Abstract
The Tail Suspension Test (TST), which measures behavioral despair, is widely used as an animal model of human depressive disorders and antidepressant efficacy. In order to identify novel genes involved in the regulation of TST performance, we crossed an inbred strain exhibiting low immobility in the TST (RIIIS/J) with two high-immobility strains (C57BL/6J and NZB/BlNJ) to create two distinct F2 hybrid populations. All F2 offspring (n = 655) were genotyped at high density with a panel of SNP markers. Whole-genome interval mapping of the F2 populations identified statistically significant quantitative trait loci (QTLs) on mouse chromosomes (MMU) 4, 6, and X. Microarray analysis of hippocampal gene expression in the three parental strains was used to identify potential candidate genes within the MMUX QTLs identified in the NZB/BlNJ × RIIIS/J cross. Expression of Gabra3, which encodes the GABAA receptor α3 subunit, was robust in the hippocampus of B6 and RIIIS mice but absent from NZB hippocampal tissue. To verify the role of Gabra3 in regulating TST behavior in vivo, mice were treated with SB-205384, a positive modulator of the α3 subunit. SB-205384 significantly reduced TST immobility in B6 mice without affecting general activity, but it had no effect on behavior in NZB mice. This work suggests that GABRA3 regulates a behavioral endophenotype of depression and establishes this gene as a viable new target for the study and treatment of human depression.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-010-9266-6) contains supplementary material, which is available to authorized users.
Introduction
Major depressive disorder (MDD) has a lifetime prevalence rate of approximately 20% and accounts for up to 80% of affective illnesses (Kessler et al. 2003). There is a significant genetic component to MDD, as twin and adoption studies indicate that the disorder has a 40–60% heritability rate (Sullivan et al. 2000). However, the complex, heterogeneous nature of depression has stymied identification of causative genes. To date, gene identification has been based primarily on a candidate gene approach informed by the hypothesized mode of action of antidepressant drugs on the monoaminergic system (Peters et al. 2004). However, the candidate gene approach is hindered by our limited understanding of the biology of depression: There are likely to be a number of unknown genes that regulate the propensity for depression in addition to those directly targeted by current pharmacological agents.
Genome-wide association studies (GWAS) offer an unbiased means to identify causative genes, but such studies generally have been unsuccessful when applied to psychiatric disorders (Craddock et al. 2005). A number of the confounding variables associated with human studies, including the effects of genetic background and the environment, can be minimized by using animal models. Several behavioral tests that approximate discrete endophenotypes of human depression have been established, including the Tail Suspension Test (TST) and the Forced Swim Test (FST) (Cryan and Mombereau 2004; Porsolt et al. 1977). In both tests, the immobility observed when an animal is faced with an inescapable stress is believed to represent behavioral despair. While no animal model can fully recapitulate the range of symptoms associated with depressive disorders, the relevance of the TST and FST to human depression has been extensively documented. First, deletion or overexpression of genes linked to human depression affects immobility in the TST and FST (Cryan et al. 2005; Urani et al. 2005). Second, antidepressant treatment reduces immobility in the TST and FST, while antipsychotic treatment has no effect (Crowley et al. 2004). Finally, factors that increase the susceptibility to depression in humans, such as amphetamine withdrawal and chronic social stress, increase immobility in the TST (Cryan and Mombereau 2004).
Inbred mouse strains differ in their propensity for behavioral despair (Jacobson and Cryan 2007). As this effect is highly heritable, the F2 offspring of an intercross between two strains can be used to map the genetic determinants of baseline TST behavior. Several groups have used quantitative trait locus (QTL) mapping to identify loci believed to regulate variability in TST performance (Lad et al. 2007; Liu et al. 2007; Tomida et al. 2009; Turri et al. 2001; Yoshikawa et al. 2002). However, these experiments have not generated overlapping loci, and the results have not been independently replicated. Furthermore, the QTLs generally range from 20 to 40 Mb in size, making candidate gene identification difficult. Indeed, with the exception of a single study, this method has not resulted in the identification of specific quantitative trait genes (Tomida et al. 2009).
We recently completed a survey of baseline behavior in 33 inbred mouse strains and found that TST baseline immobility ranges from 7 to 60% in a strain-dependent manner. In the present study, two mapping populations were created by crossing strains at opposite ends of this distribution: The low-immobility strain RIIIS/J (R3) was crossed with either the high-immobility C57BL/6J (B6) or NZB/BlNJ (NZB) strain. F2 offspring were phenotyped in the TST and genotyped at approximately 5-Mb intervals. Interval mapping conducted for each cross separately identified a significant B6-specific QTL on MMU6 and a significant NZB-specific QTL on MMUX. The genotype data from the two crosses were combined and reanalyzed, yielding a significant QTL on MMU4. The MMU4 and MMU6 loci are within the confidence intervals of previously published TST QTLs (Liu et al. 2007; Yoshikawa et al. 2002). Genome-wide gene expression data from hippocampal tissue collected from the three parental strains were used to identify genes within the significant loci that were differentially expressed between strains and were therefore good candidate quantitative trait genes. One gene located within the MMUX locus, the GABAA α3 receptor subunit (Gabra3), was expressed in both R3 and B6 hippocampus but was absent from NZB hippocampus. To confirm the in vivo significance of Gabra3, we measured TST immobility in B6 and NZB mice treated with a GABRA3-specific agonist. Our results indicate that Gabra3 regulates baseline TST behavior in mice and may represent a novel therapeutic target for the treatment of depression.
Materials and methods
Animals
Female and male RIIIS/J, C57BL/6J, and NZB/BlNJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained on a 12:12-h light:dark cycle. Mice were allowed to acclimate for at least 1 week prior to behavioral testing. Following behavioral testing, mating trios consisting of two R3 females and either a B6 or NZB male were established in order to produce B6 × R3 and NZB × R3 F1 offspring. The F1 offspring were paired in brother–sister matings. A total of 305 B6 × R3 F2 offspring (B6F2: 162 male, 143 female) and 350 NZB × R3 F2 offspring (NZBF2: 185 male, 165 female) were phenotyped in the TST.
All animal procedures were approved by the Scripps Florida Animal Care and Use Committee.
Behavioral testing
Parental, F1, and F2 intercross mice were tested for baseline immobility in the TST as previously described (Miller et al. 2008). Briefly, mice were suspended by their tail in an environmental chamber and activity was recorded by a load cell (Med Associates, St. Albans, VT). Measurements were taken in 200-ms increments for 7 min, with threshold = 3 and gain = 8 (Supplementary Table 2). Because all mice were uniformly active for the first minute, immobility was calculated by determining the percent of time spent immobile during the last 6 min of the test. All animals that climbed their tails during the TST were removed from further behavioral and genotyping analyses and were not included in the group sizes presented in Table 1.
Table 1.
Summary of TST immobility for F0, F1, and F2 mice
| Strain | Sex | Percent immobility | SD | n |
|---|---|---|---|---|
| C57BL/6J (B6) | Male | 63.9 | 0.16 | 11 |
| C57BL/6J (B6) | Female | 64.2 | 0.10 | 9 |
| NZB/BlNJ (NZB) | Male | 51.4 | 0.11 | 13 |
| NZB/BlNJ (NZB) | Female | 50.1 | – | 2 |
| RIIIS/J (R3) | Male | 5.3 | 0.09 | 14 |
| RIIIS/J (R3) | Female | 21.4* | 0.16 | 10 |
| B6F1 | Male | 13.4 | 0.12 | 28 |
| B6F1 | Female | 17.5 | 0.13 | 21 |
| NZBF1 | Male | 8.1 | 0.05 | 21 |
| NZBF1 | Female | 22.3* | 0.12 | 16 |
| B6F2 | Male | 15.3 | 0.15 | 142 |
| B6F2 | Female | 22.4* | 0.17 | 124 |
| NZBF2 | Male | 18.7 | 0.17 | 160 |
| NZBF2 | Female | 30.1* | 0.22 | 118 |
* Significantly different from other sex in strain/cross, p < 0.001
To evaluate the role of Gabra3 on TST immobility, male B6 and NZB mice received an intraperitoneal injection of vehicle, SB-205384 (2 or 4 mg/kg), or diazepam (5 mg/kg), and were given the TST 30 min after injection. SB-205384 (Tocris, Ellisville, MO) was dissolved in a mix of sterile saline, 10% DMSO, and 25% Tween-80 immediately before use. Diazepam was suspended at 5 mg/ml in a saline solution containing 2.5% ethanol (Webster Veterinary, Sterling, MA). Doses of both drugs were based upon previously published behaviorally effective levels (Cryan and Mombereau 2004; Navarro et al. 2006). One week after TST testing, general activity and anxiety were measured in the Open Field Test (OFT). Mice received the same drug treatment as in the previous week and were placed inside an open field within a sound-attenuating cubicle (Med Associates). Activity was recorded by infrared array for 10 min. General activity was measured by total distance traveled (cm), and anxiety was measured by the percent of time spent within the center 25% of the field. The effect of drug treatment on TST and OF behavior was determined by using an unpaired, unequal variance t test (JMP, SAS Software, Cary, NC).
Genotyping
DNA was obtained from a tail snip from each F2 mouse. Genotyping was performed using the Sequenom platform (Sequenom, San Diego, CA) as previously described (Pinto et al. 2007). All DNA samples were genotyped in duplicate. B6F2 offspring were genotyped for 350 SNPs, and NZBF2 offspring were genotyped for 474 SNPs. SNP locations were mapped to NCBI mouse genome build 37.1.
QTL analysis
The J/qtl and MapManager software packages were used for QTL analysis. In J/qtl, a nonparametric one QTL scan (EM model) was used to calculate a logarithm of odds (LOD) score, and 1000 permutations were run to determine the significance threshold. In MapManager, each chromosome was evaluated by interval mapping, and 10,000 permutations were used to determine suggestive and significant likelihood ratio statistic (LRS) thresholds. Confidence intervals were calculated as ± 1 LOD (4.6 LRS). Males and females were evaluated both separately and together. In addition, each intercross was evaluated separately and then SNP and phenotyping data for all F2 offspring were combined. For the combined data set, missing genotype calls were inferred, yielding 828 informative SNPs across the entire genome (approximately 1 SNP per 3 Mb). The combined data set was analyzed in both J/qtl and MapManager, as described above. All marker pairs in the combined data set were analyzed for epistatic effects using the Interaction function of MapManager at p cutoff value of 10−5 (Supplementary Table 3). No interactions were detected to be loci on different chromosomes that did not involve the highly significant QTLs on MMUX.
Gene expression analysis
Hippocampal tissue was collected from male B6, NZB, and R3 mice (12 mice per strain). RNA was extracted by phenol–chloroform phase separation and processed with an RNAeasy miniprep kit (Qiagen, Valencia, CA). Samples from four individuals per strain were pooled at equal concentrations, and 5 μg from each pool was used as a template to synthesize biotinylated cRNA (Affymetrix, Santa Clara, CA). cRNA was hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays. Data were normalized with the GCRMA algorithm using the ArrayAssist software package (Stratagene, La Jolla, CA). Any probe set with an average intensity value of less than 100 was considered below background level and removed from subsequent analysis.
Real-time PCR (RT-PCR) was used to measure the expression of candidate genes on MMUX. B6, NZB, and R3 hippocampal RNA pools (4 mice per pool) were created using samples from different mice than those that were used for microarray analysis. Two independent pools were generated for each strain and two technical replicates were run on each pool. cDNA was amplified from each pool using a Superscript III reverse transcription kit (Invitrogen). PCR primers were designed to overlap exon–exon borders of Gabra3, Gabra3, Gabrq, and Gpr50 (Supplementary Table 1). RT-PCR was performed using a SYBR® Green mix (Bio-Rad, Hercules, CA) on an ABI 7900HT (Applied Biosystems, Foster City, CA) and using Gapdh as a control. RNA relative abundance was calculated using the delta-Ct method. Statistical significance was determined by t test (GraphPad Prism, La Jolla, CA).
Sequencing
Genomic DNA from the parental strains was sequenced for coding differences. Primers (Integrated DNA Technologies, Coralville, IA) were designed to cover the splice sites and coding regions of all nine Gabra3 exons, and sequencing was performed by the University of Chicago DNA Sequencing Facility (Chicago, IL) using fluorescent dye termination. Sequences were assembled to the reference sequence NM_008067.
Results
Tail suspension of parental strains and F2 mice
Baseline TST immobility was determined for male and female B6, NZB, and R3 inbred mouse strains (Table 1; Fig. 1). B6 male and female mice exhibited similarly high TST immobility. NZB males also exhibited high immobility, but baseline TST immobility could be determined for only two NZB females, as all others climbed their tails during testing. In contrast to the B6 and NZB strains, R3 males exhibited extremely low immobility, while R3 female immobility was significantly higher than that of R3 males, but still lower than NZB or B6 mice.
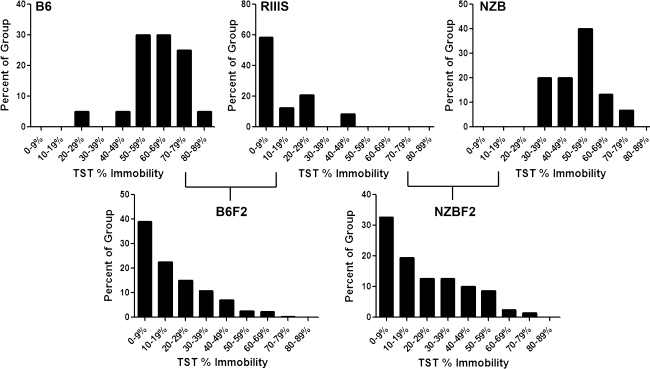
Fig. 1.
Frequency distribution of parental (F0) and F2 TST percent immobility. TST immobility scores for each strain/cross were separated into nine bins (1 = 0–9% immobility, 2 = 10–19%, 3 = 20–29%, 4 = 30–39%, 5 = 40–49%, 6 = 50–59%, 7 = 60–69%, 8 = 70–79%, 9 = 80–89%) and plotted by frequency
B6F2 and NZBF2 offspring were skewed toward the low end of the immobility scale compared to the B6 and NZB parental strains (Fig. 1, Supplementary Table 2). B6F2 mice ranged from 0 to 69% immobility (mean = 15%), but almost 50% of the group was immobile for less than 10% of the test time. NZBF2 mice exhibited an identical TST immobility range (0–68%) compared to B6F2 mice but averaged slightly higher immobility (19%). As with the parental R3 strain, females from both crosses exhibited significantly higher TST immobility than males. The tail-climbing behavior of the NZB females was not observed in the female NZBF2 offspring at a greater frequency than in females from the B6F2 cross.
QTL analysis
Interval mapping of data from all B6F2 mice identified a locus on MMU6 that reached the genome-wide significance threshold of p < 0.05 (Table 2, Figs. 2a, 3a). When only data from male mice were analyzed, a SNP on MMU2 reached p < 0.1 (Fig. 3b). In female B6F2 mice, a SNP on MMU17 exceeded the threshold for suggestive linkage (p < 0.63) (Lander and Kruglyak 1995).
Table 2.
Results from TST QTL analysis
| Cross | Data set | Chr | Position (Mb) | Marker | LOD | Confidence interval (Mb) | Effect size (%) | p value |
|---|---|---|---|---|---|---|---|---|
| B6F2 | All | 6 | 37,022,895 | rs3678202 | 4.001 | 16.92–45.38 | 6.75 | 0.021 |
| Male | 2 | 57,512,265 | rs3663027 | 3.777 | 38.38–102.72 | 11.67 | 0.064 | |
| Female | 17 | 87,480,212 | rs3689581 | 3.064 | 64.32–92.18 | 0.165 | ||
| NZBF2 | All | X | 69,924,084 | rs13483821 | 5.795 | 48.07–70.16 | 9.15 | 0.001 |
| 13 | 7,998,546 | rs6239339 | 2.948 | 0–14.84 | 0.250 | |||
| 4 | 99,636,849 | rs13477868 | 2.888 | 82.45–118.85 | 0.272 | |||
| 17 | 27,716,601 | rs4231402 | 2.875 | 0–52.80 | 0.277 | |||
| Male | 18 | 48,015,733 | rs4139897 | 3.1 | 35.85–59.87 | 0.2 | ||
| 4 | 74,880,036 | rs13477781 | 2.92 | 46.65–118.85 | 0.274 | |||
| Combined | All | X | 61,160,361 | rs13483784 | 5.474 | 45.22–74.71 | 4.25 | 0.004 |
| 4 | 99,462,176 | rs13477868 | 21.6 a (4.7) | 93.80–110.87 | 4 | 0.0001 | ||
| 5 | 81,971,499 | rs3658818 | 3.353 | 66.60–96.82 | 0.147 | |||
| 13 | 119, 532, 057 | rs3708633 | 3.065 | 116.71–end | 0.241 | |||
| Male | 2 | 61,738,719 | rs4223189 | 3.44 | 47.35–92.85 | 0.175 | ||
| 13 | 7,998,546 | rs6239339 | 3.27 | 6.09–14.54 | 0.236 | |||
| Female | X | 5,278,656 | rs13483708 | 4.923 | 0–9.78 | 1.4 | 0.013 | |
| 13 | 119,532,057 | rs3708633 | 3.45 | 116.79–end | 0.17 |
aQTL derived from MapManager analysis, value represents LRS. An equivalent LOD score, calculcated using a 1 LOD = 4.6 LRS conversion factor, is provided in parenthesis
Bold values represent genome-wide significance (p < 0.05)
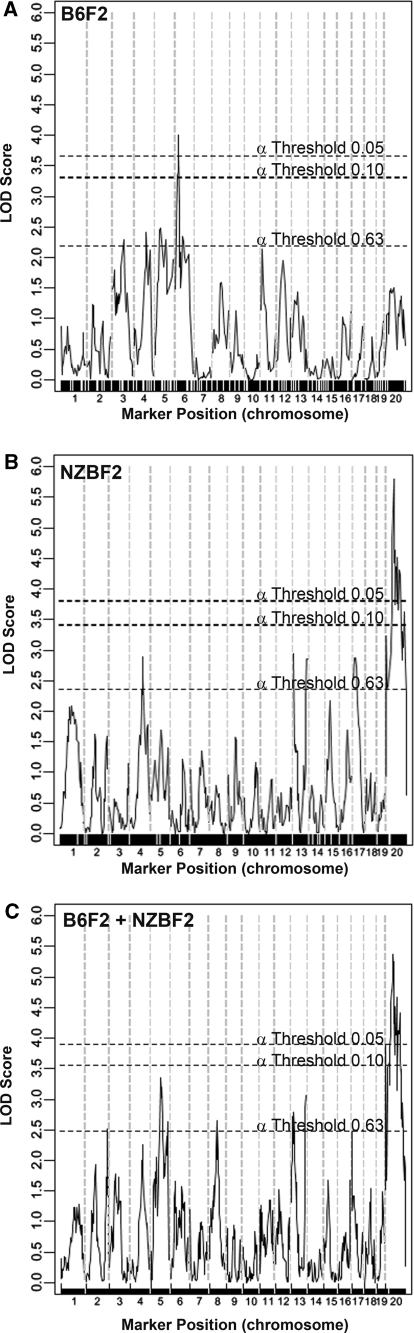
Fig. 2.
Interval mapping for B6F2, NZBF2, and B6F2 + NZBF2. LOD scores generated by J/qtl identified a significant (p < 0.05) peak on MMU6 in the B6F2 cross with an effect size of 7% (a), and a significant peak on MMUX in the NZBF2 cross with an effect size of 9% (b). Combining the genotyping data from both crosses narrowed the size of the MMUX QTL (c)
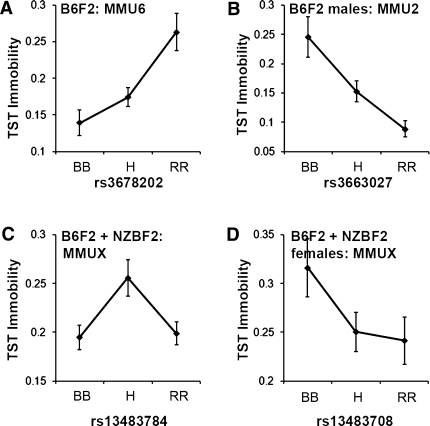
Fig. 3.
Effect plots for significant QTLs in the total B6F2 population (a), B6F2 males (b), the total population of the combined cross (c), and combined cross females (d). In general, homozygosity for the RIIIS allele (RR) was associated with low TST immobility, while heterozygosity (H) or homozygosity for the B6 (BB) or NZB (NN) alleles was associated with higher TST immobility. One exception is the MMU6 QTL in the total B6F2 population (A). Error bars = SEM
In the NZBF2 data set, a locus on MMUX exceeded the significance threshold for both J/qtl and MapManager (p < 0.05) (Figs. 2b, 3c). Both algorithms also identified loci on MMU4, MMU13, and MMU17 that exceeded the suggestive threshold. J/qtl analysis of the data from NZBF2 males alone identified a second suggestive locus on MMU4 and one on MMU18.
Combining the B6F2 and NZBF2 groups also produced a significant association with the MMUX locus (Fig. 2c) and elevated the B6F2 MMU4 locus above the MapManager significance threshold, although this locus was not identified by J/qtl. J/qtl analysis identified a female-specific QTL on MMUX with p = 0.13 (Fig. 3d).
RNA expression of QTL candidate genes
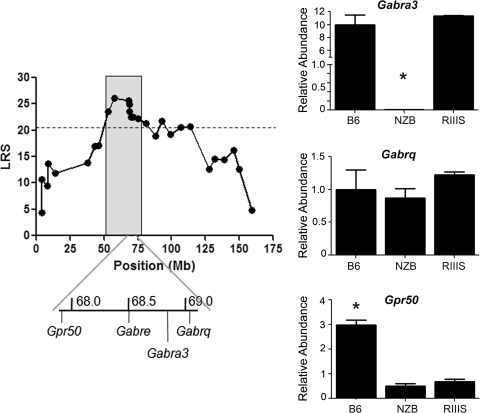
The hippocampus has been shown to play a key role in depression and the response to antidepressants (Miller et al. 2008; Santarelli et al. 2003). Therefore, we analyzed hippocampal expression of genes lying within the QTL confidence intervals for strain-specific differences. One gene within the NZBF2-specific MMUX QTL, Gabra3 (GABAA receptor α3 subunit), was expressed in both B6 and R3 hippocampus but was not expressed in NZB hippocampus (probe set 1436957_at). RT-PCR was used to confirm this strain-specific expression difference and to measure the expression of several adjacent candidate genes, including Gabre, Gabrq, and Gpr50 (Fig. 4). As predicted by the microarray data, Gabra3 expression was high in B6 and R3 mice but absent in NZB mice. Gabrq was expressed at similar levels in all three strains, Gabre expression was not detected in the hippocampus of any of the strains, and Gpr50 expression was higher in B6 hippocampus than in NZB or R3 hippocampus. The Gabra3 5′ UTR, exons, and intron/exon splice sites were sequenced in B6, NZB, and R3 but no strain-specific SNPs were identified. The Gabra3 promoter region could not be sequenced due to a highly repetitive structure (Mu and Burt 1999).
Fig. 4.
Analysis of candidate quantitative trait genes. LRS scores generated by MapManager indicated that the NZBF2 MMUX QTL had a confidence interval of 21 Mb. This region contains a cluster of GABAA receptor subunit genes, Gabre, Gabra3, and Gabrq, along with another gene, Gpr50, that has been suggested to be an affective disorder QTL candidate in humans. RT-PCR analysis of these four genes in hippocampal tissue from B6, NZB, and R3 mice indicated that expression of Gabra3 was significantly reduced in NZB compared to B6 and R3. Gabre was not expressed at a detectable level. Error bars = SEM. * p < 0.05
Effect of SB-205384 on TST and OFT behavior
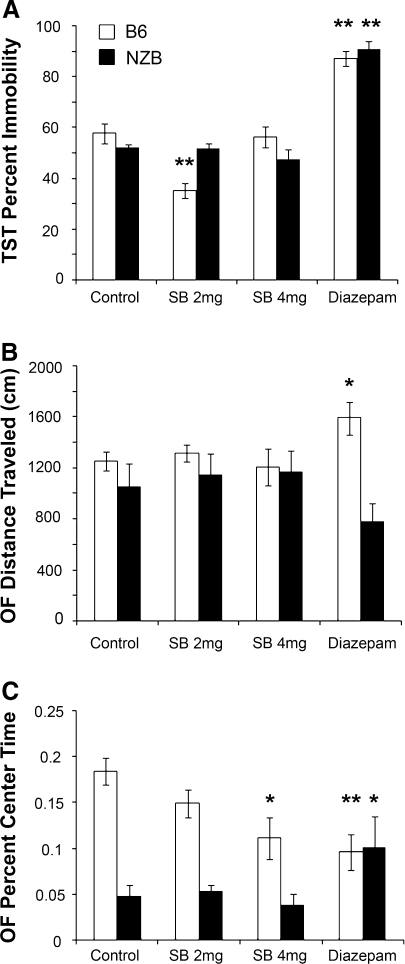
NZB and B6 mice were treated with SB-205384, a GABAA receptor α3 subunit agonist, or with diazepam, a nonspecific GABAA receptor agonist (Meadows et al. 1998). At the lower dose, SB-205384 significantly (p < 0.0001) reduced TST immobility in B6 (n = 14) but not in NZB (n = 9) mice compared to vehicle-treated mice (B6, n = 12; NZB, n = 9) (Fig. 5a). The higher dose of SB-205384 had no effect on TST immobility in either strain. Diazepam significantly (p < 0.0001) increased TST immobility in both B6 (n = 5) and NZB mice (n = 7).
Fig. 5.
Effect of GABRA3 agonist SB-205384 and GABAA agonist diazepam on TST and OFT behavior. a SB-205384 (2 mg/kg) significantly reduced TST immobility in B6 but not in NZB mice. Diazepam significantly increased TST immobility in both strains. b SB-205384 had no effect on distance traveled by either strain in the open field. Diazepam significantly increased distance traveled by B6 but not by NZB mice. c SB-205384 (4 mg/kg) significantly decreased center time in the open field in B6 mice; diazepam significantly decreased percent of time spent in the center of the field by B6 mice and increased the amount of time spent in the center of the field by NZB mice. Error bars = SEM. * p < 0.03; ** p < 0.001
In the OFT, neither dose of SB-205384 affected total distance traveled in either B6 or NZB mice (Fig. 5b). Diazepam significantly increased distance traveled in B6 mice relative to controls (p = 0.03) and slightly, but not significantly, reduced distance traveled in NZB mice. In B6 mice, the higher dose of SB-205384 significantly decreased center time (p < 0.01), while the lower dose had no effect (Fig. 5c). Neither dose of SB-205384 affected NZB center time. Diazepam significantly decreased B6 center time but significantly increased NZB center time (p < 0.01).
Discussion
In this study, two F2 hybrid crosses were used to identify genetic loci underlying a behavioral despair phenotype in mice. A B6-specific QTL on MMU6 and an NZB-specific QTL on MMUX were identified. Additional significant loci on MMU4 and MMUX were identified by combining genotyping data from both crosses. Loci on MMU2, MMU4, MMU5, MMU13, MMU17, and MMU18 exceeded the suggestive linkage threshold. Microarray expression data from the hippocampus of the parental strains were used to identify the GABAA receptor α3 subunit gene, Gabra3, as a strong candidate gene driving the NZB-specific MMUX association. In vivo pharmacological manipulation of Gabra3 confirmed a regulatory role for this gene in mouse TST performance.
The selection of parental strains was informed by a large inbred mouse strain survey of TST behavior. We hypothesized that F2 intercrosses using parental strains representing the high and low ends of the phenotypic spectrum might yield significant QTL results and that mapping efficiency could be enhanced by maximizing the genetic difference in the parental strains. Therefore, a low-immobility strain, RIIIS/J, was crossed with two high-immobility strains, C57BL/6J and NZB/BlNJ. The strains were derived from three separate ancestral branches, allowing for greater genetic diversity in the offspring (Beck et al. 2000). Interestingly, TST behavior of the F2 offspring from both crosses was strongly skewed toward low immobility; more than half of the B6F2 offspring exhibited less than 10% immobility and more than half of the NZBF2 offspring exhibited less than 20% immobility. In contrast, B6 × C3H F2 hybrids (Yoshikawa et al. 2002) and B6 × DBA recombinant inbred strains (Lad et al. 2007) exhibit a more even distribution of TST immobility. The results presented here may be explained by highly penetrant R3 alleles, particularly those identified by mapping the combined cross.
QTL analysis identified several significant loci, providing the first replication of previously identified behavioral despair QTLs. The B6F2 locus on MMU6, which accounts for 7% of TST variability, mapped to a region within the confidence interval of a previously reported QTL identified by two-locus interaction analysis of a B6 × C3H (Table 3) (Yoshikawa et al. 2002). To identify candidate genes, whole-genome expression data was examined for genes within this region that exhibited differential expression in the hippocampus. Several genes within the confidence interval exhibited a more than twofold difference between B6 and R3, including Copg2, a gene that lies within a human autism QTL (Bonora et al. 2002), Ptn, which regulates catecholamine synthesis (Ezquerra et al. 2004), and Mkrn1, a ubiquitin ligase. A SNP database (http://cgd.jax.org/ImputedSNPData/imputedSNPs.htm) was searched for genes containing coding SNPs in B6 mice, and these SNPs were examined in R3 and NZB mice (Szatkiewicz et al. 2008). Four genes, Cttnbp2, Rbm28, Olfr461, and Zfp398, contained nonsynonymous coding changes; of these, Cttnbp2 expression is restricted to brain areas relevant to TST behavior, including the hippocampus and amygdala (symatlas.gnf.org). Within the B6F2 male-specific MMU2 locus, the expression of Camk1d, which is activated by calcium and calmodulin signaling, is fourfold higher in B6 hippocampus than in R3 hippocampus.
Table 3.
Summary of published genome-wide significant TST QTLs from F2 and RI crosses
| Chr | Position | SNP ID | Cross | Group |
|---|---|---|---|---|
| 2 | 55,229,057 | rs6201706 | B6 × RIIIS (males) | Miller 2010 |
| 3 | 34 cM | NA | DeFries high/low activity | Turri 2001 |
| 4 | 78,698,065 | rs13477796 | BXD recombinant inbred (B6 × DBA) | Lad 2007 |
| 4 | 99,462,176 | rs13477868 | (B6 × RIIIS) + (NZB × RIIIS) | Miller 2010 |
| 4 | 128,974,322 | D4mit203 | B6 × C3H | Yoshikawa 2002 |
| 4 | 132,599,443 | D4mit204 | NMRI × 129S6 | Lui 2006 |
| 5 | 14 cM | NA | DeFries high/low activity | Turri 2001 |
| 5 | 73,166,580 | D5mit134 | CS strain (NBC × SII) | Tomida 2009 |
| 5 | 104,491,174 | D5mit41 | NMRI × 129S6 | Lui 2006 |
| 6 | 37,003,047 | rs3678202 | B6 × RIIIS | Miller 2010 |
| 6 | 53,125,925 | D6mit183 | B6 × C3H | Yoshikawa 2002 |
| 8 | 104,548,706 | D8mit242 | B6 × C3H | Yoshikawa 2002 |
| 11 | 45,561,177 | D11mit271 | B6 × C3H | Yoshikawa 2002 |
| 11 | 50 cM | NA | DeFries high/low activity | Turri 2001 |
| 12 | 83,437,864 | D12mit158 | NMRI × 129S6 | Lui 2006 |
| 13 | 7,998,546 | rs6239339 | (B6 × RIIIS) + (NZB × RIIIS) | Miller 2010 |
| 14 | 50,072,764 | D14mit257 | B6 × C3H | Yoshikawa 2002 |
| 15 | 3,229,130 | rs13459176 | BXD recombinant inbred (B6 × DBA) | Lad 2007 |
| 18 | 72,045,787 | D18mit186 | NMRI × 129S6 | Lui 2006 |
| 19 | 40 cM | NA | DeFries high/low activity | Turri 2001 |
| X | 4,876,764 | rs13483708 | (B6 × RIIIS) + (NZB × RIIIS) (females) | Miller 2010 |
| X | 60,167,851 | rs13483784 | NZB × RIIIS | Miller 2010 |
| X | 118,100,412 | DXmit172 | B6 × C3H | Yoshikawa 2002 |
Analysis of the NZBF2 cross yielded a highly significant QTL on MMUX with an effect size of 9%. This region is syntenic to the Xq28 region that has been associated with affective disorders in multiple human populations (Baron et al. 1987; Craddock et al. 2005). The MMUX locus has been reported to interact with the MMU11 TST locus identified by Yoshikawa et al. (2002), and both the MMUX and MMU11 loci contain clusters of genes for GABAA receptor subunits, as does an MMU5 TST QTL identified by Liu et al. (2007) (Table 3).
The association of three different GABAA subunit gene clusters with TST performance, derived from three separate F2 crosses (NZB × R3, B6 × C3H, and NMRI × 129S6), strongly suggests a role for GABA signaling in the regulation of behavioral despair. GABAA receptors are membrane Cl− channels composed of α, β, and γ subunits. Neuron-specific expression patterns have been determined for five of the six known α subunits (Fritschy et al. 1992). The α1 and α3 subunits are expressed most prominently in the brain and exhibit differential staining patterns; notably, the α3 subunit is preferentially expressed by serotonergic neurons, which are the target of existing antidepressant drug therapies (Gao et al. 1993). Gabra3 has also recently been linked to the regulation of dopaminergic signaling (Yee et al. 2005a, b), and a polymorphism in GABRA3 has been associated with bipolar disorder (Massat et al. 2002).
We compared expression of the MMUX GABAA receptor subunit genes Gabra3, Gabre, and Gabrq, in hippocampal tissue from the NZB, B6, and R3 strains. Expression of an adjacent gene, Gpr50, was also examined because it has been associated with bipolar disorder in humans (Thomson et al. 2005). A strong candidate gene would be expected to exhibit similar levels of expression in B6 and R3 mice, as this QTL was not detected in the B6F2 cross, but have different expression levels between NZB mice and the B6 and R3 strains; of the genes examined, only Gabra3 met these criteria.
Considering the strain-specific differences in Gabra3 expression and the role that the α3 subunit plays in monoaminergic signaling, we hypothesized that this gene was responsible for the MMUX linkage association identified in the NZBF2 cross. To test this hypothesis, we evaluated the effect of SB-205384, a GABAA receptor agonist selective for α3-containing GABAA receptors, on TST behavior in B6 and NZB mice. If the α3 subunit regulates TST performance, B6 mice, which express Gabra3, would be expected to respond to the drug, while NZB mice, which lack Gabra3 expression, would not respond. Diazepam, a non-specific GABAA receptor agonist, was used as a control to measure the general function of GABAA receptors in NZB mice.
In B6 mice, a low dose of SB-205384 decreased TST immobility without affecting either general locomotor activity or OFT center preference. The magnitude of the effect of 2 mg/kg SB-205384 on reducing TST immobility in B6 mice was similar to the effect of chronic fluoxetine treatment (Miller et al., unpublished data), indicating that this dose of SB-205384 is an effective antidepressant treatment. At a higher dose, SB-205384 lost its effect on TST behavior but had the same effect on OFT center preference as diazepam. The dose-dependent efficacy of SB-205384 on TST immobility and OFT center preference is likely explained by the pharmacology of the drug: SB-205384 affects decay time and magnitude of the GABA-activated current for many α subunit combinations but has a tenfold greater specificity for the α3 subunit compared to other α subunits (Meadows et al. 1998). Therefore, the lower dose of SB-205384 is likely to act solely on the α3 subunit. Although the higher dose of SB-205384 is still selective for the α3 subunit, it is also able to activate receptors containing other α subunits, similar to the effects of diazepam.
In contrast to B6 mice, NZB mice showed no behavioral response in either the TST or the OFT to any dose of SB-205384, supporting a specific role for the α3 subunit in regulating TST performance. Treatment with the non-specific GABAA agonist diazepam affected TST and OFT performance in both strains, indicating that NZB mice are capable of responding to GABAergic drugs acting at subunits other than α3. The diazepam dose did contain a negligible (2.5%) amount of ethanol required to solubilize the drug that was not present in the vehicle control and SB-205384 treatment. It is possible this impacted the behavioral testing independent of the diazepam, although this is unlikely.
We resequenced the Gabra3 5′ UTR, coding regions, and intron/exon splice sites in the three parental strains in order to identify structural variants that might cause the loss of Gabra3 expression in NZB hippocampus. We found no sequence variations among the three strains. However, the mouse Gabra3 promoter contains a region of 55 GA repeats that have been shown to have an enhancer function in Gabra3 expression (Mu and Burt 1999). As deletion of these GA repeats results in a significant reduction in Gabra3 expression in vitro, it is possible that NZB mice carry a mutation that disrupts the promoter region. Unfortunately, attempts to resequence across this region in the parental strains were unsuccessful.
Gabra3 has been previously knocked out in mice and the resulting animals have been shown to exhibit low immobility in the Forced Swim Test, another test of behavior despair (Fiorelli et al. 2008). Although this does appear to stand in contrast to the data generated with SB-205384 in which increased activity of GABRA3 led to decreased immobility, the knockdown does provide further evidence that modulation of GABRA3 activity directly impacts behavioral despair. The knockout data also further underscores the genetic complexity of the behavioral despair phenotype. The knockout was phenotyped on a C57BL/6J × 129X1/SvJ hybrid background. It is likely that this unique genetic background impacted the phenotypic expression of the knockout, just as the B6 and R3 mice in this study had divergent TST profiles despite expressing similar levels of Gabra3. In this sense, the difference between the three strains tested in the present experiments can be considered a good model of the genetics of human psychiatric disorders. Recent genome-wide association studies indicate that the majority of psychiatric disorders are caused by a large number of small effect-size genes acting in a unique combination in each individual (Purcell et al. 2009; Stefansson et al. 2009).
The independent identification of Gabra3 as a candidate QTL in the present study suggests that this gene may be a viable target for antidepressant therapy. Although current antidepressant therapies focus on the monoaminergic signaling system, several lines of evidence suggest a role for GABA signaling in depression. In addition to the linkage of the GABRA3-containing Xq28 region to affective disorders, the α1 and α5 subunits have also been associated with affective disorders in linkage studies (Oruc et al. 1997; Papadimitriou et al. 1998). Depressed patients have been shown to have reduced cortical, cerebrospinal fluid (CSF), and plasma GABA levels, and treatment with either antidepressants or electroconvulsive therapy (ECT) increases cortical GABA concentrations and GABA receptor density (Sanacora et al. 1999, 2002; Tunnicliff and Malatynska 2003). In animal models of depression, several GABAA agonists have been shown to reduce putative depressive symptoms, and deletion of Gad65, which regulates GABA synthesis, affects multiple depression-like behaviors (Cryan et al. 2005; Stork et al. 2000; Tunnicliff et al. 1973). There may be a reciprocal role between the monoaminergic and GABAergic systems, as deletion of the 5HT1A receptor reduces GABA signaling in the hippocampus, while antidepressant treatment increases GABAergic signaling (Brambilla et al. 2003).
This work clearly demonstrates that F2 mapping can be combined with whole-genome gene expression analysis to successfully identify genetic determinants of complex behaviors. In addition to providing independent confirmation of previously published TST QTLs on MMU4 and MMU6 and generating several new significant genetic associations, we identified Gabra3 as a novel candidate gene and confirmed the regulatory role of Gabra3 on TST performance using in vivo pharmacological manipulation. As Gabra3 is preferentially expressed by monoaminergic neurons, it may represent a prime therapeutic target in the treatment of depression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1 F2 TST results and genotyping data (DOC 29 kb)
Supplementary Table 2 Results of locus interaction effect analysis (XLSX 1782 kb)
Supplementary Table 3 Sequences used for RT–PCR of Gabra3, Gabre, Gabrq, and Gpr50 (XLS 48 kb)
Acknowledgments
We thank Brandon Young for invaluable assistance in producing the microarray data (Genomics Core, Scripps Florida). This study was supported by NIH grant F32MH084528 to BHM.
Disclosures
B. H. Miller, L. E. Schultz, B. C. Long, and M. T. Pletcher have no conflicts of interests of financial disclosures to disclose.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Baron M, Risch N, Hamburger R, Mandel B, Kushner S, et al. Genetic linkage between X-chromosome markers and bipolar affective illness. Nature. 1987;326:289–292. doi: 10.1038/326289a0. [DOI] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bonora E, Bacchelli E, Levy ER, Blasi F, Marlow A, et al. Mutation screening and imprinting analysis of four candidate genes for autism in the 7q32 region. Mol Psychiatry. 2002;7:289–301. doi: 10.1038/sj.mp.4001004. [DOI] [PubMed] [Google Scholar]
- Brambilla P, Perez J, Barale F, Schettini G, Soares JC. GABAergic dysfunction in mood disorders. Mol Psychiatry. 2003;8:721–737. doi: 10.1038/sj.mp.4001362. [DOI] [PubMed] [Google Scholar]
- Craddock N, O’Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Valentino RJ, Lucki I. Automated tests for measuring the effect of antidepressants on mice. Pharmacol Biochem Behav. 2004;78:269–274. doi: 10.1016/j.pbb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Mombereau C. In search of a depressed mouse: utility of models for studying depression-related behavior in genetically modified mice. Mol Psychiatry. 2004;9:326–357. doi: 10.1038/sj.mp.4001457. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Mombereau C, Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci Biobehav Rev. 2005;29:571–625. doi: 10.1016/j.neubiorev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Ezquerra L, Herradon G, Nguyen T, Vogt TF, Bronson R, et al. Pleiotrophin is a major regulator of the catecholamine biosynthesis pathway in mouse aorta. Biochem Biophys Res Commun. 2004;323:512–517. doi: 10.1016/j.bbrc.2004.08.121. [DOI] [PubMed] [Google Scholar]
- Fiorelli R, Rudolph U, Straub CJ, Feldon J, Yee BK. Affective and cognitive effects of global deletion of alpha3-containing gamma-aminobutyric acid-A receptors. Behav Pharmacol. 2008;19:582–596. doi: 10.1097/FBP.0b013e32830dc0c7. [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Benke D, Mertens S, Oertel WH, Bachi T, et al. Five subtypes of type A gamma-aminobutyric acid receptors identified in neurons by double and triple immunofluorescence staining with subunit-specific antibodies. Proc Natl Acad Sci USA. 1992;89:6726–6730. doi: 10.1073/pnas.89.15.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Fritschy JM, Benke D, Mohler H. Neuron-specific expression of GABAA-receptor subtypes: differential association of the alpha 1- and alpha 3-subunits with serotonergic and GABAergic neurons. Neuroscience. 1993;54:881–892. doi: 10.1016/0306-4522(93)90582-Z. [DOI] [PubMed] [Google Scholar]
- Jacobson LH, Cryan JF. Feeling strained? Influence of genetic background on depression-related behavior in mice: a review. Behav Genet. 2007;37:171–213. doi: 10.1007/s10519-006-9106-3. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- Lad HV, Liu L, Paya-Cano JL, Fernandes C, Schalkwyk LC. Quantitative traits for the tail suspension test: automation, optimization, and BXD RI mapping. Mamm Genome. 2007;18:482–491. doi: 10.1007/s00335-007-9029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Liu X, Stancliffe D, Lee S, Mathur S, Gershenfeld HK. Genetic dissection of the tail suspension test: a mouse model of stress vulnerability and antidepressant response. Biol Psychiatry. 2007;62:81–91. doi: 10.1016/j.biopsych.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Massat I, Souery D, Del-Favero J, Oruc L, Noethen MM, et al. Excess of allele1 for alpha3 subunit GABA receptor gene (GABRA3) in bipolar patients: a multicentric association study. Mol Psychiatry. 2002;7:201–207. doi: 10.1038/sj.mp.4000953. [DOI] [PubMed] [Google Scholar]
- Meadows HJ, Kumar CS, Pritchett DB, Blackburn TP, Benham CD. SB-205384: a GABA(A) receptor modulator with novel mechanism of action that shows subunit selectivity. Br J Pharmacol. 1998;123:1253–1259. doi: 10.1038/sj.bjp.0701721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Schultz LE, Gulati A, Cameron MD, Pletcher MT. Genetic regulation of behavioral and neuronal responses to fluoxetine. Neuropsychopharmacology. 2008;33:1312–1322. doi: 10.1038/sj.npp.1301497. [DOI] [PubMed] [Google Scholar]
- Mu W, Burt DR. The mouse GABA(A) receptor alpha3 subunit gene and promoter. Brain Res Mol Brain Res. 1999;73:172–180. doi: 10.1016/S0169-328X(99)00258-2. [DOI] [PubMed] [Google Scholar]
- Navarro JF, Buron E, Martin-Lopez M. Anxiolytic-like activity of SB-205384 in the elevated plus-maze test in mice. Psicothema. 2006;18:100–104. [PubMed] [Google Scholar]
- Oruc L, Verheyen GR, Furac I, Ivezic S, Jakovljevic M, et al. Positive association between the GABRA5 gene and unipolar recurrent major depression. Neuropsychobiology. 1997;36:62–64. doi: 10.1159/000119363. [DOI] [PubMed] [Google Scholar]
- Papadimitriou GN, Dikeos DG, Karadima G, Avramopoulos D, Daskalopoulou EG, et al. Association between the GABA(A) receptor alpha5 subunit gene locus (GABRA5) and bipolar affective disorder. Am J Med Genet. 1998;81:73–80. doi: 10.1002/(SICI)1096-8628(19980207)81:1<73::AID-AJMG14>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Peters EJ, Slager SL, McGrath PJ, Knowles JA, Hamilton SP. Investigation of serotonin-related genes in antidepressant response. Mol Psychiatry. 2004;9:879–889. doi: 10.1038/sj.mp.4001502. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Vitaterna MH, Shimomura K, Siepka SM, Balannik V, et al. Generation, identification and functional characterization of the nob4 mutation of Grm6 in the mouse. Vis Neurosci. 2007;24:111–123. doi: 10.1017/S0952523807070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsolt RD, Bertin A, Jalfre M. Behavioral despair in mice: a primary screening test for antidepressants. Arch Int Pharmacodyn Ther. 1977;229:327–336. [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanacora G, Mason GF, Rothman DL, Behar KL, Hyder F, et al. Reduced cortical gamma-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 1999;56:1043–1047. doi: 10.1001/archpsyc.56.11.1043. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Mason GF, Rothman DL, Krystal JH. Increased occipital cortex GABA concentrations in depressed patients after therapy with selective serotonin reuptake inhibitors. Am J Psychiatry. 2002;159:663–665. doi: 10.1176/appi.ajp.159.4.663. [DOI] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O, Ji FY, Kaneko K, Stork S, Yoshinobu Y, et al. Postnatal development of a GABA deficit and disturbance of neural functions in mice lacking GAD65. Brain Res. 2000;865:45–58. doi: 10.1016/S0006-8993(00)02206-X. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, et al. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19(3):199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Thomson AM, Dunbar DR, Grassie MA, et al. Sex-specific association between bipolar affective disorder in women and GPR50, an X-linked orphan G protein-coupled receptor. Mol Psychiatry. 2005;10:470–478. doi: 10.1038/sj.mp.4001593. [DOI] [PubMed] [Google Scholar]
- Tomida S, Mamiya T, Sakamaki H, Miura M, Aosaki T, et al. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat Genet. 2009;41:68–695. doi: 10.1038/ng.344. [DOI] [PubMed] [Google Scholar]
- Tunnicliff G, Malatynska E. Central GABAergic systems and depressive illness. Neurochem Res. 2003;28:965–976. doi: 10.1023/A:1023287729363. [DOI] [PubMed] [Google Scholar]
- Tunnicliff G, Wimer CC, Wimer RE. Relationships between neurotransmitter metabolism and behaviour in seven inbred strains of mice. Brain Res. 1973;61:428–434. doi: 10.1016/0006-8993(73)90551-9. [DOI] [PubMed] [Google Scholar]
- Turri MG, Datta SR, DeFries J, Henderson ND, Flint J. QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr Biol. 2001;11:725–734. doi: 10.1016/S0960-9822(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Urani A, Chourbaji S, Gass P. Mutant mouse models of depression: candidate genes and current mouse lines. Neurosci Biobehav Rev. 2005;29:805–828. doi: 10.1016/j.neubiorev.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Yee BK, Keist R, von Boehmer L, Studer R, Benke D, et al. A schizophrenia-related sensorimotor deficit links alpha 3-containing GABAA receptors to a dopamine hyperfunction. Proc Natl Acad Sci USA. 2005;102(47):17154–17159. doi: 10.1073/pnas.0508752102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Watanabe A, Ishitsuka Y, Nakaya A, Nakatani N. Identification of multiple genetic loci linked to the propensity for “behavioral despair” in mice. Genome Res. 2002;12:357–366. doi: 10.1101/gr.222602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 F2 TST results and genotyping data (DOC 29 kb)
Supplementary Table 2 Results of locus interaction effect analysis (XLSX 1782 kb)
Supplementary Table 3 Sequences used for RT–PCR of Gabra3, Gabre, Gabrq, and Gpr50 (XLS 48 kb)