Abstract
Acrolein is an α,β-unsaturated aldehyde that is a major environmental pollutant, as well as a product of cellular metabolism. DNA bases react with acrolein to form two regioisomeric exocyclic guanine adducts, namely γ-hydroxy-propanodeoxyguanosine (γ-OH-PdG) and its positional isomer α-hydroxy-propanodeoxyguanosine (α-OH-PdG). The γ-OH-PdG isomer adopts a ring-opened conformation with minimal structural perturbation of the DNA host duplex. Conversely, the α-OH-PdG isomer assumes a ring-closed conformation that significantly disrupts Watson-Crick base-pair alignments within the immediate vicinity of the damaged site. We have employed a combination of calorimetric and spectroscopic techniques to characterize the thermodynamic origins of these lesion-induced structural alterations. Specifically, we have assessed the energetic impact of α-OH-PdG centered within an 11-mer duplex by hybridizing the adduct-containing oligonucleotide with its complementary strand harboring a central base N [where N = C or A], yielding a pair of duplexes containing the nascent lesion (α-OH-PdG·C) or mismatched adduct (α-OH-PdG·A), respectively. Our data reveal that the nascent lesion is highly destabilizing, while its mismatched counterpart partially ameliorates α-OH-PdG-induced destabilization. Collectively, our data provide energetic characterizations of the driving forces that modulate error-free versus error-prone DNA translesion synthesis. The biological implications of our findings are discussed in terms of energetically probing acrolein-mediated mutagenicity versus adduct-induced genotoxicity.
Keywords: acrolein adducts, thermodynamic stability, calorimetry, mutagenicity, genotoxicity
INTRODUCTION
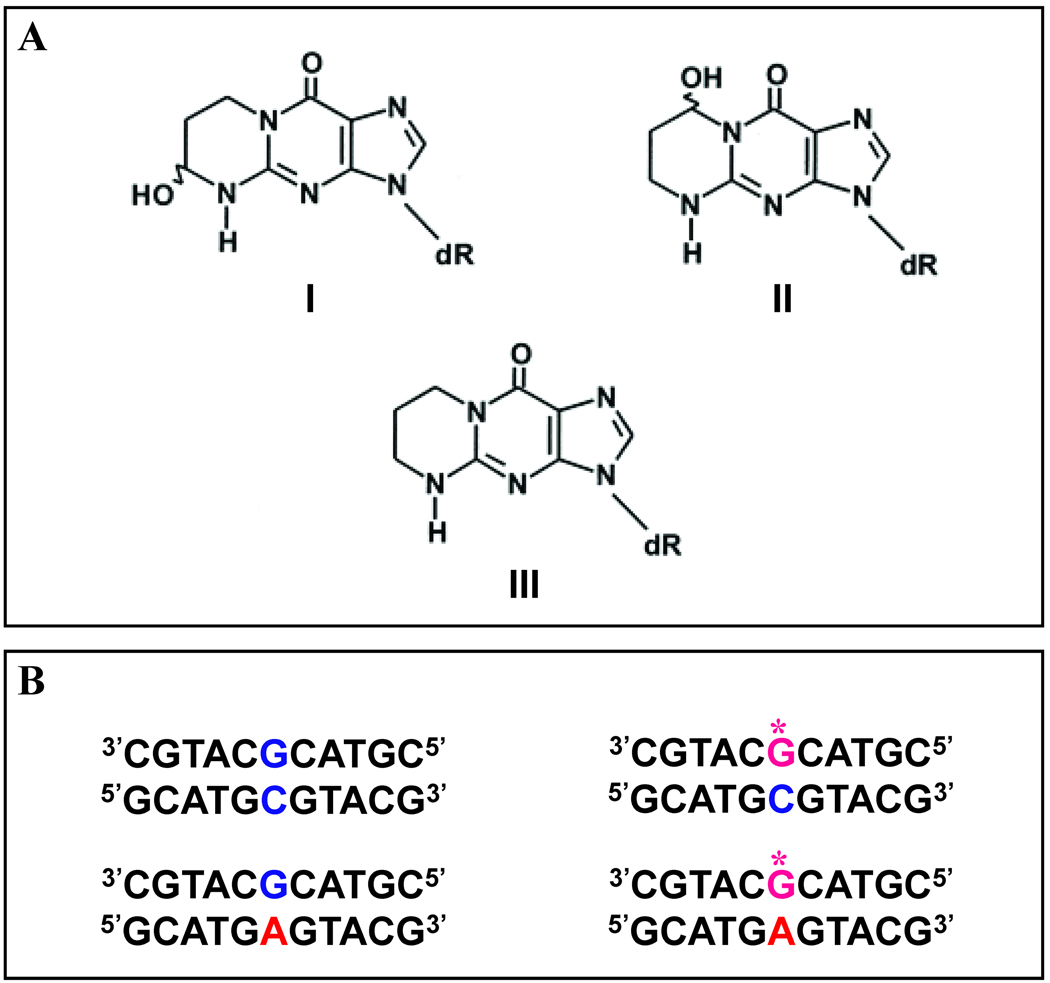
Acrolein represents one of the most widely distributed environmental pollutants and is an endogenous product of lipid peroxidation.1 This electrophilic compound is highly reactive towards specific sites within biomacromolecules (e.g. DNA bases), potentially resulting in deleterious effects to the host cell.2 Numerous reports relate acrolein to the harmful effects of tobacco smoke and industry-generated pollutants. Such results have led to chemoprevention initiatives on these compounds.3 Recent biophysical studies have probed the impact of this electrophile at the molecular level, with particular attention focused on its properties as a DNA damaging agent, as well as the consequences of such damage on replication and repair. The bifunctional acrolein electrophile reacts with deoxyguanosine bases, resulting in the formation of two regioisomeric propanodeoxyguanine adducts, namely the major gamma-hydroxy-propanodeoxyguanosine (γ-OH-PdG) and its positional isomer alpha-hydroxy-propanodeoxyguanosine (α-OH-PdG).4 The chemical structures of α-OH-PdG (I), γ-OH-PdG (II), and the unsubstituted chemically stable analog 1,N2-propanodeoxyguanosine (III) are presented in Figure 1.
Figure 1. Chemical Structures of Acrolein Adducts and DNA Host Duplexes.
A. The regioisomers of hydroxy-propanodeoxyguanosine designated as α-OH-PdG (I) and γ-OH-PdG (II) compared with the stable analog PdG (III). B. The exocyclic damaged G (α-OH-PdG) represented by G* is embedded as the central base within an 11-mer deoxyoligonucleotide that is hybridized with 5’GCATGCGTACG3’ or 5’GCATGAGTACG3’ to yield the α-OH-PdG·C or α-OH-PdG·A duplexes, respectively. The 11-mer host duplexes designated as the G·C Parent and G·A Mismatch appear on the left, while the damaged α-OH-PdG·C and α-OH-PdG·A duplexes designated as G*C and G*A appear on the right.
Initial studies on the impact of acrolein DNA adducts have been hampered by the inherent instability of the nascent lesions, which has precluded investigations at the molecular level. Consequently, prior efforts to characterize these adducts have relied on extensive use of the stable analog (hereby designated as PdG) that shares the exocyclic ring but lacks the hydroxyl group of the naturally occurring acrolein-derived adducts. The PdG analog is routinely employed as a representative model lesion for elucidation of the biological,5,6 structural,7–9 and thermodynamic properties10 of acrolein adducts, furnishing valuable information to delineate the overall impacts of acrolein adducts at the DNA level. Solution structures of DNA duplexes containing the PdG analog generally support the proposition that these exocyclic adducts primarily disrupt base pairing at the damaged site, while maintaining the overall B-DNA conformation.7–9 Duplexes harboring a PdG lesion form regular right-handed helices with Watson-Crick hydrogen-bonding observed throughout the canonical base pair alignments, including those residues flanking the lesion site. Despite the wealth of structural information gleaned from such studies, the nascently generated hydroxylated forms comprising α-OH-PdG and γ-OH-PdG possess unique isomer-specific properties that cannot always be inferred on the basis of prior findings with the PdG analog.
Recent advances in nucleic acid synthetic techniques have resulted in the successful production of chemically stable γ-OH-PdG and α-OH-PdG11–16 adducts. Availability of the natural acrolein adduct has generated renewed interest in characterizing the lesion-specific biophysical properties of both regioisomers.17–21 Elucidation of solution structures for γ-OH-PdG17 and α-OH-PdG22 has provided insights into the structural origins of acrolein-induced perturbations of duplex DNA, and the potential consequences of such alterations on DNA replication. The γ-OH-PdG regioisomer adopts an open-ring conformation that is fully accommodated within the B-DNA helix resulting in minimal structural perturbation.17 Consequently, this adduct is neither genotoxic nor mutagenic when bypassed by polymerases and generally undergoes error-free replication.13 Nonetheless, this regioisomer is capable of cross-linking with proteins,23 a property that is characteristic of the deleterious effects posed by acrolein adducts. In contrast with its positional isomer, structural evidence suggests that the α-OH-PdG regiomer adopts a closed conformation, which disrupts canonical base pairing at the damaged site,22 yet maintains the overall B-DNA conformation in a manner consistent with previous findings on the PdG model system. Moreover, recent studies reveal that this isomer effectively blocks polymerase-mediated DNA synthesis, a property that accounts for its genotoxicity.4
The in vivo toxicity of acrolein has been the subject of intense debate and considerable controvery.24 While a number of studies suggest that acrolein is mutagenic,25 the experimental variability observed amongst in vivo assays has precluded unequivocal demonstration of acrolein-mediated mutagenicity.26 Investigations have conclusively demonstrated that acrolein interacts with DNA bases in vitro, resulting in the formation of 1,N-2-dG exocyclic adducts. The extent of genotoxicity and miscoding properties may be a function of the polymerase and sequence contexts employed. Consequently, the resultant exocyclic damaged G-bases might be miscoded during replication, inducing G•C → T•A transversions and G•C → A•T transitions. Harmful effects at the DNA level are only part of a host of cellular mechanisms that may contribute to the potential mutagenicity and/or carcinogenicity of acrolein, whether derived from tobacco, industrial, or endogenous metabolic sources.
The present study evaluates the energetic impact of the α-OH-PdG acrolein lesion, including the modulation of adduct-induced perturbations as a function of opposite base identity. Our ultimate goal is to define the biological implications of these energetic consequences by assessing the ability of the replication/repair machinery to recognize, bypass and/or repair such defects. Our study is designed to elucidate the energetic origins of α-OH-PdG-induced perturbations on the double helical structure22,27 and to evaluate the forces associated with the ability of α-OH-PdG to base pair with dC and/or dA. To this end, we have pursued a combined calorimetric and spectroscopic approach to thermodynamically characterize α-OH-PdG-containing duplexes employing identical parent sequences and similar solution conditions to those reported in parallel structural studies.22,27 We evaluate the energetic origins underlying opposite base effects by comparing the thermodynamic stability of acrolein adduct-containing duplexes relative to their corresponding canonical and “undamaged” mismatched duplexes. Correlation of our energetic data with the structural impacts of this lesion facilitates identification of structural-energetics properties that may account for the consequences of acrolein-mediated DNA damage in vivo in terms of DNA replication, mutagenesis, and genotoxicity.
MATERIALS AND METHODS
Oligonucleotide Synthesis and Purification
An 11-mer deoxyribonucleotide harboring a central acrolein adduct was prepared as described elsewhere.16,20 Four 11-mer duplexes were formed by annealing a single strand comprised of the sequence d(CGTACXCATGC) in which X = G or α-OH-PdG with its corresponding complementary strand d(GCATCNGTACG) in which N = C or A. The resultant parent, mismatch, and adduct-containing duplexes are designated as G·C, G·A, α-OH-PdG·C, and α-OH-PdG·A, respectively.
Circular Dichroism
The global conformation of the G·C parent and G·A mismatch duplexes as well as the impact of the α-OH-PdG adducts on DNA structure were assessed by circular dichroism (CD) spectroscopy. Circular dichroism spectra were acquired at 0 °C on an AVIV Model 400 CD spectropolarimeter (Aviv Biomedical, Inc., Lakewood, NJ) employing a 1.0 mm quartz cuvette and DNA total strand concentration (CT) of 50.0 µM. The global conformation of each duplex was evaluated by recording the molar ellipticity over the wavelength range of 200 to 350 nm at 0.5 nm increments following signal averaging for 10 seconds. The resultant CD spectra were buffer subtracted and concentration normalized to yield molar ellipticity.
Duplex Thermal and Thermodynamic Stability
Optical Methods
The thermal and thermodynamic stability of the duplexes was evaluated by temperature-dependent spectroscopic techniques. Temperature-dependent UV melting experiments were performed on an Aviv Model 14 UV/Vis spectrophotometer (Aviv Biomedical Inc., Lakewood, NJ) employing a minimum of five DNA duplex standards spanning a total strand concentration (CT) range of 1.0 µM – 50.0 µM. Samples in quartz cuvettes of 0.1 to 1.0 cm path length were heated in the thermostatted sample compartment over the temperature range of 0 – 95.0 °C. The absorbance at 260 nm was recorded at 0.5 °C increments following signal integration for 10 sec to monitor the hyperchromicity change as a function of temperature. The van’t Hoff duplex dissociation enthalpies (ΔHVH) have been determined by both shape analysis (ΔHVHshape) as described elsewhere,28 and by evaluating the concentration-dependence of the transition temperature (Tm) via application of Equation 1:
| (1) |
Substituting the numerical value of 2 for the molecularity (n) of each duplex, the van’t Hoff duplex dissociation enthalpy (ΔHVHslope ) may be calculated from the resultant slope.
Differential Scanning Calorimetry
A model-independent duplex dissociation enthalpy for the thermally induced order-disorder transition was derived from differential scanning calorimetry (DSC). DNA standards at a duplex concentration of 100 µM were scanned at a programmed rate of 1.0 °C·min−1 in a VP-DSC (MicroCal, LLC, Northampton, MA) over the temperature range of 0 – 95.0 °C at 0.1 °C increments. The DSC profiles were buffer baseline subtracted, concentration normalized, and the resultant endotherm integrated following assignment of pre- and post-transition baselines. An average calorimetric enthalpy (ΔHcal) was calculated from at least six independently analyzed melting profiles for the G·C parent, G·A mismatch, α-OH-PdG·C, and α-OH-PdG·A duplexes.
Thermodynamic Analysis Employing a Combination of Optical and Calorimetric Approaches
In order to derive a self-consistent dissociation free energy (ΔG), the effective molecularity (neff) for each duplex was calculated by substituting the calorimetrically determined enthalpy (ΔHcal) for the value of ΔH in Equation 1 and solving for n as described previously.29–31 The dissociation free energy (ΔG) may therefore be calculated by combining the calorimetrically measured duplex dissociation enthalpy (ΔH) with the neff and transition temperature (Tm) via application of Equation 2:
| (2) |
The duplex dissociation entropy (ΔS) was determined via application of the standard thermodynamic relation in Equation 3:
| (3) |
The thermodynamic parameters calculated at a reference temperature of 25.0 °C assumes a zero heat capacity (i.e., ΔCp ~ 0). Under conditions of non-zero heat capacity, the thermodynamic parameters extrapolated to a common reference temperature (T) may be obtained by incorporating the respective values of ΔCp and neff into the following relations for ΔH (Equation 4), ΔS (Equation 5), and ΔG (Equation 6):
| (4) |
| (5) |
| (6) |
The heat capacity corrected data is evaluated in terms of α-OH-PdG adduct-induced differential destabilization (Δ ΔG, Δ ΔH, and ΔTΔS) relative to either the parent G·C or the mismatch G·A duplexes at common reference temperatures.
RESULTS AND DISCUSSION
Experimental Strategy
We have selected a family of 11-mer DNA host duplexes in which the core sequence duplicates that of the 13-mer oligonucleotides used to characterize the impact of single base bulges,31 mismatches,29 and lesions.10,29,30,32 The identical 11-mer sequence has been utilized in parallel structural studies of a comparable number of lesions and defects,17,33 including acrolein adducts.17,22,27 As depicted in Figure 1B, the α-OH-PdG adduct is embedded as the central residue within the 11-mer oligonucleotide and hybridized with complementary strands containing dG or dA as the counterbase. The resultant duplexes model the nascent lesion/error-free product (α-OH-PdG·C) and the most prominent miscoding synthesis replication product (α-OH-PdG·A), respectively. In the sections that follow, we describe the energetic impact of α-OH-PdG on DNA duplex stability and discuss the utility of such investigations in interpreting the biological consequences of acrolein adducts at the replication and repair levels.
Impact of Acrolein Adducts on Global DNA Conformation
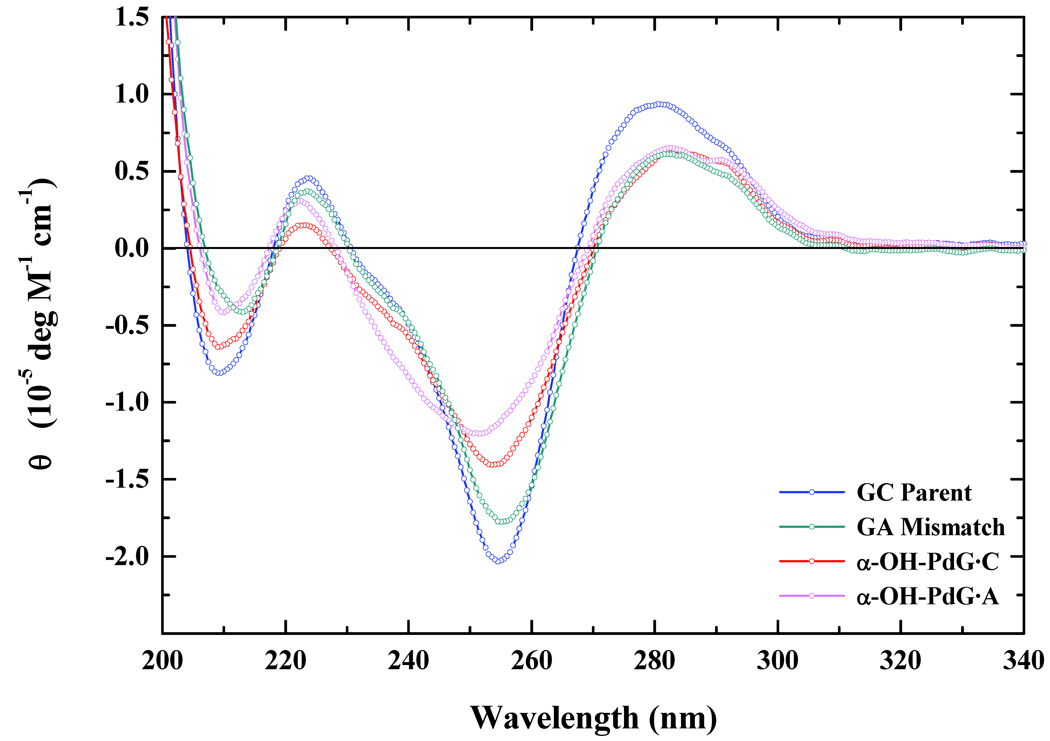
The impact of α-OH-PdG on global DNA conformation may be evaluated by comparing CD-spectra of the Watson Crick parent G·C, the corresponding mismatch G·A, the α-OH-PdG·C, and the α-OH-PdG·A duplexes as illustrated in Figure 2. The acrolein adduct is either paired with dC to form the nascent lesion (α-OH-PdG·C), or paired with dA to mimic an error-prone polymerase-mediated mutagenic intermediate (α-OH-PdG·A). Examination of the CD spectra reveal that all four duplexes exhibit structural features characteristic of the B-DNA conformation, as evidenced by the typical minima at ~ 240–260 nm and maxima at ~ 270–280 nm. The overall magnitudes of the ellipticities are reduced for the lesion-containing duplexes, particularly for α-OH-PdG·A. Our CD data are generally consistent with NMR studies on the stable PdG analog7–9 and the α-OH-PdG lesion paired with dC.22 The solution conformation of α-OH-PdG paired with dA is presented in the companion paper.27 Despite local perturbations observed in the closed-ring forms of PdG and α-OH-PdG, the overall B-DNA conformation is generally maintained. In fact, differences noted for α-OH-PdG·A are minimized relative to the corresponding G·A mismatch, an indication that these additional structural alterations can be attributed partially to the mismatch per se.
Figure 2. Circular Dichroism Spectra of Duplexes Harboring the Acrolein Adduct.
Comparison of normalized circular dichroism spectra expressed in the form of molar ellipticity for the α-OH-PdG·C (red) and α-OH-PdG·A (magenta) duplexes relative to the corresponding G·C Parent (blue) and G·A Mismatch (navy) 11-mers.
Impact of Acrolein Adducts on DNA Duplex Energetics: α-OH-PdG Reduces Thermal and Thermodynamic Stability
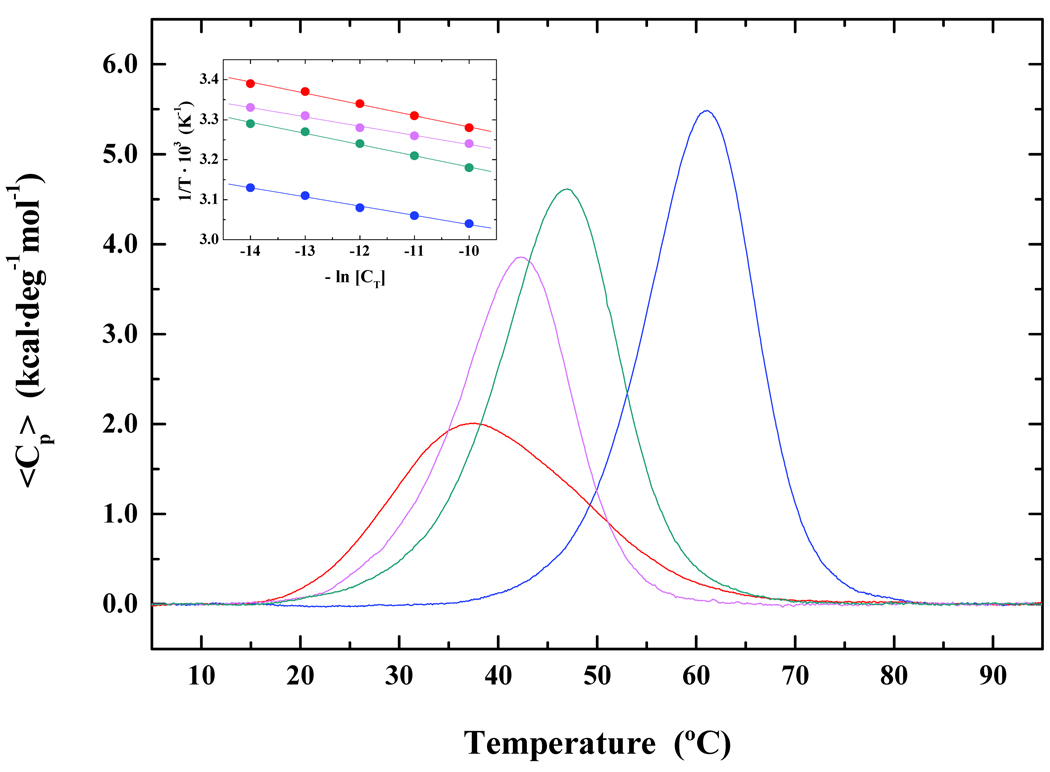
The impact of acrolein adducts on DNA duplex energetics has been investigated primarily via spectroscopic approaches,10 revealing that the PdG lesion reduces the thermal and thermodynamic stability of the host duplex. This study represents the first calorimetric assessment of the α-OH-PdG-induced impact on DNA duplex dissociation energetics. Figure 3 presents a comparative analysis of the calorimetrically measured excess heat capacity profiles for the parent G·C, mismatch G·A, α-OH-PdG·C, and α-OH-PdG·A duplexes from which we derive the respective dissociation enthalpies (ΔHcal). The inset of Figure 3 presents a van’t Hoff analysis of the optically-derived concentration and temperature-dependent dissociation profiles from which we derive ΔHvH for each duplex.
Figure 3. Impact of an Acrolein Adduct on Thermodynamic Stability of the DNA Host Duplex.
Comparison of excess heat capacity profiles determined for the α-OH-PdG·C (red) and α-OH-PdG·A (magenta) duplexes relative to the corresponding G·C parent (blue) and G·A mismatch (navy) 11-mers. The inset furnishes van’t Hoff analyses of the optically-derived concentration-dependent dissociation profiles for these four duplexes. The “undamaged” G·A mismatch and adduct-containing 11-mers paired with dA or dC significantly disrupt cooperative duplex dissociation with differential destabilization ranked as follows: G·A < α-OH-PdG·A < α-OH-PdG·C.
Our combined optical and calorimetric data are generally consistent with prior observations on PdG in that the α-OH-PdG adduct reduces both the thermal and thermodynamic stability of the host duplex, albeit with a magnitude significantly higher than that of PdG (refer to Table I). Specifically, energetic destabilization imparted by the nascent α-OH-PdG lesion (i.e., ΔTm= −23.0 °C and ΔΔG = −8.2 kcal·mol−1) is nearly double that measured previously for PdG (i.e., ΔTm = −12.9 °C and ΔΔGvH = −3.8 kcal·mol−1). Despite the similar DNA duplex sequences employed in both studies, there are a number of experimental variables that may contribute to the disparities noted between these sets of thermodynamic data. These variables include differences in adduct chemical structures (i.e., PdG versus α-OH-PdG), oligonucleotide length (i.e., 13-mer versus 11-mer), ionic strength (i.e., 1.0 M versus 0.1 M), and flanking residues (i.e., GXG versus CXC).
Table I.
A. Thermodynamic Dissociation Parameters of Canonical, Mismatch, and Adduct-Containing Duplexes. B. Impact of α-OH-PdG and Opposite Base on the Differential Thermodynamic Destabilization.
| A | Duplex Acronym | Tm (°C) |
ΔG (kcal·mol−1) |
ΔHcal (kcal·mol−1) |
TΔS (kcal·mol−1) |
|---|---|---|---|---|---|
| Canonical G·C | 60.5 | 13.2 | 79.3 | 66.1 | |
| Mismatch G·A | 47.0 | 10.5 | 70.0 | 59.5 | |
| α-OH-PdG·C | 37.6 | 5.0 | 47.4 | 42.4 | |
| α-OH-PdG·A | 42.2 | 6.4 | 58.2 | 51.8 | |
| B | Residue Substitution | ΔTm (°C) |
ΔΔG (kcal·mol−1) |
ΔΔH (kcal·mol−1) |
ΔTΔS (kcal·mol−1) |
| G·C to α -OH-PdG·C | − 22.9 | − 8.2 | − 31.9 | − 23.7 | |
| G·C to α -OH-PdG·A | − 18.3 | − 6.8 | − 21.1 | − 14.3 | |
| G·C to G·A | − 13.5 | − 2.7 | − 9.3 | − 6.6 | |
| G·A to α -OH-PdG·A | − 4.8 | − 4.1 | − 11.8 | − 7.7 | |
| α -OH-pdG·C to α -OH- PdG·A |
+ 4.6 | + 1.4 | − 10.8 | − 9.4 | |
A. Standard deviations for Tm, ΔG, ΔH, and TΔS are within 0.1 °C, 0.2 kcal·mol−1, 1.0 kcal·mol−1, and 1.0 kcal·mol−1, respectively. The thermodynamic parameters are reported at 25.0 °C following extrapolation assuming ΔCp = 0. B. The values of ΔTm, ΔΔG, ΔΔH, and ΔTΔS are calculated by subtracting the respective dissociation parameters of the specified duplexes.
Studies on DNA damage including abasic sites30, mismatches34, and bulges31, corroborate the fact that the impact of lesions and defects are highly sequence-context dependent. Moreover, lesions embedded within GXG contexts (i.e., purine-purine) are generally less destabilizing than those within CXC constructs (i.e., pyrimidine-pyrimidine). Given an identical duplex sequence, a G·A mismatch within the GGG/CAC triplet is less destabilizing (i.e., ΔTm = −6.2 °C and ΔΔG = −1.3 kcal·mol−1)10 than the corresponding A·G mismatch within the GAG/CGC domain (i.e., ΔTm = −11.2 °C and ΔΔG = −6.8 kcal·mol−1).29 In fact, algorithms predicting the energetic impact of a G·A mismatch within the identical 11-mer duplex sequence studied herein estimate a ΔΔG of 1.5 kcal·mol−1 in favor of GGG/CAC relative to GAG/CGC for 0.1 M ionic strength.35,36 Collectively, our findings allow us to rationalize the apparent disparities associated with the relative impacts of PdG versus α-OH-PdG, while underscoring the importance of systematically evaluating the impact of a lesion embedded within multiple sequence environments.
The α-OH-PdG Lesion Disrupts the Cooperativity of Duplex Dissociation
The cooperativity of duplex dissociation in the absence and presence of a lesion yields insight into the overall impact of such a modification on normal communication along the double helical structure. This assessment allows one to evaluate the veracity of the two-state assumption, namely that the ratio of the van’t Hoff and calorimetric enthalpies approaches unity. The cooperativity ratios and effective molecularities for each of the duplexes studied herein are summarized in Table II. Inspection of the data reveals that the canonical G·C and mismatch G·A duplexes fulfill the general criteria of “two-state” dissociation (i.e. ΔHVH/ΔHcal ~ 1.0). Conversely, the adduct-containing duplexes depart significantly from idealized melting behavior in a manner that is opposite-base dependent with ratios of 1.4 and 1.6 determined for α-OH-PdG·A and α-OH-PdG·C, respectively. These findings suggest that the deleterious effects of an exocyclic α-OH-PdG adduct propagates beyond the immediate vicinity of the damaged site. Our results are consistent with published data on a number of other lesions and adducts in that damaged sites are notorious for causing deviations from idealized bimolecular dissociation behavior due to lesion-induced perturbations and consequent disruption of the canonical duplex structure.29,32,37,38 As such, duplex modification generally results in the population of additional states while breaching the chain communication required for ideal melting behavior. These populated states may represent biologically relevant targets that might impose additional challenges in terms of replication and repair.
Table II.
A. Van’t Hoff and Calorimetric Duplex Dissociation Enthalpies of the Canonical G·C, Mismatch G·A, α-OH-PdG·C, and α-OH-PdG·A. B. DNA Host Duplex Cooperativity Assessed by the Ratio of van’t Hoff Enthalpies (ΔHvHshape/ΔHvHslope), van’t Hoff/Calorimetric Enthalpies (ΔHvHslope/ΔHcal), and Effective Molecularities (neff).
| A | Duplex Acronym | ΔHvH shape (kcal·mol−1) |
ΔHvH slope (kcal·mol−1) |
ΔHcal (kcal·mol−1) |
|---|---|---|---|---|
| Canonical G·C | 89.4 | 86.4 | 79.3 | |
| Mismatch G·A | 72.8 | 67.1 | 70.0 | |
| α-OH-PdG·C | 59.1 | 72.5 | 47.4 | |
| α-OH-PdG·A | 70.1 | 82.8 | 58.2 | |
| B | Duplex Acronym |
ΔHvHshape / ΔHvHslope | ΔHvHslope / ΔHcal | neff |
| Canonical G·C | 1.0 | 1.1 | 1.9 | |
| Mismatch G·A | 1.1 | 1.0 | 2.0 | |
| α-OH-PdG·C | 0.8 | 1.5 | 1.6 | |
| α-OH-PdG·A | 0.8 | 1.4 | 1.6 | |
The van’t Hoff dissociation enthalpies (ΔHvh shape and ΔHvh slope) are calculated from UV melting profiles, the calorimetric dissociation enthalpy (ΔHcal) is determined from the excess heat capacity profile, and the effective molecularity (neff ) is calculated by substituting the respective parameters into Equation 1 (Refer to Materials and Methods).
Impact of Heat Capacity Changes (ΔCp) on α-OH-PdG-induced Perturbations in Duplex Dissociation Energetics
The thermodynamic parameters reported in Table I are reflective of non-heat capacity corrected duplex dissociation enthalpies, thereby facilitating direct comparison with published values that generally neglect ΔCp effects. Since nucleic acid association/dissociation processes are accompanied by measurable ΔCp effects, such corrections should be applied when extrapolating duplex dissociation energetics to common reference temperatures (e.g., 25 °C). While incorporating heat capacity corrections, one must be cognizant of the fact that oligonucleotide duplexes pose a significant challenge in terms of the uncertainties associated with accurate determination of ΔCp via traditional extrapolations of pre- and post-transition baselines for the respective heat capacity profiles.39 We recently determined an estimate of ΔCp for canonical 12 and 13-mer duplexes comprised of a similar sequence context to the 11-mer duplex employed herein.31 Assuming an average ΔCp of 70 cal·mol−1·deg−1·bp−1, the experimentally measured calorimetric enthalpies and entropies reported for the 11-mer duplexes in Table I are adjusted by a ΔCp of ~0.77 cal·mol−1·deg−1 via application of Equations 4, 5, and 6, as described in Materials and Methods. The resultant heat capacity corrected dissociation enthalpies, entropies, and free energies for the parent G·C, mismatch G·A, α-OH-PdG·C, and α-OH-PdG·A duplexes are summarized in Table III. Comparison and evaluation of the duplex dissociation energetics in Tables I and III reveals that incorporation of the ΔCp-correction partially compresses the data, yet does not dramatically impact the differential ΔΔG values or their trends as illustrated in Figure 4.
Table III.
A. Thermodynamic Dissociation Parameters of Canonical, Mismatch, and Adduct-Containing Duplexes Extrapolated to 25.0 °C Following ΔCp Corrections. B. Impact of α-OH-PdG and Opposite Base on the ΔCp-Corrected Differential Thermodynamic Destabilization.
| A | Duplex Acronym |
ΔG (kcal·mol−1) |
ΔHcal (kcal·mol−1) |
TΔS (kcal·mol−1) |
|---|---|---|---|---|
| Canonical G·C | 12.6 | 53.2 | 40.6 | |
| Mismatch G·A | 9.9 | 53.8 | 43.9 | |
| α-OH-PdG·C | 7.1 | 38.1 | 31.0 | |
| α-OH-PdG·A | 8.2 | 45.5 | 37.4 | |
| B | Duplex Acronym |
ΔΔG (kcal·mol−1) |
ΔΔH (kcal·mol−1) |
ΔTΔS (kcal·mol−1) |
| G·C to α-OH-PdG·C | − 5.5 | − 15.1 | − 9.6 | |
| G·C to α-OH-PdG·A | − 4.4 | − 7.6 | − 3.3 | |
| G·C to G·A | − 2.7 | + 0.6 | + 3.3 | |
| G·A to α-OH-PdG·A | − 1.7 | − 8.3 | − 6.6 | |
| α-OH-pdG·C to α-OH-PdG·A |
+ 1.1 | + 7.4 | + 6.3 | |
A. The thermodynamic parameters are reported at 25.0 °C following ΔCp correction as described in Materials and Methods. B. The values of ΔTm, ΔΔG, ΔΔH, and ΔTΔS are calculated by subtracting the respective reference duplex dissociation parameter from that of the damaged and/or mismatched duplex.
Figure 4. Impact of α-OH-PdG and Opposite Base on non-ΔCp-corrected (light blue) and ΔCp-corrected (dark blue) Thermodynamic Parameters.
Differential thermodynamic destabilization of the G·C Parent sorted by decreasing order of energetic impact via replacement of dG by α-OH-PdG and/or dC by dA as specifically noted. The adduct-induced impacts on dissociation free energies are expressed as - ΔΔG to improve clarity and sorted on the basis of decreasing ΔCp-corrected ΔΔG. Replacement of dC with dA alleviates the overall energetic impact of α-OH-PdG irrespective of ΔCp-corrections, as noted by the observation that - ΔΔG < 0 for α-OH-PdG·C → α-OH-PdG·A.
Energetic Impacts of Lesions: Sequence-Context and Opposite Base Dependence
The importance of investigating sequence context and opposite base dependent energetic impacts of lesions and defects has long been recognized.29–32,34,37 In fact, investigators have invoked the well established “A-rule” as a major determinant underlying the degree of mutagenicity elicited by dG lesions and adducts,40 which is strongly dependent on sequence context.40–42 This realization underscores the necessity of probing specific lesion impacts within various sequence environments, particularly when evaluating in vivo effects amongst a multitude of sequences in the genome. Such evaluations are necessary to delineate lesion-induced properties that are often overlooked due to the averaging of sequence context variations, thereby effectively masking elucidation of measurable effects in vivo.
While the present study does not specifically address the impact of acrolein adducts within multiple sequence environments, we evaluate the energetic penalty induced by a nascent lesion and explore how such damage is perceived by polymerases during replication. The sequence context selected in this study is part of a continuing effort to characterize the impact of exocyclic and oxidative lesions, both energetically10,29–32,37,43,44 and structurally.17,45–47 In the sections that follow, we compare the relative stabilities of the G·C parent, G·A mismatch, α-OH-PdG·C, and α-OH-PdG·A duplexes to gain specific insights regarding potential genotoxic and/or mutagenic effects in vivo. We further evaluate α-OH-PdG within the context of other exocyclic or oxidative lesions previously characterized in terms of sequence-context dependent energetic impacts.
Impact of Counter Base and Flanking Residues on the Magnitude of Acrolein-Mediated Duplex Destabilization
Our studies on the energetic impact of α-OH-PdG are generally consistent with the findings of previous reports on PdG in that both lesions significantly destabilize the respective DNA host duplexes. The primary difference is that the magnitude of thermal and thermodynamic destabilization observed for the PdG analog is essentially insensitive to the identity of the base positioned opposite the adduct.10 In fact, published data reveals that there is minimal impact when exchanging the counter bases dC and dA opposite PdG, as both duplexes exhibit nearly identical thermal destabilization with relative ΔTm differences of ~0.9 °C. In terms of PdG-induced thermodynamic destabilization of the parent duplex, an interesting observation is that the free energy cost due to a G·A mismatch (i.e., G·C → G·A) is equivalent to that observed for the PdG·C → PdG·A “mutation” (i.e., ΔΔG = −1.3 kcal·mol−1).10 Such findings support the hypothesis that error-prone synthesis is not energetically favorable relative to error-free synthesis and vice-versa.
In contrast with the impact observed for a PdG analog as a function of opposite base, our data reveal that the α-OH-PdG adduct actually stabilizes the resultant duplex when the damaged base is paired with dA versus dC. Specifically, there is an alleviation of the α-OH-PdG-induced impact in that the α-OH-PdG·C → α-OH-PdG·A conversion is favorable (i.e., ΔΔG = +1.4 kcal· mol−1). Conversely, replacement of the central canonical G·C base pair with a G·A mismatch destabilizes the parent 11-mer duplex (i.e., ΔΔG = −2.7 kcal·mol−1). The thermodynamic destabilization relative to the parent 11-mer ranks as follows: α-OH-PdG·C > α-OH-PdG·A > G·A mismatch. The apparent disparities noted when comparing the energetic impacts of α-OH-PdG versus PdG may simply reflect differences in the sequence contexts employed, namely that the PdG analog is embedded within dGs in a 13-mer duplex, whereas the α-OH-PdG adduct is positioned within dCs in an 11-mer duplex. These results underscore the importance of conducting parallel measurements on the corresponding undamaged counterparts to ensure internal consistency. In subsequent sections, we discuss the utility of comparing lesion-containing duplexes with their corresponding “error-free” and “error-prone” model duplexes to characterize the energetic costs of DNA damage. Such correlations may furnish insight into the overall fate of an adduct in terms of DNA replication, repair, and consequent mutagenicity and/or genotoxicity.
Impact of Exocyclic and Oxidative Lesions: Energetically Stabilizing, Destabilizing, or Neutral Relative to “Undamaged” Mismatch Counterparts
Prior studies characterizing the impact of PdG on duplex stability have revealed that a G·A mismatch is only mildly destabilizing (i.e., ΔΔG = −1.3 kcal·mol−1) when embedded within dGs (i.e., GXG).10 Moreover, substitution of dG by a damaged base (i.e., PdG) does not aggravate the impact of a mismatch any further, as noted by the near equivalence of both the undamaged G·C to G·A and damaged G*C to G*A mutations (i.e., ΔΔG = −1.3 kcal·mol−1). The latter may be rationalized in terms of flanking residue stacking energies that are recognized to play a role on DNA duplex stability, particularly in the presence of lesions or defects. In contrast with these observations, studies on the impact of 8-oxodG29 reveal that a corresponding G·A mismatch embedded within dCs (i.e., CXC) is highly destabilizing (i.e., ΔΔG = −6.8 kcal·mol−1) relative to the parent duplex (i.e.., G·C to G·A). Incorporation of the lesion into a G·A mismatch reduces the overall energetic penalty by 3.4 kcal·mol−1 (i.e., G·C to G*·A). Conversely, the presence of an 8-oxodG lesion minimally destabilizes the resultant mismatched duplex by only -1.4 kcal·mol−1 (i.e., G*C to G*A). Consequently, mutation of G·A to G*·A is in fact stabilizing (i.e., ΔΔG = +3.4 kcal·mol−1), a finding which supports the proposal that energetics may furnish important information regarding the potential mutagenicity of a lesion or defect Such a scenario implies that the magnitude of lesion-induced energetic attenuation of a mismatch may be correlated with the propensity for mispairing during polymerase-mediated synthesis.
A unified view that arises from the diversity of lesions and sequence contexts explored to date is one in which a damaged/mismatched base pair invariably destabilizes a given DNA host duplex. Since sequence contexts appear critical in determining the hierarchy of free energy changes that encompass canonical to damaged/mismatched duplexes, the overall data is consistent with recent findings on the deleterious effects of acrolein adducts, namely that in vivo mutagenicity is not a predominant cause of the relationship commonly invoked between tobacco use and cancer.26 Presumably, sequence context variations in vivo effectively average the impacts of this adduct on DNA replication based on the observation that mutagenicity does not occur at alarming ratios. Therefore, our finding that α-OH-PdG·C is dramatically destabilized is entirely consistent with the genotoxic nature of this lesion in that it blocks replication both in vivo and in vitro. Moreover, our observation that α-OH-PdG·A is destabilizing relative to the “undamaged” G·A mismatch is consistent with the proposal that such an adduct is not highly mutagenic. The implication of such a finding is that in this particular case, the “A-rule” does not represent an energetic advantage for polymerase insertion opposite damaged relative to undamaged dG. Collectively, the stabilization of α-OH-PdG·A relative to α-OH-PdG·C may not be rationalized in terms of a specific preference for dA insertion opposite this adduct during DNA replication in vivo.
Structural Consequences of the α-OH-PdG-Induced Thermodynamic Impact on Duplex Stability
Despite a significant reduction in thermal and thermodynamic stability, structural studies reveal that the α-OH-PdG-containing duplexes adopt regular B-DNA right-handed helical conformations that are mildly distorted at the central base pair by the presence of this lesion. The glycosidic torsion angles of all undamaged residues are consistent with an anti orientation, while sugar conformations appear in the C1'-exo/C2'-endo range, and standard WC alignments are preserved in the non-damaged base pairs of the duplex (refer to companion paper27). The torsion angle of the adduct is in syn orientation at the lesion site, which positions the exocyclic 1,N2-hydroxylpropyl ring in the major groove of the duplex with its Hoogsteen edge facing the counter base. The α-OH-PdG conformer perturbs the local duplex structure, thereby hindering hydrogen bonding with the counter base.
In contrast with the findings deduced from parallel structural studies, we observe a significant α-OH-PdG-induced energetic destabilization within an identical duplex sequence and similar solution conditions. Although the NMR data suggest a local perturbation of duplex conformation that is consistent with our CD measurements, the enthalpic and entropic losses in conjunction with disruption of cooperative duplex dissociation are typical of a destabilization that propagates beyond the immediate vicinity of the damaged site. Specifically, a local destabilization may account for a free energy reduction of approximately 3.0 kcal·mol−1, whereas our data reveal that the α-OH-PdG lesion induces a free energy cost nearly three times this value (i.e., ΔΔG = −8.2 kcal·mol−1). The origins of this destabilization are enthalpic in nature and reflect a ΔΔH contribution of −32 kcal·mol−1. Significantly, these trends are maintained albeit compressed in our heat capacity corrected energetic data (i.e., ΔΔG = −5.5 kcal·mol−1 and ΔΔH = −15 kcal·mol−1). The bulky exocyclic α-OH-PdG lesion perturbs the normal arrangement of bases within the helical structure, which is consistent with the observed enthalpic destabilization and weakened base pairing properties.
The challenge of reconciling our energetic data with available solution structures resides in the finding that an α-OH-PdG damaged base induces minimal perturbation of the regular right-handed helix. Specifically, NMR data reveal that the α-OH-PdG lesion adopts a syn conformation for the glycosidic torsion angle with the exocyclic ring protruding into the major groove where there is sufficient space to accommodate the α-OH group without causing further perturbations of the structure. This modest perturbation of global DNA structure contrasts with the impact of other adducts that adopt minor groove aligned, intercalated, or cross-linked conformations, all of which are highly distorting. In the absence of a more egregious structural impact that reflects the relatively large energetic penalty imparted by the acrolein adduct and significant departure from two-state melting behavior, other events associated with counterion and/or solvent effects must be invoked to rationalize the apparent discontinuity in structure-energetic correlations. In the following section, we discuss the biological implications of our energetic data in view of current debates regarding the overall consequences of acrolein exposure or endogenous production within the cell.
BIOLOGICAL IMPLICATIONS
Energetic Preferences May Dictate the Biological Fate of an Adduct
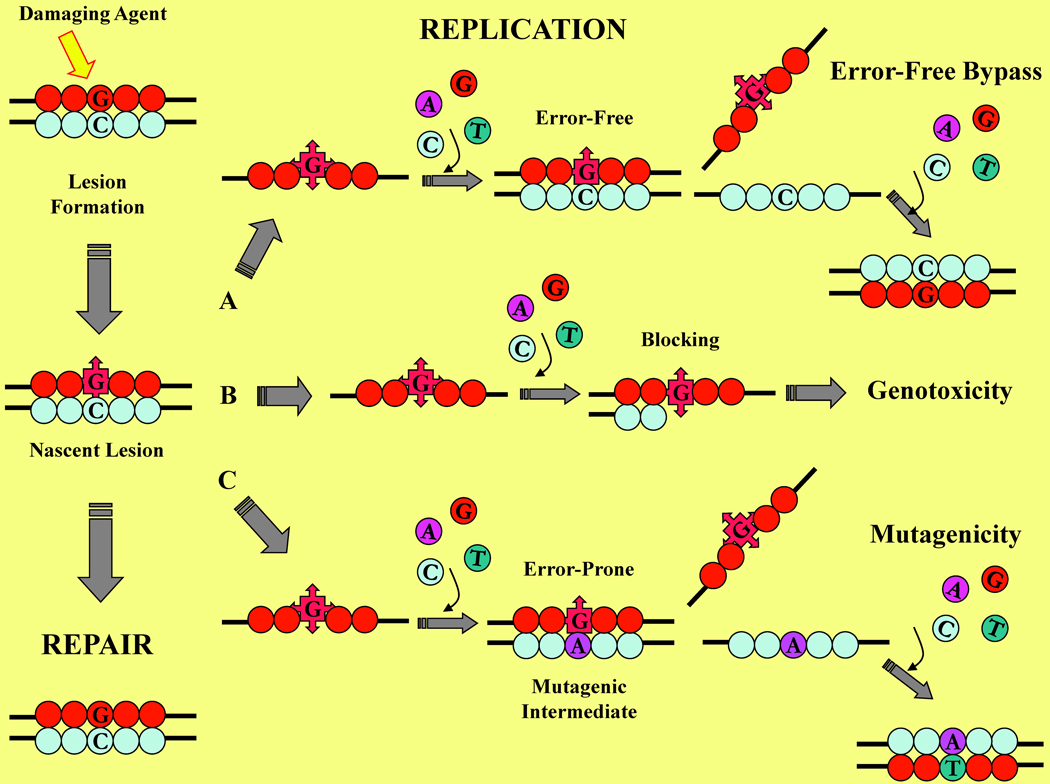
Living cells are subject to attack by an array of endogenous and exogenous DNA-damaging agents that continually threaten genome integrity. While specialized repair systems are under constant surveillance to remove the resultant damaged bases, egregious lesions oftentimes escape repair. Figure 5 furnishes a simplified schematic representation of the various processes involved in DNA replication and damage repair. Successful repair results in the restoration of normal bases as depicted in the vertical pathway on the left side of Figure 5. Persistent lesions are encountered by the replication machinery and may result in a number of outcomes as illustrated in the horizontal pathways (A, B, and C) of Figure 5. Lesions may be accommodated by translesion synthesis polymerases, which contain larger active sites and generally promote error-free synthesis as illustrated in pathway A. A considerable body of experimental evidence suggests that highly destabilizing lesions drastically disrupt the double helical geometry and block normal polymerase-mediated replication as depicted in pathway B. The resultant outcome is synthesis arrest that may be lethal to the cell as a direct consequence of blocking lesions manifesting their genotoxicity. Conversely, lesions may favor miscoding during synthesis providing the damaged mispairs are stabilized relative to their non-damaged counterparts and/or lesion-containing nascent base pairs. In the case of error-prone synthesis depicted in pathway C, mutagenesis and carcinogenesis ensues as a result of adduct-induced miscoding properties.
Figure 5. Schematic Representation Illustrating the Fate of a Damaged dG within Genomic DNA.
Following exposure to a damaging agent, a nascent lesion may be removed by the cellular repair machinery as depicted in the vertical pathway on the left. When an egregious lesion escapes repair, the resultant damaged strand undergoes replication via one of the following horizontal pathways: (A) Error-free synthesis resulting in restoration of the canonical duplex: (B) Blocking synthesis that stalls within the vicinity of the lesion site, resulting in DNA synthesis arrest and consequent genotoxicity; or, (C) Error-prone synthesis via nucleotide misincorporation, forming a mutagenic intermediate that yields a mutant product (e.g., G to T substitution).
Our characterization of the thermal and thermodynamic properties of the α-OH-PdG adduct reveals that this lesion is less destabilizing when paired with dA relative to dC. These data suggest that lesion-induced destabilization is partially alleviated by its base pairing ability with dA. In fact, the parallel structural study of the α-OH-PdG·A duplex27 reveals that the lesion paired opposite dA adopts a conventional B-DNA conformation with local perturbations confined to the damaged site. In principle, these findings suggest that in the absence of repair, the damaged base may serve as a mutagenic template by allowing dA insertion during replication, thereby yielding G → T transversion mutations. Based on the observation that the duplex harboring an α-OH-PdG·A mispair is stabilized relative to α-OH-PdG·C, one might therefore invoke the widely acknowledged “A-rule” in which a damaged dG mispaired with an incoming dATP results in G → T transversions. However, our findings reveal that the thermodynamic stability of the α-OH-PdG·A duplex is significantly lower than that of the corresponding “undamaged” mismatch G·A duplex. Consequently, the apparent absence of an energetic advantage suggests that the α-OH-PdG adduct does not elicit greater mutagenicity relative to a canonical dG per se. In fact, the “error-free” model α-OH-PdG·C duplex is highly destabilizing, and one might reasonably anticipate that this adduct arrests DNA synthesis in conjunction with its reported acrolein-mediated genotoxicity properties.
Our energetic data are generally consistent with in vivo studies15 demonstrating that the α-OH-PdG adduct is genotoxic rather than mutagenic. Specifically, α-OH-PdG extensively blocks DNA replication, yet primarily codes for dC incorporation with a low frequency of base substitution mutations when bypassed in translesion synthesis. An alternate study4 has reported that whereas the γ-OH-PdG regioisomer lacks mutagenicity, the α-OH-PdG adduct is somewhat mutagenic. Replication of the α-OH-PdG adduct in xeroderma pigmentosum A cells induces 10% base substitutions, primarily G → T transversions.20 When polymerase η replication bypass of DNA containing α-OH-PdG is assayed, this lesion poses a stronger block to replication than the γ-OH-PdG adduct, a finding that closely resembles the results for PdG in which the exocyclic adduct remains in a permanent ring-closed conformation. Functional in vivo and in vitro assays exploring the translesion synthesis of α-OH-PdG with the exonuclease deficient Klenow fragment20 and pol η4 demonstrate that this acrolein adduct is highly blocking, a finding that is consistent with the significant thermodynamic destabilization observed when the adduct is paired opposite dC, as reported herein. The polymerase assay suggests that replication blocking activity may arise from the inability of α-OH-PdG to participate in canonical Watson−Crick base pairing,4 which therefore impairs the critical steps of incorporation and extension in DNA synthesis. The variability of biological observations with respect to both polymerase type and cell system poses a challenge for deducing correlations between structure, function, and energetics. Collectively, our findings assist in rationalizing the apparent disparities associated with the impact of acrolein adducts, while underscoring the importance of studying lesion effects in multiple sequence environments.
Energetics as a Probe of Mutagenicity and Genotoxicity
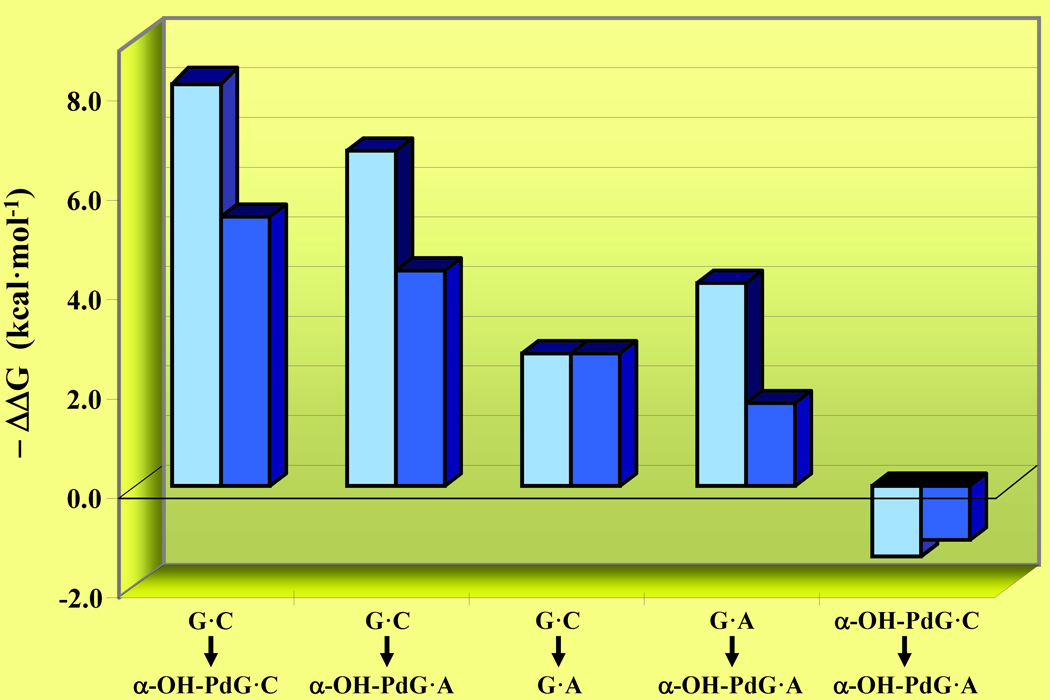
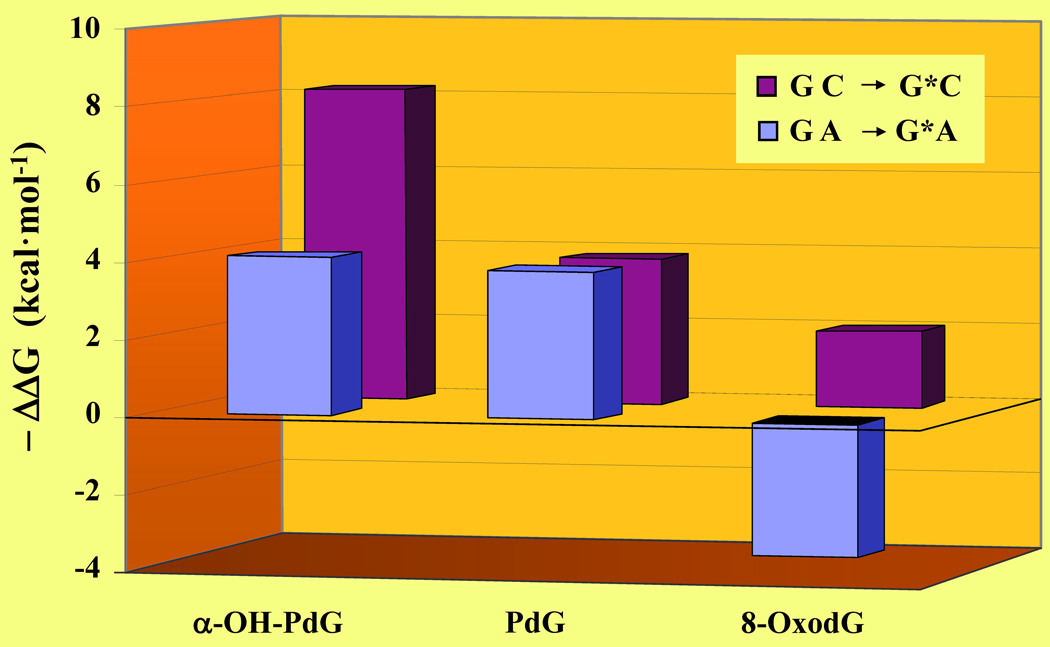
Lesion-induced thermodynamic impacts and overall sequence context effects may be exploited as qualitative probes for resolving biological consequences at the molecular level. Figure 6 illustrates the differential impacts of PdG, α-OH-PdG, and 8-oxodG relative to the corresponding undamaged DNA host duplexes. It is relevant to note that the α-OH-PdG adduct is embedded within an 11-mer duplex, whereas PdG and 8-oxodG are positioned as the central residue in a 13-mer duplex. Moreover, the PdG lesion is flanked by dGs while α-OH-PdG and 8-oxodG are flanked by dCs. The lesion-induced impacts are sorted on the basis of decreasing differential destabilization of the damaged duplexes to improve overall clarity. We hypothesize that a higher lesion-induced energetic impact (i.e., ΔΔGGC→G*C) may be correlated with an increased potential for cytotoxicity. Conversely, a mismatched lesion-containing duplex that is stabilized relative to a standard G·A mismatch (i.e., ΔΔGGA→G*A) is actually more error-prone during synthesis. In this respect, we can employ the ΔΔG values to ascribe an empirical score that describes the propensity of a lesion to promote mutagenicity or elicit genotoxic effects.
Figure 6. Lesion-Induced Energetic Impacts as a Probe of Cytotoxicity and Mutagenicity.
Comparison of the G to G* modification within the “nascent” and mismatch duplexes as monitored by the ΔΔG of G·C → G*·C (burgundy) versus G·A → G*·A (blue) for the α-OH-PdG, PdG,10 and 8-oxodG29 lesions. Although the thermodynamic parameters are derived from DNA duplexes of distinct sequence context under different solution conditions, each data set is obtained via direct comparison with its respective reference duplex (i.e., ΔΔGGC to G*C = ΔGG*C − ΔGGC and ΔΔGGA to G*A = ΔGG*A − ΔGGA). The lesion-induced impacts on duplex dissociation free energies are expressed as - ΔΔG to improve clarity and sorted on the basis of decreasing ΔΔG. Amongst these lesion-containing duplexes, 8-oxodG·A is thermodynamically stabilized relative to the corresponding “undamaged” G·A mismatch.
According to this model, both PdG and α-OH-PdG are highly destabilizing and presumably more cytotoxic than 8-oxodG. Conversely, duplexes harboring an 8-oxodG lesion are stabilized relative to an A-mismatch and therefore poised for mutagenesis. Given identical experimental conditions in vivo, an 8-oxodG damaged base is more likely to generate GT → TA transversion mutations than PdG or α-OH-PdG. Our finding that exocyclic adducts (i.e., PdG, α-OH-PdG) are more destabilizing relative to oxidatively damaged bases (i.e., 8-oxodG) is consistent with the genotoxic properties elicited by these lesions. Significantly, 8-oxodG has been associated with an array of disorders ranging from accelerated aging to carcinogenesis, all of which may be ascribed to its high mutagenicity levels. The latter is a direct consequence of the propensity exhibited by this lesion to adopt alternate Hoogsteen conformation that supports dATP insertions, thereby favoring miscoding during lesion bypass and ultimately undergoing GT → TA transversions.
Future studies are clearly warranted to systematically examine the thermodynamic properties of lesion-containing duplexes under a controlled number of variables to compare canonical and mismatched duplexes in all possible sequence contexts. The resultant data will assist ongoing structural, dynamics and biochemical efforts to identify the origins of carcinogenicity and genotoxicity of environmental and metabolically generated hazardous compounds. Experimental strategies that systematically evaluate the energetic impacts of lesions and adducts can therefore provide mutagenicity and cytotoxicity “scores” to facilitate initial predictions on potential hot spots for the deleterious effects of such damaging agents.
Implications for Repair
A large body of biochemical and genetic outcomes can be rationalized in terms of our energetic findings. In this context, our data are consistent with the biological consequences of acrolein-adduct formation drastically impacting the cell, as this lesion blocks DNA-polymerase synthesis and arrests DNA replication. In the absence of contravening mechanisms that abrogate the deleterious effects of acrolein adducts, cell death quickly ensues. Several studies reveal that propano-dG adducts may be recognized and repaired by specific nucleotide excision repair (NER) proteins.48 Considering the underlying assumption that these specialized repair systems recognize domains of “instability” within the genome in order to initiate lesion/adduct removal and repair,37 the highly destabilizing free energies measured herein for the nascent α-OH-PdG lesion supports the contention that the NER pathway may contribute in the overall maintenance of genome integrity when challenged by acrolein adduct formation. Such intervention by the cellular repair machinery is particularly relevant when considering that acrolein adducts are not solely by-products of environmental pollutants, but also are formed in significant amounts endogenously as the result of lipid peroxidation processes.
CONCLUDING REMARKS
Our energetic studies reveal that the presence of an α-OH-PdG lesion destabilizes the parent canonical duplex, with an overall magnitude of destabilization that is opposite base dependent. Specifically, pairing the acrolein adduct with dA in lieu of dC partially ameliorates lesion-induced destabilization of the α-OH-PdG·C- duplex. Our finding that the α-OH-PdG lesion is highly destabilizing is consistent with its reported blocking activity during translesion synthesis. Under specific conditions that are dependent on the cell and polymerase system, α-OH-PdG may adopt syn-conformations that either support error-free synthesis or potentially undergo mutagenesis. Our ongoing pH-dependent energetic studies in conjunction with structural data support the formation of stable syn-conformations, regardless of the dA or dC counter base. Collectively, our studies provide insights into the energetic and conformational preferences of α-OH-PdG as a function of opposite base, the modulation of which may ultimately dictate the ability of this acrolein adduct to elicit genotoxic properties by blocking polymerase-mediated synthesis or mutagenic effects due to its propensity to mispair with dA.
REFERENCES
- 1.Stevens JF, Maier CS. Molecular Nutrition & Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung FL, Young R, Hecht SS. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 3.Hecht SS, Kassie F, Hatsukami DK. Nature Reviews Cancer. 2009;9:476–488. doi: 10.1038/nrc2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Chem Res Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 5.Benamira M, Marnett LJ. Mutat Res. 1992;293:1–10. doi: 10.1016/0921-8777(92)90002-k. [DOI] [PubMed] [Google Scholar]
- 6.Moriya M, Zhang W, Johnson F, Grollman AP. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouchakdjian M, Eisenberg M, Johnson F, Grollman AP, Patel DJ. Biochemistry. 1991;30:3262–3270. doi: 10.1021/bi00227a014. [DOI] [PubMed] [Google Scholar]
- 8.Kouchakdjian M, Eisenberg M, Live D, Marinelli E, Grollman AP, Patel DJ. Biochemistry. 1990;29:4456–4465. doi: 10.1021/bi00470a028. [DOI] [PubMed] [Google Scholar]
- 9.Kouchakdjian M, Marinelli E, Gao XL, Johnson F, Grollman A, Patel D. Biochemistry. 1989;28:5647–5657. doi: 10.1021/bi00439a047. [DOI] [PubMed] [Google Scholar]
- 10.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1992;31:12096–12102. doi: 10.1021/bi00163a019. [DOI] [PubMed] [Google Scholar]
- 11.Khullar S, Varaprasad CV, Johnson F. J Med Chem. 1999;42:947–950. doi: 10.1021/jm980605u. [DOI] [PubMed] [Google Scholar]
- 12.Nechev LV, Harris CM, Harris TM. Chem Res Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 13.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. J Biol Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 14.Nechev LV, Kozekov ID, Brock AK, Rizzo CJ, Harris TM. Chem Res Toxicol. 2002;15:607–613. doi: 10.1021/tx010181y. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Johnson F. Chem Res Toxicol. 2002;15:236–239. doi: 10.1021/tx015568f. [DOI] [PubMed] [Google Scholar]
- 16.Huang YH, Torres MC, Iden CR, Johnson F. Bioorganic Chem. 2003;31:136–148. doi: 10.1016/s0045-2068(02)00525-4. [DOI] [PubMed] [Google Scholar]
- 17.de los Santos C, Zaliznyak T, Johnson F. J Biol Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 18.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. J Biol Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 19.Yang IY, Johnson F, Grollman AP, Moriya M. Chem Res Toxicol. 2002;15:160–164. doi: 10.1021/tx010123c. [DOI] [PubMed] [Google Scholar]
- 20.Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- 21.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaliznyak T, Bonala R, Attaluri S, Johnson F, de los Santos C. Nucl Acids Res. 2009;37:2153–2163. doi: 10.1093/nar/gkp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanderVeen LA, Harris TM, Jen-Jacobson L, Marrlett LJ. Chem Res Toxicol. 2008;21:1733–1738. doi: 10.1021/tx800092g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besaratinia A. Chem Res Toxicol. 2009;22:751–753. doi: 10.1021/tx900098u. [DOI] [PubMed] [Google Scholar]
- 25.Wang HT, Zhang S, Hu Y, Tang MS. Chem Res Toxicol. 2009;22:511–517. doi: 10.1021/tx800369y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SI, Pfeifer GP, Besaratinia A. Cancer Res. 2007;67:11640–11647. doi: 10.1158/0008-5472.CAN-07-2528. [DOI] [PubMed] [Google Scholar]
- 27.Zaliznyak T, Lukin M, El-khateeb M, Bonala R, Johnson F, de los Santos C. Biopolymers. 2009 doi: 10.1002/bip.21366. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marky LA, Breslauer KJ. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 29.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1995;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 30.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 31.Minetti CASA, Remeta DP, Dickstein R, Breslauer KJ. Nucl Acids Res. 2009 doi: 10.1093/nar/gkp1036. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1998;37:12507–12512. doi: 10.1021/bi981090b. [DOI] [PubMed] [Google Scholar]
- 33.Lin Z, de los Santos C. J Mol Biol. 2001;308:341–352. doi: 10.1006/jmbi.2001.4587. [DOI] [PubMed] [Google Scholar]
- 34.Allawi HT, SantaLucia J. Biochemistry. 1998;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 35.Markham NR, Zuker M. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 36.Markham NR, Zuker M. Nucl Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plum GE, Breslauer KJ. Ann NY Acad Sci. 1994;726:45–56. doi: 10.1111/j.1749-6632.1994.tb52796.x. [DOI] [PubMed] [Google Scholar]
- 38.Plum GE, Gelfand CA, Breslauer KJ. IARC Sci Publ. 1999:169–177. [PubMed] [Google Scholar]
- 39.Chalikian TV, Volker J, Plum GE, Breslauer KJ. Proc Natl Acad Sci USA. 1999;96:7853–7858. doi: 10.1073/pnas.96.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibutani S, Takeshita M, Grollman AP. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 41.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Biochemistry. 2001;40:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 42.Mekhovich O, Tang MS, Romano LJ. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- 43.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biopolymers. 1996;38:439–445. doi: 10.1002/(SICI)1097-0282(199604)38:4%3C439::AID-BIP1%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Gelfand CA, Plum GE, Mielewczyk S, Remeta DP, Breslauer KJ. Proc Natl Acad Sci USA. 1999;96:6113–6118. doi: 10.1073/pnas.96.11.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaliznyak T, Bonala RR, Lukin M, Johnson F, De los Santos C. Chem Res Toxicol. 2008;21:104. [Google Scholar]
- 46.Cullinan D, Korobka A, Grollman AP, Patel DJ, Eisenberg M, de los Santos C. Biochemistry. 1996;35:13319–13327. doi: 10.1021/bi9605705. [DOI] [PubMed] [Google Scholar]
- 47.Zaliznyak T, Lukin M, Johnson F, de los Santos C. Biochemistry. 2008;47:4606–4613. doi: 10.1021/bi7022514. [DOI] [PubMed] [Google Scholar]
- 48.Johnson KA, Fink SP, Marnett LJ. J Biol Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]