Essential tremor (ET), a neurologic disorder characterized by postural and action tremor, is one of the most common adult-onset motor disorders.1,2 While linkage regions have been identified in Icelandic and North American families, the causative gene remains elusive.1–4 Recently, a sequence variant (rs9652490 G allele) of the LINGO1 gene was found to be associated with ET in the first genome-wide association study in European and American populations.5 Here we determined the linkage disequilibrium (LD) of single nucleotide polymorphisms (SNPs) 100 kb up/downstream of the implicated LINGO1 SNP and analyzed the SNP in a case control study in an Asian population.
We included consecutive patients with ET who presented to a tertiary referral center and examined by movement disorders neurologists following a methodology as previously described.6 We followed the diagnostic criteria of classic ET based on the recommendations of the Consensus Statement of the Movement Disorders Society in 1998.7 Patients with at least one affected first-degree relative were classified as familial ET. Controls of similar gender, race, and age as ET patients and physically examined by the authors at the Health Screening Unit of the same hospital were included. Informed consent from all the study subjects was taken and the work received approval from the institutional ethics committee.
The LINGO1 SNP (rs9652490) was screened on MALDI-TOF mass spectrometry using the Sequenom MassARRAY™ system (San Diego, CA) (details described in appendix e-1 on the Neurology® Web site at www.neurology.org).
A total of 923 subjects (733 controls and 190 ET) examined by the authors were included in the analysis. The median age of the controls (56.7 years) was 15 and 4 years older than the median age at onset (41.0 years) and age of patients with ET (52.6 years). A total of 75 (39.5%) patients with ET reported a positive family history in at least 1 first-degree relative. There was a trend toward a higher frequency of the allele G of marker rs9652490 in ET (0.263 vs 0.218, OR = 1.28, χ2 = 3.29, p = 0.068). For the genotypic test, ORAG = 0.84, OR GG = 2.43, χ2 = 13.53, p = 0.001, and for recessive inheritance (allele G – [GG vs AA+AG]), χ2 = 12.72, p = 0.00036, ORG = 2.58. The population attributable risk of genotype GG = 18.8%. There is no evidence of heterogeneity between the results from our population and genotype data from the American and European populations (I2 = 0, p = 0.66). A combined meta-analysis revealed an OR = 1.48, p = 9.1 × 10−10 (table). Interestingly, analysis of only those with familial ET revealed a much more robust association (0.32 vs 0.22, OR = 1.69, χ2 = 7.27, p = 0.007). For the genotypic test, ORAG = 1.23, OR GG = 3.26, χ2 = 10.55, p = 0.005. Analysis of sporadic ET did not reveal any association (0.225 vs 0.218, OR = 1.04, p = 0.8). LD variation surrounding this SNP was largely similar between Caucasians and Asians (figure e-1). The maximum FST (fixation index) value for the SNP is only 0.0002 or 0.2% between Chinese (19.8%) and Germans (23.3%).
Table Summary of published literature
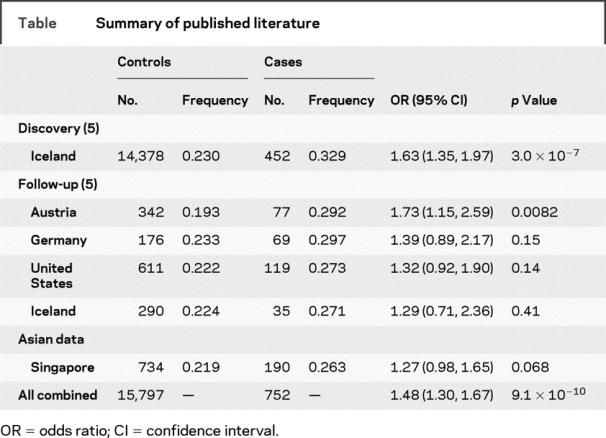
We have found negligible LD variation involving multiple SNPs 100 kb up/downstream of the functional LINGO1 SNP (rs9652490) between Asians and Caucasians. The frequency of the G allele (0.32) in our Asian patients with familial ET and the OR of 1.66 were practically identical to the discovery data set from Iceland, but higher than most of the other follow-up datasets (table). These studies have either analyzed both familial and sporadic ET as a group or did not provide information on the specific group of patients with ET (table e-1). The similarity of our result with the Icelandic discovery dataset could be explained by the fact that the majority (69.4%) of their patients with ET reported a positive family history. It would be interesting to determine how the overall effect size of the association will be affected if only familial cases are examined in Caucasians. In the LINGO1 discovery study,5 the 14,393 Icelandic controls were not screened for ET and in the 2 follow-up studies in Austria and Germany, the median age of controls was about 20–25 years younger than patients with ET. It is unclear how these would have influenced the reported effect size difference.
We have demonstrated negligible LD variation in the vicinity of the LINGO1 SNP (rs9652490) between Asians and Caucasians. This sequence variant (rs9652490 G allele) is associated with risk of familial ET. An individual with two G alleles has more than 3 times risk of having ET. Replication studies in other populations are warranted.
Supplementary Material
Supplemental data at www.neurology.org.
Supported by grants from the National Medical Research Council, Duke-NUS Graduate Medical School, Singapore Millennium Foundation, and SingHealth Foundation.
Disclosure: Dr. E.-K. Tan has received speaker honoraria from Boehringer Ingelheim and has received research support for clinical trials in movement disorders sponsored by Eisai, Schering Plough, and Allergan. Dr. Teo, Dr. Prakash, R. Li, H.-Q. Lim, and Dr. Angeles report no disclosures. Dr. L.-C. Tan has received funding for travel from Novartis, GlaxoSmithKline, Boehringer Ingelheim, and Medtronic; serves on the editorial board of Movement Disorders; has received speaker honoraria from Boehringer Ingelheim; receives research support for clinical trials sponsored by Eisai (PI), Schering Plough (PI), and Allergan (coinvestigator); and receives research support from the National Medical Research Council, Singapore [NMRC/0995/2005 (PI)]. Dr. Au, Dr. Yih, and Dr. Zhao report no disclosures.
Received April 7, 2009. Accepted in final form June 16, 2009.
Address correspondence and reprint requests to Dr. Eng-King Tan, Department of Neurology, Singapore General Hospital, Outram Road, Singapore 169608; gnrtek@sgh.com.sg
&NA;
- 1.Deng H, Le W, Jankovic J. Genetics of essential tremor. Brain 2007;130:1456–1464. [DOI] [PubMed] [Google Scholar]
- 2.Louis ED. Essential tremor. Lancet Neurol 2005;4:100–110. [DOI] [PubMed] [Google Scholar]
- 3.Higgins JJ, Pho LT, Nee LE. A gene (ETM) for essential tremor maps to chromosome 2p22-p25. Mov Disord 1997;12:859–864. [DOI] [PubMed] [Google Scholar]
- 4.Tan EK, Schapira AH. Hunting for genes in essential tremor. Eur J Neurol 2008;5:889–890. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, Steinberg S, Petursson H, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet 2009;41:277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan EK, Prakash KM, Fook-Chong S, et al. DRD3 variant and risk of essential tremor. Neurology 2007:6;68:790–791. [DOI] [PubMed] [Google Scholar]
- 7.Deuschl G, Bain P, Brin M. Consensus statement of the Movement Disorder Society on Tremor: Ad Hoc Scientific Committee. Mov Disord 1998;13 suppl 3:2–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


