Abstract
Rationale
Transcriptional profiling can detect subclinical heart disease and provide insight into disease etiology and functional status. Current microarray-based methods are expensive and subject to artifact.
Objective
To develop RNA sequencing methodologies using next generation massively parallel platforms for high throughput comprehensive analysis of individual mouse cardiac transcriptomes. To compare the results of sequencing- and array-based transcriptional profiling in the well-characterized Gαq transgenic mouse hypertrophy/cardiomyopathy model.
Methods and Results
The techniques for preparation of individually bar-coded mouse heart RNA libraries for Illumina Genome Analyzer II resequencing are described. RNA sequencing showed that 234 high abundance transcripts (>60 copies/cell) comprised 55% of total cardiac mRNA. Parallel transcriptional profiling of Gαq transgenic and non-transgenic hearts by Illumina RNA sequencing and Affymetrix Mouse Gene 1.0 ST arrays revealed superior dynamic range for mRNA expression and enhanced specificity for reporting low-abundance transcripts by RNA sequencing. Differential mRNA expression in Gαq and non-transgenic hearts correlated well between microarrays and RNA sequencing for highly abundant transcripts. RNA sequencing was superior to arrays for accurately quantifying lower-abundance genes, which represented the majority of the regulated genes in the Gαq transgenic model.
Conclusions
RNA sequencing is rapid, accurate, and sensitive for identifying both abundant and rare cardiac transcripts, and has significant advantages in time- and cost-efficiencies over microarray analysis.
Keywords: RNA sequencing, microarray, gene regulation, Gq
Introduction
Signaling factors that mediate cardiac hypertrophy and heart failure do so in large part by altering gene expression. Accordingly, physiological hypertrophy, pathological hypertrophy and heart failure are associated with distinct transcriptional signatures in human disease and experimental mouse models 1, 2. The ability to detect early changes in myocardial gene expression is essential to understanding pathophysiological mechanisms in experimental models, and is predicted to provide crucial diagnostic and prognostic information in human heart disease 3.
We developed techniques for broad, accurate, and inexpensive characterization of individual mouse cardiac transcriptional signatures using massively parallel resequencing of heart cDNA libraries using the Illumina Genome Analyzer II. The digital readouts enable parallel quantification and annotation of myocardial transcripts, including mRNAs expressed at levels below 1 copy per cell. Here, we describe the components of a molecular and bioinformatic “RNA sequencing pipeline” optimized for comparative evaluation of transcript signatures in genetic mouse heart models. We compare transcriptional profiling by Illumina mRNA sequencing with Affymetrix microarray RNA in mouse hearts with a well characterized transcriptional signature of pathological hypertrophy, the Gαq transgenic mouse 4, 5, to illustrate the advantages of RNA sequencing over array-based platforms.
Methods
Mouse models
Generation of the Gαq-40 transgenic mouse line has been described previously 4, 5. Four pairs of 8 week old male nontransgenic FVB/N and Gαq-40 transgenic mouse hearts were used for RNA sequencing studies. Echocardiographic and cardiac catheterization studies were performed using standard methods 4, 6.
Preparation and quantification of total myocardial RNA
Total RNA was isolated from flash-frozen mouse hearts using Trizol (Invitrogen), as per the manufacturer's directions, except that isopropyl alcohol precipitation of RNA was allowed to proceed for 30 min at room temperature. RNA was quantified via NanoDrop or UV spectrometer and integrity (28S:18S ratio) was assessed on 1% agarose.
Reverse transcription and preparation of cDNA
Preparation of cDNA fragments from poly(A)+ RNA was modified from a previously described protocol 7. Four μg of total cardiac RNA was twice oligo(dT) selected using the Dynabead mRNA purification system (Invitrogen). Two hundred ng of cardiac mRNA was fragmented to ~200 nt by heating to 94 C for 2.5 min in 40 mmol/L Tris acetate pH 8.2, 100 mmol/L potassium acetate, 30 mmol/L magnesium acetate, and immediately chilled on ice. After purification on Ambion NucAway columns, 100 ng of fragmented cardiac mRNA was reverse-transcribed using random hexamers, followed by second-strand cDNA synthesis using the Just cDNA double-stranded cDNA synthesis kit (Stratagene catalog #200453).
Construction of barcoded short-read libraries for Illumina sequencing
cDNAs were end-repaired using the End-It End-Repair kit (Epicentre Biotechnologies, #ER0270) and 3′ A-overhangs added using 3′-5′ exo- Klenow polymerase (New England Biolabs #M0212) and 0.2 mmol/L dATP. Illumina adapters with T-overhangs and customized to include three nt ‘barcodes’ were ligated to the cDNA at 10:1 molar excess using the Promega LigaFast kit (#M8221); different barcoded adapters were ligated to individual mouse heart cDNAs. Following column-purification (Qiagen) to remove excess unligated adapter, DNA in the 200-400 bp range was isolated via gel purification (Qiagen) on 2% low-melting agarose and amplified with 11 cycles of Phusion polymerase (New England Biolabs #F531)-mediated PCR (10 sec 98 C, 30 sec 65 C, 30 sec 72 C), using oligonucleotides complementary to Illumina sequencing adapters. Barcoded sequencing adapters and PCR oligonucleotides are described in Supplemental Table IX. The final, amplified libraries were again column-purified and quantified using PicoGreen (Quant-It, Invitrogen).
Sequencing and data processing
Four barcoded libraries were combined in equimolar (10 nmol/L) amounts and diluted to 4 pmol/L for cluster formation on a single Illumina Genome Analyzer II flowcell lane. Basecalling of DNA clusters was performed using Illumina's processing pipeline software (version 1.5) and 36-nt sequences, with quality scores, were obtained in Illumina's SCARF text format. UNIX functions were used to sort the sequences according to barcode, remove the 4 barcoding nucleotides, and convert sequence data to FASTQ format (example shell script given in Supplemental Methods). After barcode removal, the 32 base mRNA sequence reads were mapped to transcripts annotated in NCBI release 37 of the mouse genome using the publicly available packages Bowtie (release 0.12.0) (http://bowtie-bio.sourceforge.net/index.shtml) 8, TopHat (release 1.0.12) (http://tophat.cbcb.umd.edu/) 9, and Cufflinks (release 0.7.0) (http://cufflinks.cbcb.umd.edu/) 10. TopHat and Cufflinks map known and novel splice junctions, use annotation files to compute which aligned sequences map to the known transcriptome, and take into account transcript isoform diversity (alternative splicing). Cufflinks may be used with gene annotation files to calculate overall gene expression in terms of Reads Per Kb of exon per Million mapped reads (RPKM), a parameter previously defined in 7. We used the default options supplied with these software packages in our analyses (example shell script given in Supplemental Methods).
Annotation files in gtf format (http://mblab.wustl.edu/GTF22.html), including mRNAs, ESTs and noncoding elements such as ribosomal RNAs and miRNA precursors, were downloaded from Ensembl (ftp://ftp.ensembl.org/pub/current_gtf/mus_musculus/). We used the Cufflinks module (above) with annotation files from which ESTs, rRNAs and miRNA precursors had been manually removed, to focus on sequence reads mapping to coding genes. We analyzed only those RNA elements that had expression signals in at least 2 of 4 biological replicates.
The gtf annotation files from Ensembl contain many separate transcripts (identified with ENST labels) which contribute to the sequence defined for each gene (identified with ENSG labels). In order to assess alternative splicing events, we used the Cufflinks module with these same gtf annotation files, but forced calculation of RPKM values for separate transcripts (ENSTs) rather than for entire genes (ENSGs) (example shell script given in Supplemental Methods). The presence of alternatively spliced products was evaluated by manually examining Cufflinks output files for ENST entries corresponding to an ENSG entry of interest.
Comparative analysis of RNA sequencing and microarray results
To compare gene expression data obtained with RNA sequencing to the most current microarray technology, RNA from the same samples was analyzed on Affymetrix Mouse Gene 1.0 ST arrays at the Multiplexed Gene Analysis core of Washington University. Microarray data were analyzed using Partek Genomics Suite v6.4 (Partek, St Louis, MO) as previously described 11, 12; RNA sequencing data (gene symbols and RPKM values) were imported into Partek and analyzed similarly. Biological Networks Gene Ontology (BiNGO; 13) was used to assign genes into gene-ontology categories.
Reverse-transcription quantitative PCR
One microgram of total cardiac RNA was reverse-transcribed into cDNA using oligo-dT priming (qScript Flex cDNA kit, Quanta Biosciences). One-twentieth of each preparation was used for each individual qPCR, using TaqMan probes (Applied Biosystems), detailed in Supplemental Methods.
Statistical Methods
Paired and unpaired data were compared with Student's t-test. Multi-group comparisons used Bonferroni correction. Correlation coefficients and linear regressions for comparison of microarray and RNA sequencing data were calculated with GraphPad Prism (San Diego, CA). P-values and false discovery rates for gene expression data were calculated using Partek Genomics Suite 6.4 software (Partek, St Louis, MO). P value of <0.05 was defined as significant unless indicated otherwise.
Results
RNA sequencing of the normal and pathological hypertrophied heart transcriptomes
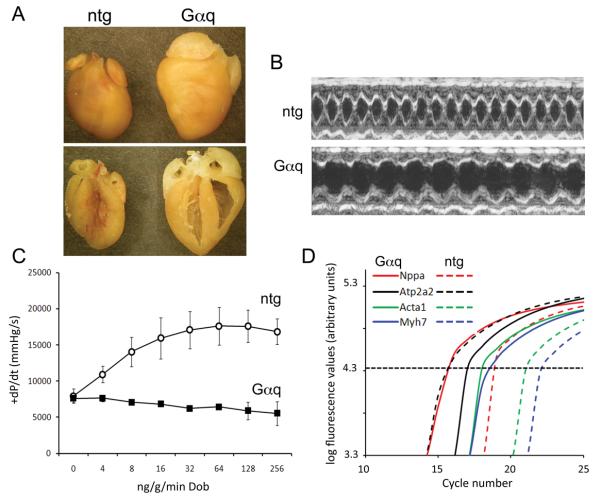
Microarray profiling has been widely used in mouse studies, but is relatively expensive and requires special equipment and expertise. Since our laboratory has found that high throughput human DNA sequencing on next-generation massively parallel systems is fast, highly reproducible between technical replicates, accurate, and inexpensive 14, we explored the utility of deep mRNA sequencing in experimental mouse cardiac models. For initial proof-of-concept studies, we RNA sequenced four pairs of 8 week old male nontransgenic FVB/N and Gαq transgenic mouse hearts. Gαq is the essential signaling transducer of pressure overload hypertrophy 15. Cardiac-specific overexpression of Gαq at 4-5 times endogenous levels intrinsically activates pathological hypertrophy signaling in a manner that closely mimics pressure overload in mice. Thus, Gαq transgenic mice exhibit cardiac hypertrophy, ventricular enlargement, diminished ejection performance, and characteristic increased expression of fetal cardiac genes 4, 6, 16-18. Figure 1 illustrates these features of the Gαq transgenic mouse model. Previous analyses of mRNA expression in the cardiac Gαq transgenic model used RNA dot blotting 4, 19-22 or Incyte microarrays 17, 18, neither of which provides a comprehensive examination of gene expression. Thus, we compared array- and sequencing-based methods of comprehensive transcriptome profiling in Gαq mouse hearts.
Figure 1. Phenotype of the adult Gαq-40 transgenic heart.
(A) Formalin-fixed intact hearts (upper panel) and four-chamber views (lower panel). (B) Representative M-mode echocardiograms. (C) Response to graded infusion of dobutamine during cardiac catheterization (mean ± SEM, n=3 each genotype). (D) Representative RT-qPCR fluorescence curves for ntg and Gαq cardiac gene expression (dotted line indicates Ct).
The few RNA sequencing studies published to date have typically used large numbers of sequence reads to map the transcriptomes at very high resolution (e.g. 23, 24). Since one or more sequencing lanes are devoted to a single sample, this can be very expensive. Furthermore, this degree of resolution is not required for comparative analysis of transcriptomes in a case-control study design (as with nontransgenic verses transgenic, or normal verses diseased). For this reason, we developed procedures to individually DNA-barcode RNA sequence libraries and pool multiple libraries into a single sequencing lane, and segregate the sequence data post hoc according to barcode. To assess the efficacy of barcoding, library pooling, and sorting of the sequence reads, we tracked a rare DNA marker polymorphism (SNP) in 24 sequencing libraries individually barcoded, pooled, and sequenced in a single Illumina GA II lane. (The marker SNP was present in two of the 24 sequencing libraries, as determined by Sanger sequencing.) Deconvolution of the barcoded, pooled DNA sequence correctly identified both libraries containing the marker SNP. To ensure that this technique was applicable to quantitative analysis of transcriptome levels by RNA sequencing, we individually barcoded a Gαq and a nontransgenic heart cDNA library, combined them, and resequenced them in a single Illumina GA II lane. Here, high expression of the transgene (Gnaq) and other characteristic molecular markers of this model (Myh7, Acta1) marked the Gαq library, which was correctly classified as such according to barcode.
For comparative transcriptome profiling of Gαq and nontransgenic hearts, four barcoded libraries were analyzed in a single sequencing lane. The total number of barcoded RNA sequence reads from the eight cardiac libraries was 18.2 million, of which 10.5 million (57.8%) aligned to NCBI release 37 of the mouse genome (Supplemental Table I). Using the criterion that an RNA element must be detected in at least 2 of 4 biological replicates, these sequences mapped to 14,109 different annotated RNA elements (included noncoding RNAs and miR precursors), of which 11,180 were coding mRNAs. The remaining sequences corresponded largely to ribosomal and transfer RNA. 10,844 coding mRNAs were detected in nontransgenic hearts, while 10,775 coding mRNAs were detected in Gαq hearts. 10,437 mRNAs were common to both nontransgenic and Gαq hearts.
Gene expression values in nontransgenic hearts ranged from ~1 copy per cell (corresponding to an RPKM value <3 7) for Casp8 (caspase 8) to 3873 copies per cell for mt-Co1 (mitochondrial cytochrome C oxidase subunit I) (Supplemental Table II), consistent with myocardium being mitochondrial-rich 25 and having low rates of caspase-mediated apoptosis 26. The most abundant transcript in Gαq hearts (2,604 copies per cell) was Nppa encoding atrial natriuretic factor, consistent with the cardiomyopathic phenotype of this model 4, 16, 27. Gαq was the 24th-most abundant transcript in Gαq hearts, expressed at 574 copies per cell (Supplemental Table III.) Complete gene expression data for each individual heart are in Supplemental Table IV.
Recent PMAGE (polony multiplex analysis of gene expression) analysis indicated that approximately 200 unique heart mRNAs were expressed at >60 copies/cell, and that this small fraction (<1%) of the ~25,000 mouse gene transcripts comprised ~65% of total cardiac mRNA 28. Our RNA sequencing confirms this surprising observation, identifying 234 very high abundance mRNAs expressed at or greater than 60 copies per cell, which comprised 55% of the total myocardial mRNA complement of normal hearts (Supplemental Table II). In Gαq hearts, 236 mRNAs were expressed at or greater than 60 copies per cell, representing 52% of total myocardial mRNA. 206 of these 236 very high abundance Gαq heart mRNAs were among the 234 very high abundance transcripts in nontransgenic hearts (88% concordance), indicating a relatively modest impact of the hypertrophy gene expression program on the most prevalent cardiac transcripts.
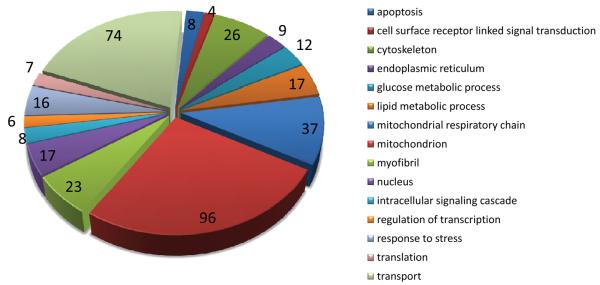
Gene-ontology analysis of the 234 most abundant cardiac mRNAs reveals a preponderance of mitochondrial, transport, cytoskeletal, and contractile genes (Figure 2, Supplemental Table V). Among the abundant transcripts, those differentially expressed in Gαq hearts were Nppb (BNP), Acta1 (α-skeletal actin), Ankrd1, Atp2a2 (SERCA2a), Myom2, Mybpc3, Hspb6, Idh3b, Pdha1, Hadha, Acadvl, Ndufv2, Acaa2, Fhl2, i.e. several members of the “fetal cardiac gene program” that is non-specifically regulated in myocardial disease 29. Thus, quantitative analysis of the cardiac transcriptome by RNA sequencing shows that a relatively few, highly abundant cardiac transcripts encoding homeostatic genes account for the majority of cardiac mRNAs, but that measuring the relative expression of these abundant transcripts provides little insight into cardiac status 1.
Figure 2. Gene ontology (GO) analysis of highly expressed genes in nontransgenic mouse heart.
The 234 highest-expressing genes in nontransgenic mouse hearts (≥60 copies per cell) were classified into 15 GO categories using BiNGO 50. Size of the pie slice corresponds to the number of matches to a given GO category (shown at the edge of each slice). A total of 360 matches were made to the 15 categories shown. Classification of each gene is given in Supplemental Table III.
RNA sequencing reveals greater depth of the normal and hypertrophy mRNA signature
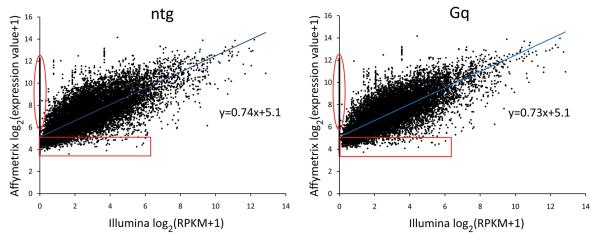
To directly compare results of RNA sequencing and conventional microarray transcript profiling of mouse hearts, we performed parallel microarray surveys of mRNA expression on the same eight mouse cardiac RNA samples studied above using Affymetrix Mouse Gene 1.0 ST arrays with probe sets for 25,175 annotated genes. The arrays reported signals for all ~25,000 probe sets, compared to only 11,180 coding cardiac mRNAs to which RNA sequencing reads were mapped. Thus, unfiltered array data generated “results” for 125% more RNAs than were present by RNA sequencing. To determine the reason for this discrepancy, expression levels of all mRNAs detected by either or both methods (~20,000 genes with matching gene symbols between the array platform and RNA sequencing gene annotation files) were compared for each of the eight hearts (Figure 3; Supplemental Figure I). The correlation between mRNA expression levels reported by array and RNA sequencing was generally good (Spearman correlation coefficient, r = 0.811 ± 0.006, mean ± SEM, n=8 hearts). However, the regression lines do not go through the y-axis origin, but rather intercept the y-axis (microarray data) at ~5. Also, the slopes of the regression lines are significantly less than one (0.74±0.011, P=1×10−6), indicating compression of expression values by microarray analysis. Finally, a number of mRNAs for which RNA sequencing showed no reads (Illumina sequencing values (x-axis) of 0) were reproducibly indicated as highly expressed by the microarray studies (Affymetrix array value (y-axis) of ~6 to ~12; circumscribed in red on Figure 3). These types of problems have been attributed to background hybridization and false positive hybridization on microarrays 23. Indeed, background hybridization on microarrays complicates establishing the correct threshold for mRNA detection 11, 30, 31. In our own data set, if we use the y-axis value at the regression line x-axis intercept to establish a threshold value for the microarray data, then hundreds of low abundance mRNAs (851 in the nontransgenic sample and 757 in the Gαq sample shown in Figure 3 [red boxes]) detected by RNA sequencing would be incorrectly filtered. Thus, unfiltered microarray data report expression levels for mRNAs that are not really there (vertical ovals on Figure 2), but correcting for this problem by increasing the threshold for mRNAs calling eliminates genuine low abundance mRNAs within the hybridization noise (horizontal squares in Figure 3).
Figure 3. Correspondence of gene expression determined by RNA sequencing vs microarray, for individual hearts.
Gene expression determined by RNA sequencing (Illumina) is plotted against gene expression determined by microarrays (Affymetrix). Values shown are log2(RPKM+1) for RNA sequencing (x-axis) and log2(Affymetrix signal units+1) for microarrays. A value of 1 was added to both RPKM and Affymetrix signal units to avoid taking the log of 0. Red circles highlight genes reported to be expressed by microarrays, but not by RNA sequencing. Red boxes show genes expressed below a typical cutoff level for microarray analysis. One nontransgenic heart and one Gq heart are shown; plots are representative of those for all 8 hearts (shown in Supplemental Figure I).
Case-control design and “fold-expression” reporting does not correct the limitations of microarray-based mRNA profiling
We considered that non-specific microarray hybridization signals in a case-control comparison might cancel each other out when the data are reported as the “fold-change”, rather than as absolute transcript expression values. Accordingly, we compared the results of our microarray and RNA sequencing data expressed as fold-regulation between Gq and non-transgenic mouse hearts. Microarray analysis of Gαq transgenic heart mRNA identified 841 differentially expressed transcripts (FDR<0.03, P<0.001, fold-change ≥1.3; 382 upregulated and 459 downregulated) (Supplemental Table VI). At the same P-value cutoff and fold-change threshold, RNA sequencing identified differential expression of only 125 genes; 77 upregulated and 48 downregulated (Supplemental Tables VI and VII). Of the 752 genes reported as differentially regulated by microarrays, but not by RNA sequencing, 45 were not detected in any hearts by RNA sequencing, and the vast majority (n=674) were present at or less than 20 copies per cell, i.e. in the group where we had shown that microarrays provide the least confident data. Unfortunately, one of the disadvantages of the “fold-change” analytical approach is that these critical absolute expression data are not evident, making it difficult to assess the validity of individual results as a function of absolute mRNA levels.
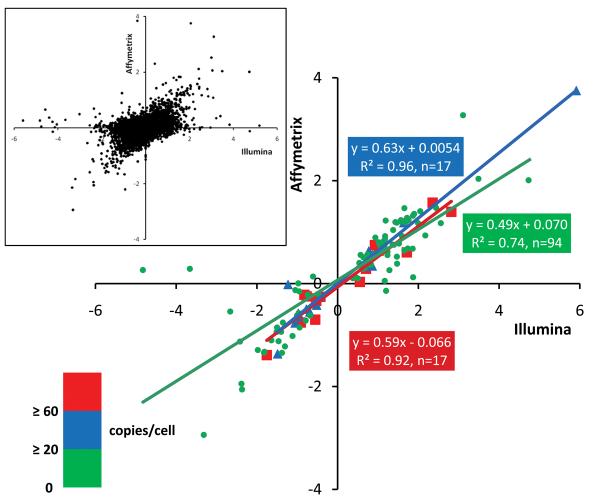
To better understand the striking disparities between microarray and RNA-sequencing based differential mRNA profiling, we plotted the fold expression values provided either by microarray or RNA sequencing of only the regulated mRNAs (i.e. those whose Gαq/nontransgenic expression was P<0.001 by RNA sequencing), stratified by mRNA abundance in normal hearts. Thus, three individual regression lines were generated, separately reporting fold mRNA expression change in Gαq hearts for very high abundance transcripts (>60 copies/cell, n=14), common transcripts (20-60 copies/cell, n=17), and low abundance transcripts (<20 copies/cell, n=94) (Figure 4). The correlation of “fold-regulation” between RNA sequencing and microarray data was good for the very high abundance and common mRNAs (Pearson r2=0.92 and 0.96, respectively), but was suboptimal for low abundance mRNAs, even though there were more data points in this group (r2=0.74). Furthermore, the slopes of all three linear regressions were very shallow (0.49-0.63), again reflecting a compressed range of expression data for microarray results in comparison to RNA sequencing.
Figure 4. Fold-changes in gene expression determined by RNA sequencing vs microarray, in Gq-overexpressing compared to normal hearts.
Fold-change in gene expression determined by RNA sequencing (Illumina) is plotted against fold-change in gene expression determined by microarrays (Affymetrix), using a log2 scale, for the 125 significantly regulated genes defined in Table 1. Red squares denote genes expressed at or above 60 copies/cell in nontransgenic hearts, blue triangles denote genes expressed between 20-60 copies/cell, and green circles denote genes expressed at or less than 20 copies/cell. Inset: comparison of fold-changes in all genes detected using RNA sequencing and microarrays, regardless of significance.
Quantification of mRNA counts by RNA sequencing is critically dependent upon correct assignment of sequence reads to their proper genes, i.e. on accurate annotation. Thus, mis-identification might explain some discrepancies between RNA sequencing and microarrays. To assess this possibility, we performed directed RT-qPCR for ten transcripts representing a range of absolute expression values and relative regulation in Gαq and nontransgenic hearts (Figure 5), and compared the results to those we obtained on the same RNA samples by sequencing and arrays (Table 1). When all three methods reliably detected mRNAs in both experimental groups, the increase in mRNA content in Gαq vs nontransgenic hearts, described as “fold-expression”, was similar between RT-qPCR and RNA sequencing, but these values were compressed by microarray (Table 1). These data suggest that mis-identification of sequence reads is not responsible for differences in fold-expression reported from RNA sequencing and microarrays. However, RNA sequencing detected transcripts from both Gαq and nontransgenic hearts for only five mRNAs, and RT-qPCR was either below the threshold for accuracy (Ct ≥35) or could not detect a further four transcripts. On the other hand, RT-qPCR and microarrays agreed for detection of Tuba1a, which was not observed by RNA sequencing. This single instance likely represents a mis-annotation artifact. We also examined the degree of inter-sample variability in the three assay techniques, comparing expression of genes in Table 1 between the 4 nontransgenic, or the 4 Gαq, biological replicates. RT-qPCR exhibited the greatest variance, with the lower-abundance genes recording higher variation. RNA sequencing had less variance, and microarrays showed the least variance (due to data range compression). In short, RNA sequencing has less variance than RT-qPCR without the data compression inherent in microarrays.
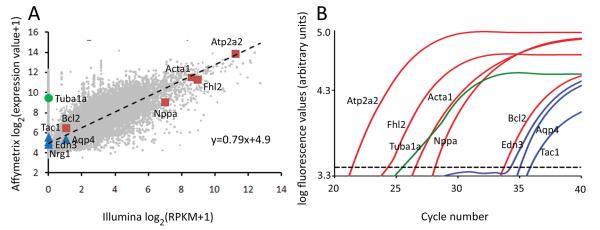
Figure 5. Comparison of gene expression by microarray, RNA sequencing and RT-qPCR.
(A) Gene expression in nontransgenic hearts, determined by RNA sequencing, plotted against gene expression determined by microarrays. Values shown are log2(RPKM+1) for RNA sequencing (x-axis) and log2(Affymetrix signal units+1) for microarrays. Light gray, all expressed genes with regression line as in Figure 3. Red squares, genes regulated by both arrays and sequencing; blue triangles, genes regulated on arrays but poorly detected by sequencing; green circle, highly expressed on arrays but not detected by sequencing. (B) Representative TaqMan qPCR traces for genes shown in (A). Dotted line = fluorescence threshold for Ct determination.
Table 1. Comparison of gene regulation measured by microarray, RNA sequencing, and RT-qPCR methods.
Fold-change indicates the difference in gene expression observed between Gq and ntg samples for a particular method.
| RNA sequencing | Microarray | RT-qPCR | ||||
|---|---|---|---|---|---|---|
| Gene | ntg copies/cell |
Gq copies/cell |
Fold-change | Fold- change |
Fold- change |
|
| Nppa | 43 | 2604 | 60.2 | 13.4 | 98.8 | |
| Acta1 | 127 | 897 | 7.0 | 2.6 | 7.9 | |
| Atp2a2 | 811 | 458 | −1.8 | −1.2 | −2.2 | |
| Fhl2 | 267 | 50 | −3.4 | −2.6 | −4.0 | |
| Bcl2 | 0.4 | 2 | 5.4 | 2.8 | 4.6 | |
| Edn3 | nd | 1.6 | 3.1 | 2.5 | Ct > 35 | |
| Nrg1 | nd | 1.0 | 2.9 | nd | Ct > 35 | |
| Aqp4 | 0.4 | nd | −1.6 | −11.7 | Ct > 35 | |
| Tac1 | nd | nd | −1.7 | −4.0 | Ct > 35 | |
| Tuba1a | nd | nd | 1.3 | 1.1 | ||
nd = not detected.
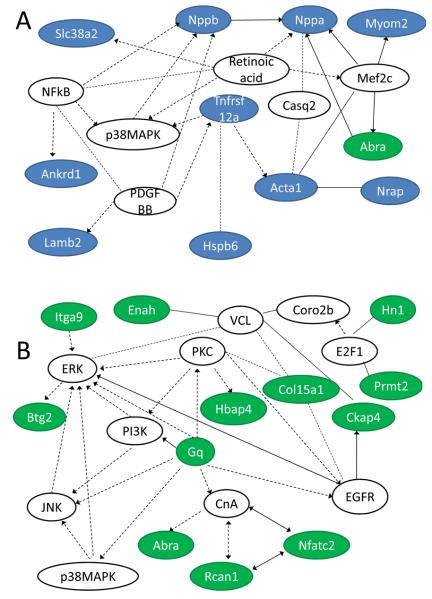
The utility of RNA sequencing is to derive greater understanding of gene regulation pathways in normal and diseased tissue. We used Ingenuity Pathways Analysis software (http://www.ingenuity.com) to examine possible signaling relationships between the regulated genes in Gαq hearts (Figure 4, Supplemental Table VII). Seventy-two of 77 Gαq-upregulated genes and 42 of 48 Gαq-downregulated genes were assigned to signaling networks (Supplemental Table VIII). Interestingly, separating the data into low- and high- abundance transcripts generated networks with a distinctly different focus (Figure 6 and Supplemental Table VIII). The most intriguing networks focused on cellular growth/proliferation and on cell death. A signaling pathway involving members of these two networks suggests relationships between Gnaq, Rcan (regulator of calcineurin), Nfatc2, and Abra (actin-binding Rho activating protein), as well as signals involving the EGF receptor and the MAP kinase family (Figure 6).
Figure 6. Signaling networks of Gαq-regulated transcripts.
Ingenuity Pathways Analysis software (http://www.ingenuity.com) was used to depict potential signaling pathways between (A) high-abundance and (B) low-abundance, Gαq-regulated gene products. Lines with arrowheads, molecule acts on a target; lines without arrowheads, binding only. Solid lines, direct interaction; dotted lines, indirect interaction. Blue background, high-abundance genes; green background, low-abundance genes; white background, member of signaling pathway but not regulated in Gαq hearts.
Discussion
Here, we describe methods for RNA sequencing and transcript analysis in mouse cardiac models, and demonstrate that RNA sequencing has advantages for quantitative analysis of cardiac-expressed transcripts in normal and hypertrophied mouse hearts.
The idea that transcriptional signatures can provide information into the nature and/or cause of cardiac disease is decades old, but has yet to completely fulfill its promise. The Nidal-Ginard and Chien laboratories were among the first to describe prototypical transcriptional changes in heart disease 29, 32. There followed the description of a handful of “fetal genes” whose expression was strikingly increased in cardiac hypertrophy and/or failure 33-35. Measures of these few transcripts by Northern blotting, and more recently real-time quantitative PCR, became standard as early markers of cardiac pathology 1, 2. In-depth analysis of the transcriptome has revealed specific transcriptional signatures for physiological versus pathological hypertrophy, and for early versus late heart failure 3.
RNA sequencing takes advantage of massively parallel next generation DNA sequencing platforms that are increasingly available at academic institutions and industry. After development, optimization, and implementation of these techniques, preparation of bar-coded cDNA sequencing libraries from cardiac RNA took one individual 3 days to complete, the Illumina sequencing took 2 days, and sequence alignment and initial analysis were completed over a 2 day period. The total cost for sequencing eight mouse heart mRNA libraries (two Illumina GA II sequencing lanes) was $2100, including reagents and instrument time. By comparison, the Core turnover for Affymetrix microarray results was 2 weeks, at a cost of $3600 for eight cardiac mRNA profiles. Most importantly, RNA sequencing provided accurate quantitative digital data on lower abundance transcripts where most of the gene regulation was found, but that were measured less reliably on microarrays.
The arrays appear to be reporting changes in relative gene expression for large numbers of mRNAs that are expressed at absolute levels below those that are optimal for array-based measurements, 20 copies per cell. In contrast, RNA sequencing appeared exquisitely sensitive to changes in the abundance of these rare transcripts, as 94 of the 125 Gαq-regulated mRNAs reported by this technique were present at less than 20 copies per cell in the control, nontransgenic hearts. Thus, the major disparity between the microarray transcriptional profiling and RNA sequencing occurs with mRNAs expressed at low levels: microarrays falsely reported regulation of the rare transcripts, but also failed to detect changes in almost one quarter (36 of 125) of significantly regulated low abundance mRNAs. We do note that whole-organ transcriptomes, such as we have studied here, are not cell-type specific and thus some low abundance mRNAs (especially at or below 1 copy/cell) may not originate from cardiac myocytes. However, cardiac myocyte-specific transgenesis in mouse hearts can provide cell-autonomous data in the in vivo context, and even after physiological modeling, that is not possible with cultured cells. This is especially important since elucidation of new signaling pathways from RNA sequencing studies will likely involve transcripts expressed at lower abundance, in accordance with our observation that high- and low-abundance Gαq-regulated transcripts generated different signaling networks.
Another potential advantage of RNA sequencing over traditional microarrays is the ability to easily identify alternatively spliced transcripts. While the newer generation of Affymetrix microarrays utilizes probesets that cover multiple exons of a gene, the limited ability of array hybridization techniques to provide quantitative information on gene or exon expression complicates non-biased identification and quantification of alternatively spliced isoforms. In addition, the Tophat/Cufflinks analytical software we employ provide the ability to detect both known and novel splicing events via aligning sequence reads to the entire genome, whereas microarrays are limited by the probesets placed on the arrays. While this aspect of RNA sequencing was not the focus of the current studies, we explored the potential of this application for several cardiac-expressed genes known to undergo alternative splicing: Both transcripts for Atp2a2 (SERCA2) 36, both cardiac-expressed transcripts of the creatine transporter Slc6a8 37, three of five known splice forms of Cacna1c (Cav1.2) 38, three of five alternative transcripts of Ank2 (ankyrin B) 39, and both transcripts of Bnip3l (Nix) 18 were detected, although we failed to observe alternative splicing of Ccrk 40 that is expressed at less than 1 copy per cell. These findings suggest that RNA sequencing can readily detect alternative splicing events, but that more sequencing reads may be required to detect rare alternately spliced isoforms.
Unbiased transcriptional profiling of mouse cardiac models has increasingly been used to identify the impact of molecular perturbations on heart development, adaptation, and function, and to define specific molecular mediators of these effects 41. The potential for RNA sequencing to detect subtle regulated events is great, but the technology is relatively new, the creation and bar-coding of libraries is unfamiliar to many laboratories, and the analytical platforms can be challenging. Here, we describe an RNA-sequencing pipeline that, once it is implemented, is essentially turn-key in its operation. Although RNA sequencing is the de-facto “gold standard” technique for identifying and quantifying transcripts, to our knowledge it has not been applied to profiling of mouse cardiac genes, and so we were careful to compare the results of RNA sequencing to microarray profiling in the well-characterized Gαq model of cardiomyopathy. We found that Illumina RNA sequencing is rapid, accurate, highly sensitive for identifying both abundant and rare transcripts, and has significant advantages in time- and cost-efficiencies over Affymetrix microarray analysis. We expect that these advantages will not only be similar with any massively parallel RNA sequencing platform, but that the relative benefits of RNA sequencing over microarrays will continue to increase as the technology advances.
Novelty and Significance.
What is known?
Accurate, unbiased gene transcription profiling is necessary for understanding disease mechanisms and can assist in determining diagnosis and prognosis.
Large-scale transcriptional profiling has been performed using microarrays for several years, but expense, limited dynamic range and background correction remain problematic.
What new information does this article contribute?
Next-generation (high-throughput) sequencing of RNA was compared to microarray analysis in a case-control design, using the established Gq-overexpression mouse model of heart failure.
DNA-barcoding of individual heart samples permits multiple samples to be sequenced simultaneously, bringing the cost of RNA sequencing below that of microarrays.
RNA sequencing provides quantitative gene expression data and reveals a greater dynamic range of gene regulation.
RNA sequencing offers insight into regulation of low-abundance genes that microarrays cannot.
Summary
RNA profiling offers detailed insight into critical transcriptional regulatory mechanisms in health and disease. The value of transcriptional profiling is affected by the accuracy of the data and the sensitivity to detect changes in expression of uncommon mRNAs. RNA sequencing using massively parallel next generation platforms has the potential to enhance accuracy and depth of transcriptional signatures. We compared cardiac gene expression profiling using RNA sequencing and the current state-of-the art microarrays, applying both platforms to analysis of mRNA expression in the Gq-overexpression mouse model of cardiac hypertrophy. We describe how to individually DNA barcode RNA sequencing libraries from individual hearts for batch sequencing that reduces cost while providing digital mRNA expression data at or below the level of 1 RNA copy per cell. We found that RNA sequencing and microarrays provided comparable data on regulation of high-abundance genes, but that RNA sequencing was superior for detection and quantitation of low-abundance genes, which represent the majority of regulated genes in the Gaq model. The widespread implementation of RNA sequencing in disease studies should enhance diagnostic and prognostic profiling, facilitating a more detailed description of signaling mechanisms involving low-abundance genes that were previously missed with microarray.
Supplementary Material
Acknowledgements
None
Sources of funding
Supported by NIH R01s HL087871 and HL059888, and by UL1 RR024992 from the National Center for Research Resources.
Non-standard abbreviations and acronyms
- Ct
cycle threshold
- CV
coefficient of variation
- Ntg
non-transgenic
- PMAGE
polony multiplex analysis of gene expression
- RPKM
reads per kilobase of exon per million mapped reads
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- SAGE
serial analysis of gene expression
- TG
transgenic
Footnotes
Disclosures
The authors declare that they have no conflicts of interest relating to this manuscript.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dorn GW, II, Robbins J, Sugden PH. Phenotyping hypertrophy - eschew obfuscation. Circ Res. 2003;92:1171–1175. doi: 10.1161/01.RES.0000077012.11088.BC. [DOI] [PubMed] [Google Scholar]
- 2.Dorn GW., II The fuzzy logic of physiological cardiac hypertrophy. Hypertension. 2007;49:962–970. doi: 10.1161/HYPERTENSIONAHA.106.079426. [DOI] [PubMed] [Google Scholar]
- 3.Dorn GW, II, Matkovich SJ. Put your chips on transcriptomics. Circulation. 2008;118:216–218. doi: 10.1161/CIRCULATIONAHA.108.789933. [DOI] [PubMed] [Google Scholar]
- 4.D'Angelo DD, Sakata Y, Lorenz JN, Boivin GP, Walsh RA, Liggett SB, Dorn GW., II Transgenic Gαq overexpression induces cardiac contractile failure in mice. Proc Natl Acad Sci USA. 1997;94:8121–8126. doi: 10.1073/pnas.94.15.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorn GW, II, Tepe NM, Lorenz JN, Koch WJ, Liggett SB. Low- and high-level transgenic expression of β2-adrenergic receptors differentially affect cardiac hypertrophy and function in Gαq-overexpressing mice. Proc Natl Acad Sci USA. 1999;96:6400–6405. doi: 10.1073/pnas.96.11.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., II Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 8.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat Biotechnol. 2010 doi: 10.1038/nbt.1621. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., II Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW., II MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 14.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., 2nd Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhter SA, Luttrell LM, Rockman HA, Iaccarino G, Lefkowitz RJ, Koch WJ. Targeting the receptor-Gq interface to inhibit in vivo pressure overload myocardial hypertrophy. Science. 1998;280:574–577. doi: 10.1126/science.280.5363.574. [DOI] [PubMed] [Google Scholar]
- 16.Adams JW, Sakata Y, Davis MG, Sah VP, Wang Y, Liggett SB, Chien KR, Brown JH, Dorn GW., II Enhanced Gαq signaling: A common pathway mediates cardiac hypertrophy and apoptotic heart failure. Proc Natl Acad Sci USA. 1998;95:10140–10145. doi: 10.1073/pnas.95.17.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aronow BJ, Toyokawa T, Canning A, Haghighi K, Delling U, Kranias E, Molkentin JD, Dorn GW., II Divergent transcriptional responses to independent genetic causes of cardiac hypertrophy. Physiol Genomics. 2001;6:19–28. doi: 10.1152/physiolgenomics.2001.6.1.19. [DOI] [PubMed] [Google Scholar]
- 18.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., II Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 19.Sakata Y, Hoit BD, Liggett SB, Walsh RA, Dorn GW., II Decompensation of pressure-overload hypertrophy in Gαq-overexpressing mice. Circulation. 1998;97:1488–1495. doi: 10.1161/01.cir.97.15.1488. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Toyokawa T, Hahn H, Dorn GW., II Epsilon protein kinase C in pathological myocardial hypertrophy. Analysis by combined transgenic expression of translocation modifiers and Galphaq. J Biol Chem. 2000;275:29927–29930. doi: 10.1074/jbc.C000380200. [DOI] [PubMed] [Google Scholar]
- 21.Syed F, Odley A, Hahn HS, Brunskill EW, Lynch RA, Marreez Y, Sanbe A, Robbins J, Dorn GW., II Physiological growth synergizes with pathological genes in experimental cardiomyopathy. Circ Res. 2004;95:1200–1206. doi: 10.1161/01.RES.0000150366.08972.7f. [DOI] [PubMed] [Google Scholar]
- 22.Satoh M, Matter CM, Ogita H, Takeshita K, Wang CY, Dorn GW, II, Liao JK. Inhibition of apoptosis-regulated signaling kinase-1 and prevention of congestive heart failure by estrogen. Circulation. 2007;115:3197–3204. doi: 10.1161/CIRCULATIONAHA.106.657981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goffart S, Kleist-Retzow JC, Wiesner RJ. Regulation of mitochondrial proliferation in the heart: power-plant failure contributes to cardiac failure in hypertrophy. Cardiovasc Res. 2004;64:198–207. doi: 10.1016/j.cardiores.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 26.Dorn GW., II Apoptotic and non-apoptotic programmed cardiomyocyte death in ventricular remodelling. Cardiovasc Res. 2009;81:465–473. doi: 10.1093/cvr/cvn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorn GW, II, Brown JH. Gq signaling in cardiac adaptation and maladaptation. Trends Cardiovasc Med. 1999;9:26–34. doi: 10.1016/s1050-1738(99)00004-3. [DOI] [PubMed] [Google Scholar]
- 28.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 29.Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 30.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izumo S, Lompre AM, Matsuoka R, Koren G, Schwartz K, Nadal-Ginard B, Mahdavi V. Myosin heavy chain messenger RNA and protein isoform transitions during cardiac hypertrophy: interaction between hemodynamic and thyroid hormone-induced signals. J Clin Invest. 1987;79:970–977. doi: 10.1172/JCI112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bishopric NH, Simpson PC, Ordahl CP. Induction of the skeletal alpha-actin gene in alpha 1-adrenoceptor-mediated hypertrophy of rat cardiac myocytes. J Clin Invest. 1987;80:1194–1199. doi: 10.1172/JCI113179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockmann PT, Will DH, Sides SD, Brunnert SR, Wilner GD, Leahy KM, Wiegand RC, Needleman P. Reversible induction of right ventricular atriopeptin synthesis in hypertrophy due to hypoxia. Circ Res. 1988;63:207–213. doi: 10.1161/01.res.63.1.207. [DOI] [PubMed] [Google Scholar]
- 35.Waspe LE, Ordahl CP, Simpson PC. The cardiac β-myosin heavy chain isogene is induced selectively in α1-adrenergic receptor-stimulated hypertrophy of cultured rat heart myocytes. J Clin Invest. 1990;85:1206–1214. doi: 10.1172/JCI114554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ver Heyen M, Heymans S, Antoons G, Reed T, Periasamy M, Awede B, Lebacq J, Vangheluwe P, Dewerchin M, Collen D, Sipido K, Carmeliet P, Wuytack F. Replacement of the muscle-specific sarcoplasmic reticulum Ca2+-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction-relaxation of the heart. Circ Res. 2001;89:838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Munoz C, Rosenberg EH, Jakobs C, Salomons GS. Identification, characterization and cloning of SLC6A8C, a novel splice variant of the creatine transporter gene. Gene. 2008;418:53–59. doi: 10.1016/j.gene.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Liao P, Yong TF, Liang MC, Yue DT, Soong TW. Splicing for alternative structures of Cav1.2 Ca2+ channels in cardiac and smooth muscles. Cardiovasc Res. 2005;68:197–203. doi: 10.1016/j.cardiores.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 39.Hashemi SM, Hund TJ, Mohler PJ. Cardiac ankyrins in health and disease. J Mol Cell Cardiol. 2009;47:203–209. doi: 10.1016/j.yjmcc.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu H, Dai H, Jain K, Shah R, Hong C, Pain J, Tian B, Vatner DE, Vatner SF, Depre C. Characterization of a novel cardiac isoform of the cell cycle-related kinase that is regulated during heart failure. J Biol Chem. 2008;283:22157–22165. doi: 10.1074/jbc.M710459200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School. http:///www.cardiogenomics.org, accessed December 16, 2009.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.