papadopoulos and Papademetriou 2009; Robben et al. 2009; Sadagopan et al. 2007; Tian et al. 2009; Toma et al. 2008), whereas mutations in mt-Nd2 are reported to play a critical role in the production of mitochondrial reactive oxygen species (Gusdon et al. 2007), and a mutation in mt-Atp6 is reported in the rat to cause age-related impaired glucose tolerance, a hallmark of type 2 diabetes mellitus (Mathews et al. 1995, 1999). The identification of four to five different nonsynonymous variants between S and SHR or AS provides the opportunity to examine the effects of these variants of mt-Nd2 on levels of mitochondrial reactive oxygen species. Likewise, a distinct variation in mt-Atp6 of Glu in S to Lys in either SHR or AS could be further investigated in the context of glucose tolerance.
Significant changes are observed also within tRNA genes, providing unique opportunities to assess and validate these variants as quantitative trait nucleotides for hypertension, especially because of recent associations reported between mitochondrial tRNA genes and a number of traits including hypertension, hypercholesterolemia, hypomagnesemia, and hypertrophic cardiomyopathy (Li et al. 2009; Liu et al. 2009).
Phylogenetically, the S and SHR are a large distance from each other (Thomas et al. 2003). The S is derived from non-Wistar Sprague-Dawley rats, whereas both the SHR and the AS rat are derived from Wistar rats. Interestingly, all of the variants between S and SHR/AS detected in our study are also reported as highly conserved variants between other non-Wistar and Wistar-derived strains (Abhyankar et al. 2009). One of the variable regions among the Wistar-derived strains is at position 1137 of 16s rRNA, wherein the number of C residues ranges between 2 to 5 (Abhyankar et al. 2009). The SHR strain reported in our study has two C residues, which is different from the SHR/Mol rat which is reported to have five C residues from 1137 (Abhyankar et al. 2009). The significance of these stretches of variable numbers of C residues in Wistar-derived strains remains to be determined.
The AS rat is believed to be a subline of the genetically hypertensive (GH) rat (http://rgd.mcw.edu). This is reflected in the sequence of the mitochondrial genomes of AS and GH rats, which are identical. The stretch of four C residues described above is also identical in both AS and GH rats.
Construction of conplastic strains, wherein mitochondrial DNA from one strain is substituted with that of another strain, offers a good design by which to study a complex trait’s genetic contribution of the mitochondrial DNA independent of the nuclear DNA. From the data presented it is clear that constructing conplastic strains between S and R is a moot point. This does not mean that the mitochondrial genomes of these strains do not harbor any elements that control the trait of BP. The data collected also suggest that such elements, if any, can be traced even among hypertensive strains by constructing and testing conplastic strains between S or R as one of the parental strains and SHR or AS as the other parental strain. These designs can be expected to be valuable tools to dissect the contributions of the mitochondrial genome to mechanisms of hypertension and other complex polygenic traits.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgment
This study was funded by grants from the National Institutes of Health to BJ (HL706709 and HL020176).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
S. Kumarasamy and K. Gopalakrishnan contributed equally to this work.
References
- Abhyankar A, Park HB, Tonolo G, Luthman H. Comparative sequence analysis of the non-protein-coding mitochondrial DNA of inbred rat strains. PLoS One. 2009;4:e8148. doi: 10.1371/journal.pone.0008148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneas I, Rodrigues MV, Pauletti BA, Silva GJ, Carmona R, et al. Congenic strains provide evidence that four mapped loci in chromosomes 2, 4, and 16 influence hypertension in the SHR. Physiol Genomics. 2009;37:52–57. doi: 10.1152/physiolgenomics.90299.2008. [DOI] [PubMed] [Google Scholar]
- Chan SH, Wu KL, Chang AY, Tai MH, Chan JY. Oxidative impairment of mitochondrial electron transport chain complexes in rostral ventrolateral medulla contributes to neurogenic hypertension. Hypertension. 2009;53:217–227. doi: 10.1161/HYPERTENSIONAHA.108.116905. [DOI] [PubMed] [Google Scholar]
- Chen CS, Hiura Y, Shen CS, Iwai N. Assessment of mitochondrial DNA polymorphisms in salt-sensitive hypertension in Dahl salt-sensitive rats. Hypertens Res. 2008;31:107–115. doi: 10.1291/hypres.31.107. [DOI] [PubMed] [Google Scholar]
- Chen J, Gusdon AM, Thayer TC, Mathews CE. Role of increased ROS dissipation in prevention of T1D. Ann N Y Acad Sci. 2008;1150:157–166. doi: 10.1196/annals.1447.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lu Y, Lee CH, Li R, Leiter EH, et al. Commonalities of genetic resistance to spontaneous autoimmune and free radical-mediated diabetes. Free Radic Biol Med. 2008;45:1263–1270. doi: 10.1016/j.freeradbiomed.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicila GT, Morgan EE, Lee SJ, Farms P, Yerga-Woolwine S, et al. Epistatic genetic determinants of blood pressure and mortality in a salt-sensitive hypertension model. Hypertension. 2009;53:725–732. doi: 10.1161/HYPERTENSIONAHA.108.126649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- Cowley AW, Jr, Roman RJ, Jacob HJ. Application of genome substitution techniques in gene-function discovery. J Physiol. 2004;554:46–55. doi: 10.1113/jphysiol.2003.052613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng AY. Genetic basis of polygenic hypertension. Hum Mol Genet. 2007;16:R195–R202. doi: 10.1093/hmg/ddm126. [DOI] [PubMed] [Google Scholar]
- Deng AY, Rapp JP. Locus for the inducible, but not a constitutive, nitric oxide synthase cosegregates with blood pressure in the Dahl salt-sensitive rat. J Clin Invest. 1995;95:2170–2177. doi: 10.1172/JCI117906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukhanina OI, Dene H, Deng AY, Choi CR, Hoebee B, et al. Linkage map and congenic strains to localize blood pressure QTL on rat chromosome 10. Mamm Genome. 1997;8:229–235. doi: 10.1007/s003359900399. [DOI] [PubMed] [Google Scholar]
- Garrett MR, Dene H, Walder R, Zhang QY, Cicila GT, et al. Genome scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res. 1998;8:711–723. doi: 10.1101/gr.8.7.711. [DOI] [PubMed] [Google Scholar]
- Garrett MR, Joe B, Dene H, Rapp JP. Identification of blood pressure quantitative trait loci that differentiate two hypertensive strains. J Hypertens. 2002;20:2399–2406. doi: 10.1097/00004872-200212000-00019. [DOI] [PubMed] [Google Scholar]
- Gilibert S, Bataillard A, Nussberger J, Sassard J, Kwitek AE. Implication of chromosome 13 on hypertension and associated disorders in Lyon hypertensive rats. J Hypertens. 2009;27:1186–1193. doi: 10.1097/HJH.0b013e328329e4c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, McBride MW, Gaasenbeek M, Gilday K, Beattie E, et al. Candidate genes that determine response to salt in the stroke-prone spontaneously hypertensive rat: congenic analysis. Hypertension. 2007;50:1134–1141. doi: 10.1161/HYPERTENSIONAHA.107.095349. [DOI] [PubMed] [Google Scholar]
- Gregorova S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, et al. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008;18:509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusdon AM, Votyakova TV, Reynolds IJ, Mathews CE. Nuclear and mitochondrial interaction involving mt-Nd2 leads to increased mitochondrial reactive oxygen species production. J Biol Chem. 2007;282:5171–5179. doi: 10.1074/jbc.M609367200. [DOI] [PubMed] [Google Scholar]
- Iwai J, Heine M. Dahl salt-sensitive rats and human essential hypertension. J Hypertens Suppl. 1986;4:S29–S31. [PubMed] [Google Scholar]
- Joe B, Garrett MR. Substitution mapping: using congenic strains to detect genes controlling blood pressure. In: Raizada MK, Paton JF, Katovich MJ, Kasparov S, editors. Cardiovascular genomics. Totowa, NJ: Humana Press; 2005. pp. 41–58. [Google Scholar]
- Joe B, Garrett MR. Genetic analysis of inherited hypertension in the rat. In: Dominiczak A, Connell J, editors. Genetics of hypertension. Amsterdam, The Netherlands: Elsevier Science; 2006. pp. 177–200. [Google Scholar]
- Joe B, Saad Y, Lee NH, Frank BC, Achinike OH, et al. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet. 2009;18:2825–2838. doi: 10.1093/hmg/ddp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NH, Haas BJ, Letwin NE, Frank BC, Luu TV, et al. Cross-talk of expression quantitative trait loci within two interacting blood pressure quantitative trait loci. Hypertension. 2007;50(6):1126–1133. doi: 10.1161/HYPERTENSIONAHA.107.093138. [DOI] [PubMed] [Google Scholar]
- Lewis JL, Russell RJ, Warnock DG. Analysis of the genetic contamination of salt-sensitive Dahl/Rapp rats. Hypertension. 1994;24:255–259. doi: 10.1161/01.hyp.24.3.255. [DOI] [PubMed] [Google Scholar]
- Li R, Liu Y, Li Z, Yang L, Wang S, et al. Failures in mitochondrial tRNAMet and tRNAGln metabolism caused by the novel 4401A>G mutation are involved in essential hypertension in a Han Chinese Family. Hypertension. 2009;54:329–337. doi: 10.1161/HYPERTENSIONAHA.109.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li R, Li Z, Wang XJ, Yang L, et al. Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension. 2009;53:1083–1090. doi: 10.1161/HYPERTENSIONAHA.109.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews CE, McGraw RA, Berdanier CD. A point mutation in the mitochondrial DNA of diabetes-prone BHE/cdb rats. FASEB J. 1995;9:1638–1642. doi: 10.1096/fasebj.9.15.8529844. [DOI] [PubMed] [Google Scholar]
- Mathews CE, McGraw RA, Dean R, Berdanier CD. Inheritance of a mitochondrial DNA defect and impaired glucose tolerance in BHE/Cdb rats. Diabetologia. 1999;42:35–40. doi: 10.1007/s001250051109. [DOI] [PubMed] [Google Scholar]
- Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, et al. Chromosomal mapping of the genetic basis of hypertension and renal disease in FHH rats. Am J Physiol Renal Physiol. 2007;293:F1905–F1914. doi: 10.1152/ajprenal.00012.2007. [DOI] [PubMed] [Google Scholar]
- Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, et al. Chromosome substitution reveals the genetic basis of Dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol. 2008;295:F837–F842. doi: 10.1152/ajprenal.90341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno C, Dumas P, Kaldunski ML, Tonellato PJ, Greene AS, et al. Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol Genomics. 2003;15:243–257. doi: 10.1152/physiolgenomics.00105.2003. [DOI] [PubMed] [Google Scholar]
- Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, et al. Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics. 2007;31:228–235. doi: 10.1152/physiolgenomics.00280.2006. [DOI] [PubMed] [Google Scholar]
- Papadopoulos DP, Papademetriou V. Metoprolol succinate combination in the treatment of hypertension. Angiology. 2009;60:608–613. doi: 10.1177/0003319708326450. [DOI] [PubMed] [Google Scholar]
- Pravenec M, Hyakukoku M, Houstek J, Zidek V, Landa V, et al. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res. 2007;17:1319–1326. doi: 10.1101/gr.6548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JP, Dene H. Development and characteristics of inbred strains of Dahl salt-sensitive and salt-resistant rats. Hypertension. 1985;7:340–349. [PubMed] [Google Scholar]
- Robben JH, Fenton RA, Vargas SL, Schweer H, Peti-Peterdi J, et al. Localization of the succinate receptor in the distal nephron and its signaling in polarized MDCK cells. Kidney Int. 2009;76:1258–1267. doi: 10.1038/ki.2009.360. [DOI] [PubMed] [Google Scholar]
- Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, et al. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics. 2007;89:343–353. doi: 10.1016/j.ygeno.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad Y, Yerga-Woolwine S, Saikumar J, Farms P, Manickavasagam E, et al. Congenic interval mapping of RNO10 reveals a complex cluster of closely-linked genetic determinants of blood pressure. Hypertension. 2007;50:891–898. doi: 10.1161/HYPERTENSIONAHA.107.097105. [DOI] [PubMed] [Google Scholar]
- Saad Y, Toland EJ, Yerga-Woolwine S, Farms P, Joe B. Congenic mapping of a blood pressure QTL region on rat chromosome 10 using the Dahl salt-sensitive rat with introgressed alleles from the Milan normotensive strain. Mamm Genome. 2008;19:85–91. doi: 10.1007/s00335-007-9084-7. [DOI] [PubMed] [Google Scholar]
- Sadagopan N, Li W, Roberds SL, Major T, Preston GM, et al. Circulating succinate is elevated in rodent models of hypertension and metabolic disease. Am J Hypertens. 2007;20:1209–1215. doi: 10.1016/j.amjhyper.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Schlick NE, Jensen-Seaman MI, Orlebeke K, Kwitek AE, Jacob HJ, et al. Sequence analysis of the complete mitochondrial DNA in 10 commonly used inbred rat strains. Am J Physiol Cell Physiol. 2006;291:C1183–C1192. doi: 10.1152/ajpcell.00234.2006. [DOI] [PubMed] [Google Scholar]
- St Lezin EM, Pravenec M, Wong A, Wang JM, Merriouns T, et al. Genetic contamination of Dahl SS/Jr rats. Impact on studies of salt-sensitive hypertension. Hypertension. 1994;23:786–790. doi: 10.1161/01.hyp.23.6.786. [DOI] [PubMed] [Google Scholar]
- Stoll M, Cowley AW, Jr, Tonellato PJ, Greene AS, Kaldunski ML, et al. A genomic-systems biology map for cardiovascular function. Science. 2001;294:1723–1726. doi: 10.1126/science.1062117. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Chen CF, Jensen-Seaman MI, Tonellato PJ, Twigger SN. Phylogenetics of rat inbred strains. Mamm Genome. 2003;14:61–64. doi: 10.1007/s00335-002-2204-5. [DOI] [PubMed] [Google Scholar]
- Thomas SR, Witting PK, Drummond GR. Redox control of endothelial function and dysfunction: molecular mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2008;10:1713–1765. doi: 10.1089/ars.2008.2027. [DOI] [PubMed] [Google Scholar]
- Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, et al. Novel role of fumarate metabolism in Dahl salt-sensitive hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toland EJ, Saad Y, Yerga-Woolwine S, Ummel S, Farms P, et al. Closely linked non-additive blood pressure quantitative trait loci. Mamm Genome. 2008;19:209–218. doi: 10.1007/s00335-008-9093-1. [DOI] [PubMed] [Google Scholar]
- Toland EJ, Yerga-Woolwine S, Farms P, Cicila GT, Saad Y, et al. Blood pressure and proteinuria effects of multiple quantitative trait loci on rat chromosome 9 that differentiate the spontaneously hypertensive rat from the Dahl salt-sensitive rat. J Hypertens. 2008;26:2134–2141. doi: 10.1097/HJH.0b013e32830ef95c. [DOI] [PubMed] [Google Scholar]
- Toma I, Kang JJ, Sipos A, Vargas S, Bansal E, et al. Succinate receptor GPR91 provides a direct link between high glucose levels and renin release in murine and rabbit kidney. J Clin Invest. 2008;118:2526–2534. doi: 10.1172/JCI33293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder RY, Morgan DA, Haynes WG, Sigmund RD, McClain AM, et al. Genetic characterization of the “new” Harlan Sprague Dawley Dahl salt-sensitive rats. Hypertension. 1996;27:546–551. doi: 10.1161/01.hyp.27.3.546. [DOI] [PubMed] [Google Scholar]
- Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, et al. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 2009;19:159–165. doi: 10.1101/gr.078865.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wester-Rosenlof L, Gimsa U, Holzhueter SA, Marques A, et al. The mtDNA nt7778 G/T polymorphism affects autoimmune diseases and reproductive performance in the mouse. Hum Mol Genet. 2009;18:4689–4698. doi: 10.1093/hmg/ddp432. [DOI] [PubMed] [Google Scholar]
- Zimmerman MC, Zucker IH. Mitochondrial dysfunction and mitochondrial-produced reactive oxygen species: new targets for neurogenic hypertension? Hypertension. 2009;53:112–114. doi: 10.1161/HYPERTENSIONAHA.108.125567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.