Abstract
Viral encephalitis remains a significant cause of morbidity and mortality throughout the world. We performed microarray analysis to identify genes and pathways that are differentially regulated during reovirus encephalitis and that may provide novel therapeutic targets for virus-induced diseases of the central nervous system (CNS). An increase in the expression of 130 cellular genes was found in the brains of reovirus-infected mice at early times post infection, compared to mock-infected controls. The up-regulation of these genes was consistent with activation of innate immune responses, particularly interferon signaling. At later times post infection, when significant CNS injury is present and mice exhibit signs of severe neurologic disease, many more (1374) genes were up-regulated, indicating that increased gene expression correlates with disease pathology. Virus-induced gene expression at late times post infection was again consistent with the activation of innate immune responses. However, additional significant pathways included those associated with cytokine signaling and apoptosis, both of which can contribute to CNS injury. This is the first report comparing virus-induced cellular gene and pathway regulation at early and late times following virus infection of the brain. The shift of virus-induced gene expression from innate immune responses at early times post infection to cytokine signaling and apoptosis at later times suggests a potential therapeutic strategy that preserves early protective responses whilst inhibiting later responses that contribute to pathogenesis.
Keywords: encephalitis, gene expression, reovirus
Introduction
Reoviruses belonging to serotype 3, exemplified by the prototype strains Dearing (T3D) and Abney (T3A), induce lethal encephalitis in newborn mice. T3 reoviruses infect and injure specific regions of the mouse brain, including the hippocampus, thalamus, and cortex, and virus-induced injury within these regions is associated with the apoptotic death of infected neurons (Oberhaus et al, 1997; Richardson-Burns et al, 2002). Reovirus-induced apoptosis in the brain is triggered by extrinsic apoptotic signaling pathways involving cell surface death receptors and their apoptosis-inducing ligands. For example, Fas and FasL are up-regulated in the brains of reovirus-infected mice and Fas colocalizes in individual neurons with virus antigen and activated caspase 3 (Clarke et al, 2009). Further, neuronal apoptosis is inhibited by soluble Fas following reovirus infection of primary cortical cultures (Richardson-Burns et al, 2002). It has thus been proposed that Fas-FasL binding contributes to activation of caspase 8, the initiator caspase associated with extrinsic apoptotic signaling, both in reovirus-infected primary neurons and in the brains of reovirus-infected mice (Clarke et al, 2009; Richardson-Burns et al, 2002). Activated caspase 8 can cleave and activate caspase 3 directly, resulting in apoptotic cell death. Alternatively, caspase 8 can cleave the Bcl-2 family protein Bid, which then activates mitochondrial/intrinsic apoptotic signaling pathways through interactions with other Bcl-2 family proteins. Expression of a dominant negative form of the death receptor–associated adaptor protein FADD inhibits reovirus-induced cleavage of Bid (Kominsky et al, 2002). In addition, Bid cleavage and activation of caspase 9, the initiator caspase associated with mitochondrial apoptotic signaling, have been demonstrated in reovirus-infected HEK293 cells and in the brains of reovirus-infected mice (Clarke et al, 2009; Kominsky et al, 2002). These results indicate that both extrinsic and intrinsic apoptotic signaling pathways contribute to the ultimate death of reovirus-infected neurons and disease pathology. The reovirus protein μ1c also mediates mitochondrial apoptotic signaling pathways, although the mechanism by which this occurs has not been established (Danthi et al, 2008).
Reovirus infection of the brain is associated with the activation of a number of transcription factors, including nuclear factor (NF)-κB and c-Jun (Beckham et al, 2007; O’Donnell et al, 2005). NF-κB family members exist as heterodimers or homodimers that are sequestered in the cytoplasm by association with inhibitor kappa B α (IκBα), or other structurally related inhibitors. Agonists of NF-κB, including viral infection, trigger a signaling cascade that leads to the phosphorylation and degradation of IκBα, which allows the translocation of NF-κB to the nucleus and the transcriptional activation of target genes. Following reovirus infection, the prototypical form of NF-κB, containing the p50 and p65 subunits, is activated and translates to the nucleus both in vitro and in vivo (Connolly et al, 2000; O’Donnell et al, 2005). Further, activation of NF-κB is required for reovirus-induced activation of caspase 3 and apoptosis, although the exact mechanism by which NF-κB contributes to apoptotic signaling remains to be determined (Connolly et al, 2000; O’Donnell et al, 2005). However, the activation of NF-κB following reovirus infection is not sufficient for virus-induced apoptosis because NF-κB is activated following infection with both apoptotic and nonapoptotic strains of reovirus (Clarke et al, 2003). A second component of reovirus-induced NF-κB regulation, involving the inhibition of NF-κB at later times post infection, thus also appears to be required for virus-induced apoptosis (Clarke et al, 2003, 2005).
The transcription factor c-Jun is activated following phosphorylation by the mitogen-activated protein kinase (MAPK) c-Jun N-terminal kinase (JNK). JNK and c-Jun are activated following reovirus infection of tissue culture cells and primary cortical neurons and in the brains of reovirus-infected mice (Beckham et al, 2007; Clarke et al, 2001). Further, reovirus-induced c-Jun activation requires JNK activity in vivo (Beckham et al, 2007). JNK is also required for reovirus-induced activation of caspase 3 and apoptosis (Beckham et al, 2007; Clarke et al, 2004). The demonstration that JNK is required for reovirus-induced up-regulation of Fas (Clarke et al, 2009) suggests that JNK-induced activation of c-Jun and c-Jun–induced gene expression also contributes to viral-induced apoptosis.
In contrast to the proapoptotic effects of NF-κB and c-Jun, reovirus-induced activation of signal transducer and activator of transcription (STAT) 1 and members of the SMAD family of transcription factors appear to be protective (Beckham et al, 2009; Goody et al, 2007). Thus mice lacking the STAT1 gene demonstrate increased susceptibility to reovirus infection, with increased mortality and higher viral titers in the brain compared to wild-type animals (Goody et al, 2007). Similarly, treatment of mice with an inhibitor of transforming growth factor-β receptor 1 (TGF-β R1) resulted in increased apoptosis within the brain as demonstrated by increased cleavage of caspase 3 and poly (ADP-ribose) polymerase (PARP) (Beckham et al, 2009).
The observation that transcription factors are activated during reovirus encephalitis and that this activation affects pathogenesis suggests that virus-induced changes in gene expression likely play a major role in reovirus-induced central nervous system (CNS) disease. Reovirus-induced gene expression has been investigated in tissue culture systems (Debiasi et al, 2003; O’Donnell et al, 2006; Poggioli et al, 2002; Smith et al, 2006). However, reovirus-induced gene expression during viral encephalitis has not been studied and is likely to be influenced by the unique cell types present within the brain. Indeed, published studies of virus-induced global gene expression changes in the brain are limited despite the importance of viral CNS disease as a cause of significant human morbidity and mortality (Bordignon et al, 2008; Koterski et al, 2007; Sharma et al, 2008; Sui et al, 2003; Venter et al, 2005). In this report, we analyzed gene expression changes in the murine brain following infection with reovirus. We found that infection resulted in the up-regulation of genes associated with innate immune responses at early times post infection. At later times post infection, additional genes associated with cytokine signaling and apoptosis were up-regulated. This shift of virus-induced gene expression suggests a therapeutic strategy that maintains early protective immune responses whilst inhibiting later responses that contribute to disease pathology.
Results
Effector caspases are activated in the brains of T3D-infected mice
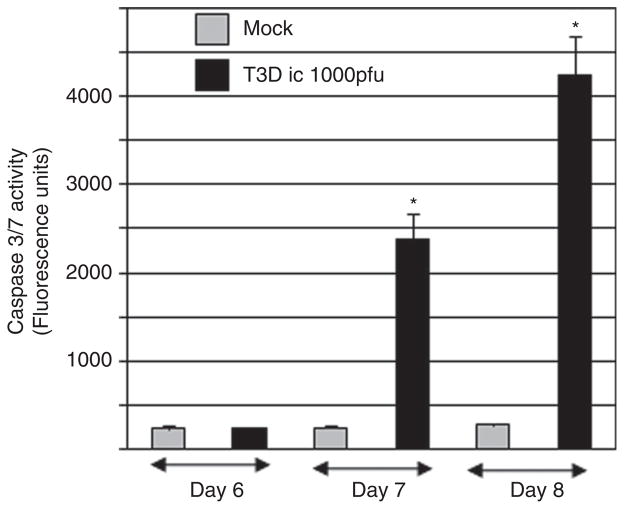
We have previously demonstrated that tissue injury in the brains of mice infected with type 3 strains of reovirus is associated with apoptosis and the cleavage of caspase 3 (Oberhaus et al, 1997; Richardson-Burns et al, 2002). However, an increase in the activity of the effector caspases 3 and 7 (caspase 3/7) following reovirus infection of the brain has not previously been shown. These caspases recognize the same peptide (DEVD) sequence. In order to determine the kinetics of caspase 3/7 activation in the brain during T3D encephalitis, we performed caspase activity assays on brain lysates from T3D-infected and mock-infected mice at 6, 7, and 8 days post infection. Figure 1 shows that caspase 3/7 activity increases in the brains of T3D-infected mice, compared to mock-infected controls, starting at day 7 post infection. At day 7 post infection, there is 6.2-fold (P < .05) more caspase 3/7 activity in reovirus-infected, compared to mock-infected, brains. Reovirus-induced caspase 3/7 activity increases through day 8 post infection when a 14.1-fold (P < .005) increase in activity is seen in reovirus-infected, compared to mock-infected, brains. T3D-infected mice typically die between days 8 and 9 post infection.
Figure 1.
Effector caspases 3 and/or 7 are activated in the brains of reovirus-infected mice. Two-day-old mice were infected with reovirus (T3D) by intracerebral (i.c.) inoculation (103 p.f.u.). Caspase 3/7 fluorogenic substrate (activity) assays were performed on lysates prepared from the brains of T3D reovirus– and mock-infected mice at days 6, 7, and 8 post infection. The graph shows the average fluorescence (caspase 3/7 activity) in reovirus-infected lysates compared to mock-infected controls. Error bars represent standard errors of the mean. Statistically significant differences in reovirus-infected compared to mock-infected animals are indicated (*). At least six individual reovirus- and mock-infected animals were used at each time point.
Reovirus-induced changes in gene expression dramatically increase at late stages post infection, corresponding with the onset of apoptotic neuronal death, tissue injury, and the presentation of neurologic symptoms
Transcription factors, including c-Jun, NF-κB, STAT1, and SMADs are activated following reovirus infection, both in vitro and in vivo (Beckham et al, 2007, 2009; Clarke et al, 2001; Connolly et al, 2000, Goody et al, 2007; O’Donnell et al, 2005). These transcription factors are also thought to contribute to reovirus-induced pathogenesis. We performed microarray analysis to determine (a) whether virus-induced transcription factor activation in the brain is associated with increased cellular gene expression; and (b) whether reovirus-induced gene expression is associated with pathogenesis. For our studies, RNA was extracted from the brains of three T3D-infected and three mock-infected mice at days 3, 6, and 8 post infection. These time points were chosen to allow a comparison of virus-induced gene expression before and after the activation of caspase 3/7 (Figure 1) and the onset of apoptotic neuronal death.
RNA extracted from the brains of virus-infected and mock-infected animals was analyzed using Affymetrix 430.2 mouse whole-genome chips. These chips contain 45,000 probe sets designed to detect the differential expression of around 39,000 individual mouse genes.
Table 1 lists the number of probe sets that identified differentially expressed genes in the brains of T3D-infected, compared to mock-infected, mice at days 3, 6, and 8 post infection. The table shows that 141 probe sets detected the differential expression of a cellular gene in the brains of reovirus-infected, compared to mock-infected, mice at 3 days post infection. A similar number of probe sets (128) detected the differential expression of a cellular gene at day 6 post infection. However, a much larger change in cellular gene expression, both in terms of the number of probes that identified increased gene expression (1582) and the fold-change of identified genes, was seen at later times (day 8 post infection), correlating with the development of clinical neurologic symptoms and an increase in the activity of caspase 3 (Figure 1). Very few probe sets detected down-regulated genes in the brains of reovirus-infected, compared to mock-infected animals at any time post infection and the magnitude of expression change for these genes was small (Table 1). These results indicate that reovirus infection of the brain results in increased gene expression. A pronounced increase in reovirus-induced gene expression at late time post infection correlates with the appearance of severe neurologic symptoms.
Table 1.
Differential gene expression in the brains of reovirus-infected compared to mock-infected micea
| Days p.i.b | Probe setsc detecting the differential expression of cellular genes (fold change) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no. | Fold change of probe sets detecting genes with increased expression |
Fold change of probe sets detecting genes with decreased expression |
|||||||
| (1–1.5) | (1.5–5) | (5–50) | (50–100) | (100+) | (1–1.5) | (1.5–5) | (5–10) | ||
| 3 | 141 | 8 | 72 | 59 | 0 | 0 | 0 | 2 | 0 |
| 6 | 128 | 11 | 51 | 62 | 2 | 1 | 0 | 1 | 0 |
| 8 | 1582 | 244 | 682 | 372 | 36 | 40 | 121 | 86 | 1 |
RNA was extracted from the brains of reovirus and mock infected mice at 3, 6, and 8 days following intracerebral (i.c.) inoculation. Gene expression was compared using Affymetrix 430.2 mouse whole-genome chips. A minimum of three reovirus-infected and mock-infected animals were used at each time point. Identified genes were differentially regulated with a false detection rate (FDR) < 0.1, P < .002.
p.i. = post infection.
A probe set is a group of oligonucleotides designed to detect the differential expression of an individual gene. The 39,000 genes on the Affymetrix 430.2 mouse whole-genome chip are represented by one or more probe sets.
Reovirus-induced cellular gene expression indicates that signaling pathways associated with the innate immune response and interferon signaling are activated in the brains of reovirus-infected mice at early times post infection
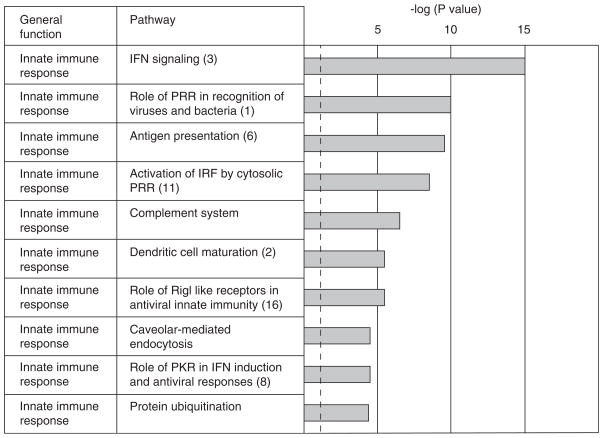
Microarray data were analyzed using pathway analysis software (Ingenuity Systems, Redwood, CA) to identify cellular pathways that were significantly represented by the pattern of differential gene expression in the brains of reovirus-infected mice. In order to identify genes that may play a role in the establishment of disease, we initially analyzed probe sets (Table 1) that identified differentially expressed genes at “early” times post infection (day 3 and/or day 6 post infection), before the onset of apoptosis and clinical symptoms. One hundred sighty-six different probe sets identified the differential expression of genes in the brains of reovirus-infected mice at day 3 and/or day 6 post infection. Ingenuity pathway analysis identified 23 cellular signaling pathways that were significantly (P < .05) represented at these early times post infection by the pattern of differential gene expression in the brains of reovirus-infected, compared to mock-infected, mice. The 10 most significant (P < 1.49e−04) of these pathways are shown in Figure 2. The majority of the pathways (9/10) identified at early times post infection were associated with the innate immune response. The most significant pathway identified was interferon signaling (P = 4.63e−16). However, several other identified pathways were associated with interferon (IFN) signaling, including (i) role of pattern recognition receptors in recognition of viruses and bacteria; (ii) activation of interferon regulatory factors by cytosolic pattern recognition receptors; (iii) role of retinoic acid inducible gene-I (RIG-I)-like receptors in antiviral innate immunity; and (iv) role of protein kinase R (PKR) in interferon induction and antiviral responses.
Figure 2.
Cellular signaling pathways associated with innate immune responses are activated in the brains of reovirus-infected mice at early times post infection. Ingenuity pathway analysis was performed on probe sets that identified differentially expressed genes at day 3 and/or day 6 post reovirus infection (Table 1). The bar chart shows the 10 most significant pathways identified and the general function of these pathways (Ingenuity Systems).
The “early” genes were also placed into functional categories using Affymetrix software (see Supplementary Table 1). For this analysis, results from different probe sets identifying the differential expression of an individual gene at day 3 or day 6 were combined and fold changes from duplicate probe sets representing the same gene at the same time point were averaged. In addition, genes where the fold change did not exceed 2.0 on any day were excluded. Supplementary Table 1 lists the Affymetrix-assigned function of the remaining 131 genes that were differentially regulated at day 3 and/or day 6 post infection. Interestingly, all of these genes (except one, see below) were also up-regulated at day 8 post infection. In addition, all the up-regulated genes showed an increase in expression at 8 days post infection compared to day 3 and/or day 6 post infection, although the magnitude of expression change varied. The one exception to this pattern of gene expression was the gene encoding lysophosphatidylcholine acetyl transferase 1, which was the only gene to show a negative fold change and the only gene that was differentially expressed at early (day 3 post infection) but not late times (day 8) post infection. In accordance with our results using Ingenuity pathway analysis, the Affymetrix-assigned function of 44 out of the 131 of the identified differentially regulated genes at early time following reovirus infection was IFN signaling. One major family of interferon-stimulated genes (ISGs) that were up-regulated in the brains of reovirus-infected mice encodes the 2′-5′oligoadenylate (2′-5′OAS) proteins. These proteins activate RNase L, which produces small RNA cleavage products from self or viral RNA that induce the production of IFN α/β through a pathway involving activation of (RIG-I), melanoma differentiation–associated gene 5 (MDA5), and their interacting protein IPS1 (IFNβ promoter stimulator protein 1) (Malathi et al, 2007). It has been shown that 2′-5′OAS genes confer resistance to infection with flaviviruses, including hepatitis C virus (Knapp et al, 2003), West Nile virus (Kajaste-Rudnitski et al, 2006), and Dengue virus (Warke et al, 2003). Interestingly, the genes encoding RIG-I (Ddx58) and MDA5 (Ifih1) are also up-regulated in the brains of reovirus-infected mice at both early and late times following infection (Supplementary Table 1). Other antiviral ISG genes up-regulated during reovirus encephalitis include members of the Myxovirus protein family (Mx), which mediates protection against influenza infection (Tumpey et al, 2007).
The promoters of differentially expressed genes at early times following reovirus infection contain significantly more binding sites for interferon regulatory factors 1 and 2 and STAT1 than controls
We performed Opossum analysis (Ho Sui et al, 2005) on genes that were differentially regulated at early times (day 3 and/or day 6) post infection (n = 52 software-selected genes). The promoters of these (target) genes were statistically more likely to contain binding sites for interferon regulatory factors (IRFs) 1 and 2 and for STAT1 than (background) genes that were not differentially expressed in the brain following reovirus infection (Table 2). IRF1, IRF2, and STAT1 regulate IFN stimulated gene expression. Binding sites for the transcription factor RXRA (retionoid × receptor alpha)-VDR (vitamin D receptor) were also present in the promoters of the early genes. This transcription factor is active during development. The up-regulation of genes containing binding sites for this transcription factor thus likely represents developmental, rather than reovirus-induced, events.
Table 2.
Identification of transcription factor (TF) binding sites in reovirus-induced genes in the brains of mice at early times post intracerebral inoculation (Opossum analysis)a
| TF | Bkgd gene hitsb |
Bkgd gene non-hitsb |
Target gene hitsb |
Target gene non-hitsb |
Bkgd TFBS hitsc |
Bkgd TFBS rated |
Target TFBS hitsc |
Target TFBS rated |
Z scoree | Fisher scoref |
|---|---|---|---|---|---|---|---|---|---|---|
| IRF2 | 643 | 14507 | 4 | 48 | 709 | 0.0006 | 6 | 0.0027 | 18.03 | 1.8e-01 |
| STAT1 | 2165 | 12985 | 9 | 43 | 2693 | 0.0016 | 12 | 0.0042 | 12.52 | 3.2e-01 |
| RXRA-VDR | 406 | 14744 | 2 | 50 | 420 | 0.0003 | 3 | 0.0011 | 10.09 | 4.1e-01 |
| IRF1 | 4631 | 10519 | 15 | 37 | 7857 | 0.0041 | 24 | 0.0071 | 9.566 | 6.6e-01 |
Probe sets that were identified as being differentially regulated (FDR < 0.1, P < .002) in T3D compared to mock-infected animals (Table 1) were analyzed for the presence of transcription factor binding sites (Opossum analysis).
Background (Bkgd) genes (15,150 mouse genes were used as background/control genes). Target genes (52 reovirus-induced genes were used as target/experimental genes). Gene hits/non-hits (the number of genes, target or background, for which a TFBS was/was not predicted within the conserved noncoding regions.
TFBS hits. The number of times a TFBS was detected within the conserved noncoding gene regions (background or target).
TFBS rate. The rate of occurrence of a TFBS within the conserved noncoding regions of the genes (target or background). The rate is equal to the number of times the site was predicted (hits) multiplied by the width of the TFBS profile, divided by the total number of nucleotides in the conserved noncoding regions of the gene set.
Z score. The likelihood that the number of TFBS nucleotides detected for the included target genes is significant as compared with the number of TFBS nucleotides detected for the background set. Z score is expressed in units of magnitude of the standard deviation. A Z score of > 10 was used as an indication of significance based on empirical studies (Opossum software).
Fisher score. The probability that the number of hits versus non-hits for the included target genes could have occurred by random chance based on the hits versus non-hits for the background set. A Fisher score of < .01 was used as an indication of significance based on empirical studies (Opossum software).
IRF2 = interferon regulatory factor 2; IRF1 = interferon regulatory factor 1; RXRA = retinoid × receptor alpha; VDR = vitamin D receptor; Bkgd = background; TF = transcription factor; TFBS = transcription factor binding site; STAT1 = signal transducer and activator of transcription 1.
Validation of microarray data using QPCR (interferon-regulated genes)
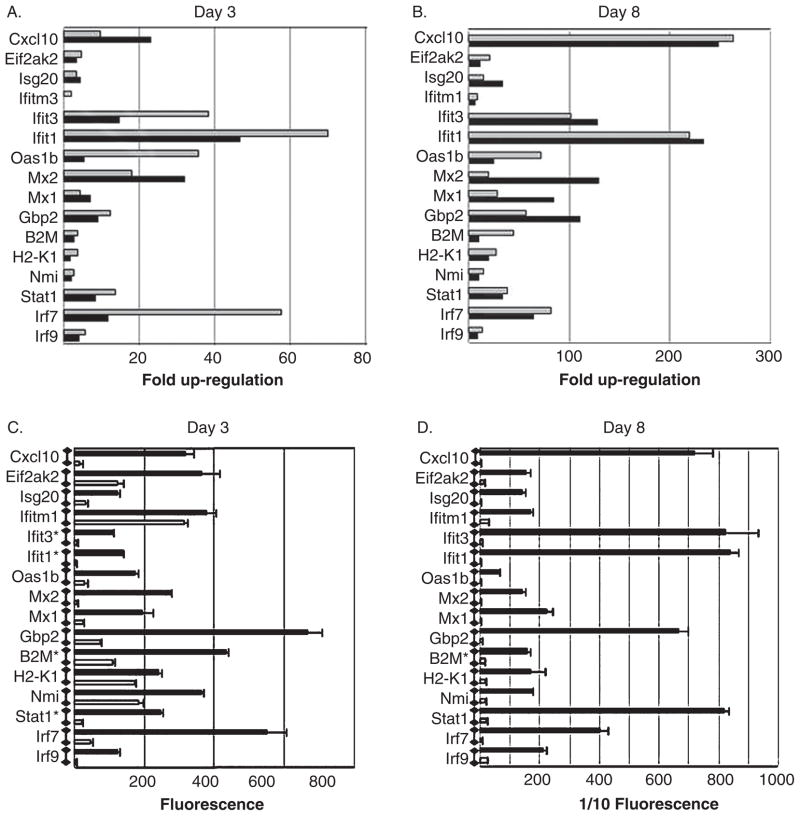
Because the majority of genes that were up-regulated at early times post infection were involved in interferon signaling, we validated our microarray data by quantitative polymerase chain reaction (QPCR) using primers designed to determine the expression of IFN-regulated genes. RNA extracted from the brains of three additional T3D reovirus–infected and mock-infected animals at day 3 and day 8 post intracerebral (i.c.) inoculation were used for QPCR validation. These animals were littermates from those used for our microarray experiment (see above). There is a high degree of consistency between results obtained using microarray analysis and QPCR for a number of IFN-regulated genes at days 3 and 8 post infection (Figure 3). The figure also shows that there is an increase in the expression change of IFN regulated genes between day 3 and day 8 post infection.
Figure 3.
Confrirmation of microarray data by quantitative PCR (QPCR): interferon-regulated genes. Two-day-old mice were infected with reovirus (T3D) by intracerebral inoculation (103 p.f.u.). RNA extracted from the brains of reovirus-infected mice was compared to mock-infected controls. (A and B) The graphs show the mean fold up-regulation of interferon associated genes at days 3 (A) and 8 (B) post infection in T3D reovirus–infected, compared to mock-infected, animals as determined by microarray analysis (light gray bars) and QPCR (dark gray bars). The P values for all reovirus-induced, interferon-regulated genes at day 3 post infection were 0.05 (Ifitm1) to 3.87e−06 (B2M) as determined by microarray analysis and 0.04 (H2-K1) to 2.00e−05 (IRF9) as determined by QPCR. The P values for all reovirus-induced, interferon-regulated genes at day 8 post infection were.001 (H2-K1) to 4.3e−09 (Ifit1) as determined by microarray analysis and 0.01 (Nmi) to 7.0e−05 (Stat1) as determined by QPCR. The graphs (C and D) show the mean fluorescence generated by an individual probe set (Affymetrix 430.2 microarray) representing each of the interferon-regulated genes shown in A and B. Error bars represent the standard error of the mean. For illustrative purposes the horizontal axis in D shows 1/10 of the actual fluorescence reading. In addition, fluorescence values for genes indicated with an asterisk are also reduced by a factor of 10. Three individual T3D reovirus–infected mice and three individual mock-infected mice were used to determine fold up-regulation by microarray analysis and three different mice from the same litters were used to determine fold up-regulation by QPCR.
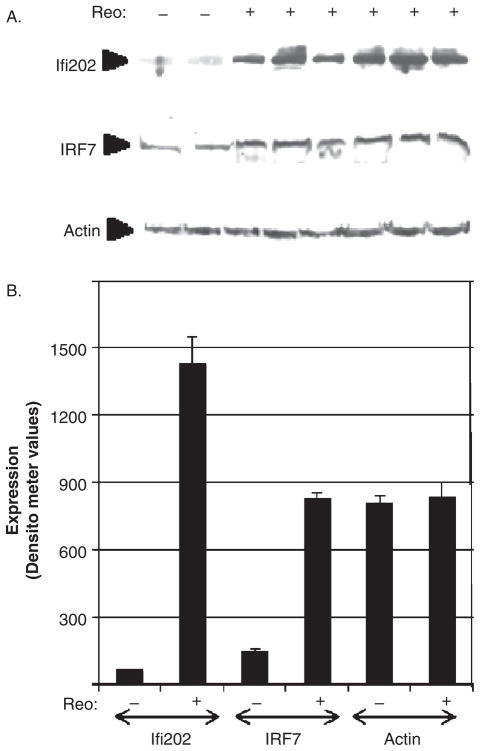
Microarray data were also validated by Western blot analysis, which showed an increase in the expression of two IFN-induced proteins IFR7 (IFN regulatory factor 7) and Ifi202 (IFN-induced protein 202) in reovirus-infected, compared to mock-infected, brains (Figure 4). These proteins were used as representative IFN-regulated proteins. IRF7 is an IFN-regulated transcription factor that has been shown to play a role in the transcriptional activation of virus-inducible cellular genes, including IFN-β, whereas Ifi202 is an IFN-regulated inhibitor of p53-mediated apoptosis and is suspected to contribute to systemic lupus erythematosus (SLE) (D’Souza et al, 2001; Xin et al, 2006).
Figure 4.
The interferon-induced proteins IRF7 and Ifi202 are increased in the brains of reovirus-infected mice compared to mock-infected controls. Two-day-old mice were infected with reovirus (T3D) by intracerebral inoculation (103 p.f.u.). Western blot analysis was performed on lysates prepared from the brains of reovirus-infected mice and mock-infected controls at 8 days post infection using antibodies directed against IRF7 (interferon regulatory factor 7) and Ifi202 (interferon-induced protein 202). (A) Western blotting indicates that IRF7 and Ifi292 are up-regulated in the brains of reovirus-infected mice compared to mock-infected controls. Actin was used as a control for protein loading. The blot is representative of three individual experiments. (B) The graph shows the average increase in expression of IRF7 and Ifi202 proteins as determined by densitometric analysis.
Reovirus-induced cellular gene expression indicates that cellular signaling pathways associated with cytokine signaling and apoptosis are activated in the brains of reovirus-infected mice at late times post infection
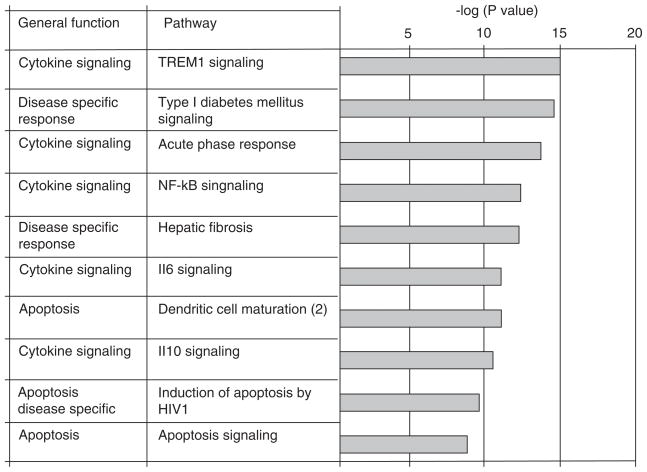
Ingenuity pathway analysis was also performed on genes that were differentially regulated in the brains of reovirus-infected animals at 8 days post infection (“late” genes). This analysis included genes that were also differentially expressed at day 3 and/or day 6 post infection. We hypothesized that genes that were differentially expressed at late times post infection might play a prominent role in disease severity and progression. One hundred and eleven pathways were significantly (P < 0.05) represented by the pattern of differential gene expression in the brains of reovirus-infected, compared to mock-infected, mice at 8 days post infection. The 17 most significant of these pathways (P < 1.53e−09) included 7 of the pathways that were also identified at early times post infection (Figure 2). Table 3 shows that for the pathways that were identified at both early and late times post infection, the ratio of the number of genes in a pathway that were differentially regulated in the brains of reovirus-infected, compared to mock-infected, mice divided by the total number of genes assigned to that pathway (Ingenuity) increases at late, compared to early, times post infection. The 10 additional pathways are shown in Figure 5 and include pathways associated with the general functions of cytokine signaling, apoptosis, and disease-specific pathways.
Table 3.
Pathway activation in the brains of reovirus-infected mice at early and late times post infection
| Early times post infection (day 3 and/or day 6) |
Late times post infection (day 8) |
|||
|---|---|---|---|---|
| P valuea | Ratiob | P value | Ratio | |
| Role of PRR in recognition of bacteria and viruses | 4.82e−11 | 10/88 (0.11) | 3.51e−22 | 33/88 (0.38) |
| Dendritic cell maturation | 1.8e−06 | 8/165 (0.05) | 6.14e−20 | 40/165 (0.24) |
| IFN signaling | 4.63e−16 | 10/29 (0.35) | 8.56e−16 | 17/29 (0.59) |
| Antigen presentation | 1.82e−10 | 7/39 (0.18) | 1.02e−14 | 16/39 (0.41) |
| Role of PKR in IFN induction and antiviral responses | 1.43e−04 | 4/47 (0.09) | 1.01e−13 | 19/47 (0.40) |
| Activation of IRF by cytosolic PRR | 1.78e−09 | 8/74 (0.11) | 2.54e−12 | 20/74 (0.27) |
| Role of RIG-I like receptors in antiviral innate immunity | 2.57e−06 | 5/52 (0.10) | 1.28e−09 | 14/52 (0.27) |
P value represents how likely is observed observation different from random chance.
The ratio is the number of genes from data that are in pathway divided by the total number of genes that are designated to the pathway by Ingenuity.
Figure 5.
Cellular signaling pathways associated with cytokine, apoptosis, and disease-specific responses are activated in the brains of reovirus-infected mice at late times post infection. Ingenuity pathway analysis was performed on probe sets that identified differentially expressed genes at day 8 post reovirus infection (Table 1). The bar chart shows the 10 most significant pathways identified (excluding pathways that were also identified at early times post infection) and the general function of these pathways (Ingenuity systems).
The promoters of differentially expressed genes at late times following reovirus infection contain significantly more binding sites for interferon regulatory factors 1 and 2, STAT1, and the NF-κB family of transcription factors than controls
We performed Opossum analysis on genes that were differentially regulated at late times post infection (n = 745 software-selected genes). The promoters of these (target) genes were statistically more likely to contain binding sites for IRF 1 and 2 and STAT1 than (background) genes that were not differentially expressed in the brain following reovirus infection (Table 4). Binding sites for the NF-κB family of transcription factors were also identified as being overrepresented in reovirus-induced genes at day 8 following infection compared to control/background genes. The transcription factors IRF1, IRF2, and STAT1 regulate interferon-induced gene expression. The NF-κB family of transcription factors play a role in the regulation of cytokine gene expression. Additional transcription factor binding sites that are present in the promoters of reovirus-induced genes at late times following intracerebral inoculation include E74-like factor 5 (ELF5) and GA-binding protein transcription factor, alpha (GABPA). ELF5 and GABPA are members of the ETS family of transcription factors, which are present throughout the body and are involved in a wide variety of functions, including the regulation of cellular differentiation, cell cycle control, cell migration, cell proliferation, apoptosis (programmed cell death), and angiogenesis. In addition, in the mouse brain, ETS factors have been shown to be important players in the specification and differentiation of dopaminergic neurons (Flames and Hobert, 2009).
Table 4.
Identification of transcription factor (TF) binding sites in reovirus-induced genes in the brains of mice at late times post intracerebral inoculation (Opossum analysis)a
| TF | Bkgd gene hitsb |
Bkgd gene non-hitsb |
Target gene hitsb |
Target gene non-hitsb |
Bkgd TFBS hitsc |
Bkgd TFBS ratec |
Target TFBS hitsc |
Target TFBS ratec |
Z scorec | Fisher scorec |
|---|---|---|---|---|---|---|---|---|---|---|
| STAT1 | 2165 | 12985 | 160 | 585 | 2693 | 0.0016 | 206 | 0.003 | 32.76 | 1.67e-07 |
| IRF2 | 643 | 14507 | 54 | 691 | 709 | 0.0006 | 62 | 0.0012 | 25.08 | 1.95e-04 |
| RelA/p65 | 4868 | 10304 | 275 | 470 | 8017 | 0.0035 | 476 | 0.0049 | 24.12 | 3.05e-03 |
| ELF5 | 12185 | 2965 | 607 | 138 | 73487 | 0.287 | 3504 | 0.0326 | 23.11 | 2.57e-01 |
| GABPA | 5381 | 9769 | 291 | 454 | 9203 | 0.004 | 513 | 0.005 | 20.46 | 2.73e-02 |
| NF-κB | 5960 | 9190 | 310 | 435 | 11447 | 0.005 | 613 | 0.0063 | 19.2 | 1.15e-01 |
| Rel | 7798 | 7352 | 409 | 336 | 18677 | 0.0081 | 951 | 0.0098 | 18.99 | 3.66e-02 |
| IRF1 | 4631 | 10519 | 259 | 486 | 7857 | 0.0052 | 418 | 0.0052 | 16.86 | 9.08e-03 |
Probe sets identified differentially regulated genes (Table 1) in T3D compared to mock-infected animals were analyzed for the presence of transcription factor binding sites (Opossum analysis).
Background (Bkgd) genes (15,150 mouse genes were used as background/control genes). Target genes (745 reovirus-induced genes were used as target/experimental genes). Gene hits/non-hits (the number of genes, target or background) for which a TFBS was/was not predicted within the conserved noncoding regions.
An explanation of the column headings can be found following Table 2.
STAT1 = signal transducer and activator of transcription 1; IRF2 = interferon regulatory factor 2; RelA/p65 = V-rel reticuloendotheliosis viral oncogene homolog A; ELF5 = E74-like factor 5; GABPA = GA-binding protein transcription factor, alpha; NF-κB = nuclear factor kappa B; Rel = cRel; IRF1 = interferon regulatory factor 1.
Validation of microarray data using QPCR (cytokine-regulated genes)
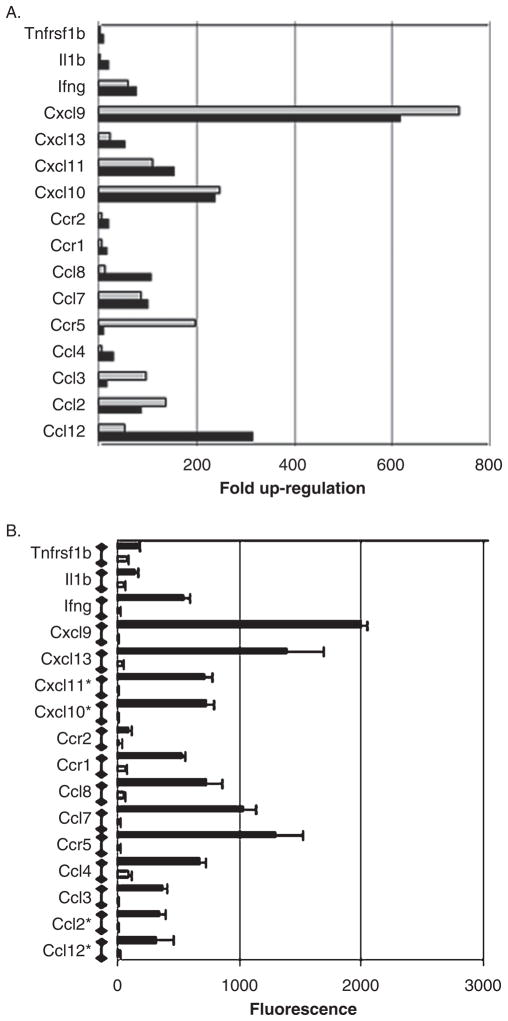
Because the majority of genes that were up-regulated at late times post infection were involved in interferon signaling (Figure 3) and cytokine signaling, we further validated our microarray data by QPCR using primers designed to determine the expression of cytokine genes. RNA extracted from the brains of three additional T3D reovirus–infected and mock-infected animals at day 8 post intracerebral (i.c.) inoculation were used for QPCR validation. These animals were littermates from those used for our microarray experiment (see Materials and Methods). There is a high degree of consistency between results obtained using microarray analysis and QPCR for a number of cytokine genes at day 8 post infection (Figure 6).
Figure 6.
Confirmation of microarray data by quantitative PCR (QPCR): cytokine genes. Two-day-old mice were infected with reovirus (T3D) by intracerebral inoculation (103 p.f.u.). RNA extracted from the brains of reovirus-infected mice was compared to mock-infected controls. (A) The graph shows the mean fold up-regulation of cytokine genes at day 8 post infection in T3D reovirus-infected, compared to mock-infected, animals as determined by microarray analysis (light gray bars) and QPCR (dark gray bars). The P values for all reovirus-induced cytokine genes at day 8 post infection were between 0.001 (Il1β) and 5.2e−08 (Cxcl9) as determined by microarray analysis and between 0.05 (Ccr5) and 3.00e−05 (Cxcl13) as determined by QPCR. Three individual T3D reovirus–infected mice and three individual mock-infected mice were used to determine fold up-regulation by microarray analysis and three different mice from the same litters were used to determine fold up-regulation by QPCR. (B) The graph shows the mean fluorescence generated by an individual probe set (Affymetrix 430.2 microarray) representing each of the reovirus-induced cytokine genes shown in A (reovirus-infected = black bars, mock-infected = gray bars). Error bars represent the standard error of the mean. Fluorescence values for genes identified with an asterisk are reduced by a factor of 10 for illustrative purposes.
Discussion
Reovirus encephalitis is associated with the activation of a number of cellular transcription factors including NF-κB (O’Donnell et al, 2005) and c-Jun (Beckham et al, 2007). Further, the activation of these transcription factors is thought to be required for virus-induced pathogenesis. Microarray analysis identified limited differential gene expression in the brain 3 and 6 days following reovirus infection. However, there was a considerable increase in differential gene expression at day 8 post infection. This increase in differential gene regulation at day 8 post infection corresponds to the activation of caspase 3/7 and the onset of clinical neurologic symptoms. The vast majority of differentially regulated genes at all times post infection were up-regulated. In order to determine the function of the differentially regulated genes, we divided the genes into two groups. The “early” genes (differentially regulated at day 3 and/or day 6 post infection) were hypothesized to play a prominent role in the establishment of disease. In contrast, the “late” genes (up-regulated at day 8 post infection) were hypothesized to play a prominent role in disease progression.
Gene expression in the brain at “early” times post reovirus infection
We identified 186 probe sets (131 genes) that identified the differential expression of cellular genes at early times post infection (Table 1 and Supplementary Table 1). All of these genes, with one exception, were up-regulated and were also up-regulated at day 8 post infection, with an increase in expression change from “early” to “late” times. Our results of gene expression changes in the brain at early time post infection with reovirus are remarkably consistent with recent reports of gene expression changes in the CNS of mice infected with dengue virus type 1 (Bordignon et al, 2008) and Venezuelan equine encephalitis virus (Sharma et al, 2008). The major pathways affected by reovirus infection at early times post infection were involved in IFN signaling, antigen presentation and processing, and complement activation. These changes are consistent with a robust innate immune response characterized in particular by genes induced by type I IFNα/β.
IFNα/β is induced in response to virus infection and contributes to the innate antiviral immune response (Randall and Goodburn, 2008). In addition to the cell autonomous effects of IFNα/β, these cytokines also modulate the immune system by activating effector cell function and promoting the development of the acquired immune response. The pivotal role of the IFN response in viral pathogenesis is not only of fundamental interest but may also have practical application in the design and manufacture of attenuated virus vaccines and the development of novel antiviral drugs.
The role of IFNα/β in reovirus infections of the brain has not previously been characterized, although previous studies have suggested a protective role for the STAT proteins in reovirus-induced encephalitis (Goody et al, 2007). The current study shows that reovirus infection of the brain results in the up-regulation of many ISG. Although modulation in IFNα/β was not detected in the brains of reovirus-infected mice by our initial criteria (false detection rate [FDR] < .1, P < .002), an increase in the levels of IFNα/β mRNA were detected by microarray analysis at 8 days post infection with a less stringent selection criteria (FDR < .1, P < .005) and by QPCR (not shown). An increase in the levels of IFNα/β mRNA was not detected in the brains of reovirus-infected mice at early times post infection. However, low levels of IFNα/β have been shown to be capable of inducing high levels of ISGs, as demonstrated in several microarray experiments (Bourne et al, 2007; Johnston et al, 2001; Sariol et al, 2007). Additionally, other authors have found the expression of ISGs in chimpanzee liver cells chronically infected with hepatitis C virus (Bigger et al, 2004) and in monkey peripheral blood mononuclear cells (PBMCs) infected with Dengue virus type 1 (Sariol et al, 2007) without detecting IFN transcripts.
IFNα/β is thought to play a protective role in reovirus-induced myocarditis, with myocarditic reoviruses (including T1L) repressing the IFNα/β signaling pathway (Zurney et al, 2009). Interestingly, in this study, T3D (which was used in the current study to induce viral encephalitis) was not found to inhibit IFNα/β signaling and was not myocarditic (Zurney et al, 2009). Repression of IFNα/β signaling during viral myocarditis is brought about by the reovirus μ2 protein and occurs through a mechanism involving nuclear accumulation of interferon regulatory factor 9 (IRF9) (Zurney et al, 2009). Consistent with this finding is that inhibition of IFNα/β signaling may be specific to infected cells, since IFNα/β produced by hematopoietic cells following peroral T1L inoculation protects mice against lethal infection (Johansson et al, 2007).
The use of mice deficient for the type I IFN receptor (IFNAR) has highlighted the importance of IFNα/β signaling in the control of viral replication in the CNS (Randall and Goodbourn, 2008). For example, in the case of Sindbis virus, an increase in viral load was reported in the CNS of wild-type, compared to IFNAR-deficient, strain, resulting in significantly increased susceptibility to viral infection (Ryman et al, 2000). Multiple studies have demonstrated the widespread ability of CNS cell types to both induce and respond to IFN. For example, neurons, macrophages/microglia, and ependymal cells from mice infected with Theilers virus (TMEV) or La Crosse virus (LACVdelNSs, a variant mutated in the non-structural (NS) IFN-antagonist gene) produce IFN (Delhaye et al, 2006). Expression of ISGs has also been reported in the CNS after infection with lymphocytic choriomeningitis virus (LCMV) (Wacher et al, 2007), West Nile virus (Wacher et al, 2007), dengue virus type 1 (Bordignon et al, 2008), and Venezualen equine encephalitis virus (Sharma et al, 2008). Additional analysis in primary neurons has also indicated that these cells respond to IFNα by expressing genes involved in the antiviral response (Wang and Campbell, 2005). In this study, the up-regulation of IFN-induced genes was dependent on STAT1.
Gene expression in the brain at “late” times post reovirus infection
Ingenuity pathway analysis was also performed on genes that were differentially regulated in the brains of reovirus-infected animals at 8 days post infection (“late” genes). The 17 most significant (P < 1.53e−09) pathways identified included 7 of the pathways that were also identified at early times post infection (Figure 2). Table 3 shows that for the pathways that were identified at both early and late times post infection, the ratio of the number of genes in a pathway that were differentially regulated in the brains of reovirus-infected, compared to mock-infected, mice divided by the total number of genes assigned to that pathway (Ingenuity) increases at late, compared to early, times post infection. The 10 additional pathways (Figure 4) include pathways associated with the general functions of apoptosis, cytokine signaling, and disease-specific pathways.
The up-regulation of mRNAs encoding proteins involved in death receptor signaling is central to all the apoptotic pathways identified as being significantly represented in the brains of reovirus-infected, compared to mock-infected, mice. These include mRNAs corresponding to Fas, caspase 8, Daxx, Bid, caspase 7, TNFR1 and TNFR2, TRAF2, RIP, and IKK. The contribution of Fas, caspase 8, and Bid to reovirus-induced apoptosis in the brain has previously been reported so the observation that the mRNAs encoding these proteins are up-regulated is confirmatory (Clarke et al, 2009). However, the mRNAs of several additional, previously unrecognized, proteins that may contribute to apoptosis was also increased in the brains of reovirus-infected, compared to mock-infected, mice. For example, Daxx, caspase 12, BAK, and p53 all have suspected roles in apoptosis. In addition, mRNA for the death receptor–associated proteins TNFR1 and TNFR2, RIP, Traf2, and IKK is up-regulated and may contribute to reovirus-induced apoptosis via activation of NF-κB.
Although much is known regarding the mechanism of apoptotic neuronal death during reovirus encephalitis, the role of other cells within the brain, including microglia, remains to be determined. These cells are the resident immune cells of the central nervous system and have a critical role in the host defense against invading microorganisms. Microglial activation is viewed as an adaptive response whereby microglia release neuroprotective factors to facilitate the recovery of injured neurons and they also phagacytose dying or damaged neurons before they release toxic agents into the surrounding area. Although initiation of immune responses by microglia is an important protective mechanism in the CNS, unrestrained inflammatory responses may result in irreparable damage to the brain, as is the case following infection of the brain with Japanese encephalitis virus. One proinflammatory mediator that is associated with microglial activation is interleukin-6 (IL6). Cellular pathways defined by Ingenuity systems that were identified as being significantly represented by the pattern of differential gene expression in the brains of reovirus-infected, compared to mock-infected, mice and that encompass IL6 responses include (i) IL10 signaling, (ii) IL6 signaling, (iii) hepatic fibrosis/hepatic stellate cell activation, (iv) acute-phase repose signaling, and (v) TREM 1 signaling. Recent reports from our laboratory have shown that microglia are activated in the brain following reovirus infection. It will thus be interesting to determine the role of microglia in IL6 up-regulation and neuronal injury during reovirus encephalitis.
These studies have identified the up-regulation of many cellular genes during viral encephalitis. These genes are consistent with the activation of specific cellular pathways. Future studies will examine the role of these genes and pathways in viral pathogenesis.
Materials and methods
Cells and virus
Reovirus strain type 3 Dearing (T3D) is a laboratory stock that has been plaque purified and passaged (twice) in L929 (ATCC CCL1) cells to generate working stocks. T3D was grown in L929 mouse fibroblasts (ATCC CL-1) that were maintained in 2 × 199 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco/BRL, Gaithersburg, MD) and 4 mM L-glutamine. T3D stocks were diluted in phosphate-buffered saline (PBS) and 2-day-old Swiss Webster mice were intracerebrally (i.c.) inoculated with 103 plaque forming units (p.f.u.) of T3D in a 10-μl volume. Mock-infected animals underwent identical i.c. inoculations using PBS-diluted L929 conditioned medium.
Caspase fluorometric assay
Activity of the effector caspases 3 and 7 was determined using a kit obtained from R&D Systems (BF1100). Briefly, brains were removed, homogenized in 1 ml lysis buffer and incubated on ice for 10 min. Fifty microliters of the resulting homogenate was then tested for protease activity by the addition of a caspase 3/7–specific peptide (DEVD) conjugated to the fluorescent reporter molecule 7-amino-4-trifluoromethyl coumarin (AFC).
Western blotting
Brains were removed from T3D-infected and mock-infected mice and were transferred to 1 ml of PBS and stored at −80°C. Vials were then thawed gently to room temperature and the PBS was aspirated off. Brains were then homogenized in 300 μl of pH 7.5, 2 mM EDTA, 10 mM EGTA, 20% glycerol, 0.1% NP-40, 50 mM β-mercaptoethanol, 100 μg/ml leupeptin, 2 μg/ml aprotinin, 40 μM Z-D-DCB [Z-Asp-2,6-dichlorobenzoyloxymethylketone], and 1 mM PMSF [phenylmethylsulfonyl fluoride], using a dounce homogenizer. Lysates were then transferred to 1.5-ml eppendorf tubes and were stored on ice until centrifugation (20,000 × g for 3 min). The supernatant was then transferred to a fresh tube containing 300 μl of 2× Laemmli buffer (125 mM Tris, pH 6.8, 4% sodium dodecyl sulfate [SDS], 10 mM β-mercaptoethanol, 20% glycerol, and 0.004% bromophenol blue). Brain lysates were boiled for 5 min and stored at −70°C. Proteins were electrophoresed overnight by SDS–polyacrylamide gel electrophoresis (PAGE) at a constant voltage of 70 V. Proteins were then electroblotted onto Hybond-C nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ). Immunoblots were blocked with 5% nonfat dry milk (NFDM) in Tris-buffered saline Tween-20 (TBST) for 2 h at room temperature before being probed with antibodies directed against IRF7 (Santa Cruz Biotechnology SC-9083, Santa Cruz, CA) and ifi202 (Santa Cruz Biotechnology SC-50358). Antibodies were diluted in 3% NFDM/TBST according to the manufacturer’s directions. All lysates were standardized for protein concentration with antibodies directed against actin (Oncogene, Cambridge, MA; catalog no. CP01) diluted 1:10,000 in 3% NFDM/TBST. Autoradiographs were quantitated by densitometric analysis using a Fluor-S MultiImager (Bio-Rad Laboratories, Hercules, CA). Statistical analysis was performed using Graphpad software (Instat).
RNA purification, microarray, and QPCR
In order to generate the most consistent data for our microarray studies, particular care was taken to standardize infections for RNA extractions. For each time point, two litters of mice that were born within 6 h of each other were used. Half of each litter was mock-infected and placed with one mother and the other half of each litter was T3D-infected and placed with the other mother. The litters used contained similar numbers of pups and all pups used were between 1.8 and 2.0 g. Intracerebral inoculations were performed between 48 and 54 h following birth. At 3, 6, and 8 days post infection, mice were sacrificed and brains from mock- and virus-infected mice were immediately placed in RNAlater and stored (short term) at −20°C. Approximately 160 mg of brain tissue (about half of brain after cerebellum removed) was homogenized in 1 ml of Qiazol (Qiagen RNeasy Lipid Tisssue Mini Kit; catalog no. 74804) for at least 20 strokes until completely emulsified. The emulsion was transferred to a clean 1.5-ml tube and set aside for at least 5 min before addition of 200 μl chloroform. The mixture was shaken for 15 s and set aside for 5 min before centrifuging at 12,000 × g for 15 min at 4°C. The upper aqueous phase was then carefully removed and transferred to a new tube. One volume of 70% ethanol (prepared with Diethylpyrocarbonate [DEPC] water) was then added and the solution was mixed before being transferred to an RNeasy column. RNA was purified following the manufacturers specifications and 0.5 to 1 μl of RNasin was added to the sample which was then stored at −80°C. Affymetrix 430.2 mouse whole-genome chips (Santa Clara, CA) were used for microarray analysis of RNA extracted from reovirus-infected and mock-infected brains. Individual Affymetrix chips were used for each reovirus-infected and mock-infected sample according to the manufacturer’s specifications. All microarray experiments and initial analysis was performed at the microarray core facility (University of Colorado–Denver). Analysis was performed using R statistical computation software and packages from Bioconductor open source software for bioinformatics (Gentleman et al, 2004). Prior to statistical analysis, two preprocessing steps involving normalization and gene filtering were performed. Raw data from array scans were processed using the Robust Multi-chip Average (RMA) normalization method to subtract a background value that is based on modeling the perfect match (PM) signal intensities as a convolution of an exponential distribution of signal and a normal distribution of nonspecific signal while ignoring the mismatch (MM) signal (Irizarry et al, 2003). After normalization, data were filtered using two criteria: (1) Affymetrix mRNA detection calls were used to exclude all probe sets with an “absent” call in all samples; and (2) transcripts that demonstrated little variation across all arrays were removed. This was performed by comparing the variance of the log-intensities for each gene with the median of all variance for the entire array. Genes not significantly more variable than the median were filtered out. Differentially regulated genes were analyzed using the Ingenuity Pathway Analysis software (IPA; Ingenuity Systems, Redwood City, CA). Ingenuity Pathways Analysis knowledge base is a curated database constructed based on scientific evidence based on hundreds and thousands of journal articles, textbooks, and other data sources. IPA software was used to define which well-characterized cell signaling pathways are most relevant during reovirus encephalitis. The significance (P values) of the association between the data set and the canonical pathway was measured by comparing the number of genes that are differentially regulated during reovirus encephalitis that participate in a pathway relative to the total number of genes in all pathway annotations stored in the Ingenuity Pathway knowledge base. Fisher’s exact test was used to calculate a P value determining the probability that the association between the genes in the data and the canonical pathway is explained by chance only.
For quantitative polymerase chain reaction (QPCR), cDNA was prepared from purified RNA by reverse transcribing 1.0 μg of each RNA sample using SABiosciences First Strand Kit (C-03; SABiosciences, Frederick, MD). The 20 μl final volume of cDNA was diluted with 1275 μl of water and added to 1275 μl of 2× SABiosciences RT2 SYBR green master mix (PA-010). Twenty-five microliters was dispensed into each well of a SABiosciences RT2 profile α/β interferon signaling (PAMM-14) or cytokine (PAMM-11) PCR array. Quantitative real-time polymerase chain reaction QRTPCR was carried out on a Bio-Rad Opticon2 machine (Bio-Rad).
Supplementary Material
Acknowledgments
This work was supported by grants 5R01NS050138 (K. L. T.) and 5R01NS051403 (K. L. T.) from the National Institute of Health and VA Merit funding (K. L. T.). K. L. Tyler is supported by the Reuler-Lewin Family Professorship. Microarrays experiments were performed at the University of Colorado microarray core, which is funded in part by Colorado CTSA grant RR025780 (NCCR).
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Beckham JD, Goody RJ, Clarke P, Bonny C, Tyler KL. Novel strategy for treatment of viral central nervous system infection by using a cell-permeating inhibitor of c-Jun N-terminal kinase. J Virol. 2007;81:6984–6992. doi: 10.1128/JVI.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham JD, Tuttle K, Tyler KL. Reovirus activates transforming growth factor beta and bone morphogenetic protein signaling pathways in the central nervous system that contribute to neuronal survival following infection. J Virol. 2009;83:5035–5045. doi: 10.1128/JVI.02433-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordignon J, Probst CM, Mosimann AL, Pavoni DP, Stella V, Buck GA, Satproedprai N, Fawcett P, Zanata SM, de Noronha L, Krieger MA, Duarte Dos Santos CN. Expression profile of interferon stimulated genes in central nervous system of mice infected with dengue virus type-1. Virology. 2008;377:319–329. doi: 10.1016/j.virol.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Bourne N, Scholle F, Silva MC, Rossi SL, Dewsbury N, Judy B, De Aguiar JB, Leon MA, Estes DM, Fayzulin R, Mason PW. Early production of type I interferon during West Nile virus infection: role for lymphoid tissues in IRF3-independent interferon production. J Virol. 2007;81:9100–9108. doi: 10.1128/JVI.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Beckham JD, Leser JS, Hoyt CC, Tyler KL. Fas-mediated apoptotic signaling in the mouse brain following reovirus infection. J Virol. 2009;83:6161–6170. doi: 10.1128/JVI.02488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, DeBiasi RL, Meintzer SM, Robinson BA, Tyler KL. Inhibition of NF-kappa B activity and cFLIP expression contribute to viral-induced apoptosis. Apoptosis. 2005;10:513–524. doi: 10.1007/s10495-005-1881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Meintzer SM, Moffitt LA, Tyler KL. Two distinct phases of virus-induced nuclear factor kappa B regulation enhance tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in virus-infected cells. J Biol Chem. 2003;278:18092–18100. doi: 10.1074/jbc.M300265200. [DOI] [PubMed] [Google Scholar]
- Clarke P, Meintzer SM, Wang Y, Moffitt LA, Richadson-Burns SM, Johnson GL, Tyler KL. JNK regulates the release of proapoptotic mitochondrial factors in reovirus-infected cells. J Virol. 2004;78:13132–13138. doi: 10.1128/JVI.78.23.13132-13138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Meintzer SM, Widmann C, Johnson GL, Tyler KL. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J Virol. 2001;75:11275–11283. doi: 10.1128/JVI.75.23.11275-11283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JL, Rodgers SE, Clarke P, Ballard DW, Kerr LD, Tyler KL, Dermody TS. Reovirus-induced apoptosis requires activation of transcription factor NF-kappaB. J Virol. 2000;74:2981–2989. doi: 10.1128/jvi.74.7.2981-2989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S, Xin H, Walter S, Choubey D. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem. 2001;276:298–305. doi: 10.1074/jbc.M007155200. [DOI] [PubMed] [Google Scholar]
- Danthi P, Coffey CM, Parker JS, Abel TW, Dermody TS. Independent regulation of reovirus membrane penetration and apoptosis by the mu1 phi domain. PLoS Pathog. 2008;4:e1000248. doi: 10.1371/journal.ppat.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBiasi RL, Clarke P, Meintzer SM, Jotte R, Kleinschmidt-Demasters BK, Johnson GL, Tyler KL. Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J Virol. 2003;77:8934–8947. doi: 10.1128/JVI.77.16.8934-8947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaye S, Paul S, Blakqori G, Minet M, Weber F, Staeheli P, Michiels T. Neurons produce type I interferon during viral encephalitis. Proc Natl Acad Sci U S A. 2006;103:7835–7840. doi: 10.1073/pnas.0602460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini MJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody RJ, Beckham JD, Rubtsova K, Tyler KL. JAK-STAT signaling pathways are activated in the brain following reovirus infection. J NeuroVirol. 2007;13:373–383. doi: 10.1080/13550280701344983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, Kelsall BL. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J Exp Med. 2007;204:1349–1358. doi: 10.1084/jem.20061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Jiang W, Chu T, Levine B. Identification of genes involved in the host response to neurovirulent alphavirus infection. J Virol. 2001;75:10431–10445. doi: 10.1128/JVI.75.21.10431-10445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A, Mashimo T, Frenkiel MP, Guenet JL, Lucas M, Despres P. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem. 2006;281:4624–4637. doi: 10.1074/jbc.M508649200. [DOI] [PubMed] [Google Scholar]
- Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC, Hill AV, Thursz MR. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4:411–419. doi: 10.1038/sj.gene.6363984. [DOI] [PubMed] [Google Scholar]
- Kominsky DJ, Bickel RJ, Tyler KL. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ. 2002;9:926–933. doi: 10.1038/sj.cdd.4401045. [DOI] [PubMed] [Google Scholar]
- Koterski J, Twenhafel N, Porter A, Reed DS, Martino-Catt S, Sobral B, Crasta O, Downey T, DaSilva L. Gene expression profiling of nonhuman primates exposed to aerosolized Venezuelan equine encephalitis virus. FEMS Immunol Med Microbiol. 2007;51:462–472. doi: 10.1111/j.1574-695X.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- O’Donnell SM, Hansberger MW, Connolly JL, Chappell JD, Watson MJ, Pierce JM, Wetzel JD, Han W, Barton ES, Forrest JC, Valyi-Nagy T, Yull FE, Blackwell TS, Rottman JN, Sherry B, Dermody TS. Organ-specific roles for transcription factor NF-kappaB in reovirus-induced apoptosis and disease. J Clin Invest. 2005;115:2341–2350. doi: 10.1172/JCI22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell SM, Holm GH, Pierce JM, Tian B, Watson MJ, Chari RS, Ballard DW, Brasier AR, Dermody TS. Identification of an NF-kappaB-dependent gene network in cells infected by mammalian reovirus. J Virol. 2006;80:1077–1086. doi: 10.1128/JVI.80.3.1077-1086.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhaus SM, Smith RL, Clayton GH, Dermody TS, Tyler KL. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J Virol. 1997;71:2100–2106. doi: 10.1128/jvi.71.3.2100-2106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggioli GJ, DeBiasi RL, Bickel R, Jotte R, Spalding A, Johnson GL, Tyler KL. Reovirus-induced alterations in gene expression related to cell cycle regulation. J Virol. 2002;76:2585–2594. doi: 10.1128/JVI.76.6.2585-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Richardson-Burns SM, Kominsky DJ, Tyler KL. Reovirus-induced neuronal apoptosis is mediated by caspase 3 and is associated with the activation of death receptors. J NeuroVirol. 2002;8:365–380. doi: 10.1080/13550280260422677. [DOI] [PubMed] [Google Scholar]
- Ryman KD, Klimstra WB, Nguyen KB, Biron CA, Johnston RE. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J Virol. 2000;74:3366–3378. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol CA, Munoz-Jordan JL, Abel K, Rosado LC, Pantoja P, Giavedoni L, Rodriguez IV, White LJ, Martinez M, Arana T, Kraiselburd EN. Transcriptional activation of interferon-stimulated genes but not of cytokine genes after primary infection of rhesus macaques with dengue virus type 1. Clin Vaccine Immunol. 2007;14:756–766. doi: 10.1128/CVI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Bhattacharya B, Puri RK, Maheshwari RK. Venezuelan equine encephalitis virus infection causes modulation of inflammatory and immune response genes in mouse brain. BMC Genomics. 2008;9:289. doi: 10.1186/1471-2164-9-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y, Potula R, Pinson D, Adany I, Li Z, Day J, Buch E, Segebrecht J, Villinger F, Liu Z, Huang M, Narayan O, Buch S. Microarray analysis of cytokine and chemokine genes in the brains of macaques with SHIV-encephalitis. J Med Primatol. 2003;32:229–239. doi: 10.1034/j.1600-0684.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M, Myers TG, Wilson MA, Kindt TJ, Paweska JT, Burt FJ, Leman PA, Swanepoel R. Gene expression in mice infected with West Nile virus strains of different neurovirulence. Virology. 2005;342:119–140. doi: 10.1016/j.virol.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Campbell IL. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J Virol. 2005;79:8295–8302. doi: 10.1128/JVI.79.13.8295-8302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warke RV, Xhaja K, Martin KJ, Fournier MF, Shaw SK, Brizuela N, de Bosch N, Lapointe D, Ennis FA, Rothman AL, Bosch I. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol. 2003;77:11822–11832. doi: 10.1128/JVI.77.21.11822-11832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, D’Souza S, Jorgensen TN, Vaughan AT, Lengyel P, Kotzin BL, Choubey D. Increased expression of Ifi202, an IFN-activatable gene, in B6. Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J Immunol. 2006;176:5863–5870. doi: 10.4049/jimmunol.176.10.5863. [DOI] [PubMed] [Google Scholar]
- Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol. 2009;83:2178–2187. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.