Abstract
p53 and ARF are well-established tumor suppressor proteins that function together in the negative regulation of cancer. Recently, both of these proteins were found to play surprising roles in autophagy. Autophagy (“self-eating”) is a critical response of eukaryotic cells to metabolic and other stress. During this process, portions of the cytosol are sequestered into characteristic double membrane vesicles that are delivered to the lysosome for degradation, leading to the release of free amino acids and subsequent survival. The mechanisms whereby p53 and ARF control autophagy are only now becoming elucidated. An emerging question is whether we can develop metabolic poisons that preferentially destroy tumor cells depending on their reliance on autophagy for survival, and on their p53 and ARF status.
p53 and ARF in cancer
The Tp53 and Ink4a/ARF genes encode the well-known tumor suppressor proteins p53 and ARF, which respond to oncogenic, genotoxic and other stresses to induce growth arrest or apoptosis. These two proteins participate in a signaling module that is subject to frequent mutational inactivation in cancer. In this signaling pathway, oncogene activation leads to transcriptional upregulation of ARF. Induced ARF then binds and inhibits the E3 ubiquitin ligase for p53 (MDM2), which results in stabilized p53 (see [1] for review). ARF is itself subject to transcriptional repression by p53, and tumor cells with mutant p53 frequently express high levels of ARF [2–4]. Both of these proteins have recently been found to be intimately linked to metabolic pathways, and to play roles in autophagy. Interestingly, the relationship between p53 and ARF in autophagy is as complex as it is in cancer: in the stressed cell, both proteins play positive roles in autophagy. Conversely, in the unstressed cell, p53 negatively regulates this process, in part due to its ability to negatively regulate ARF (Figure 1).
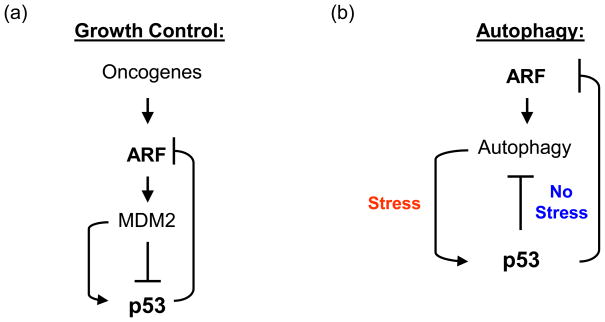
Figure 1. The p53-ARF axis in cancer and autophagy.
A) In the p53-ARF-MDM2 pathway in cancer, oncogenes transcriptionally induce ARF, which binds and inhibits MDM2, leading to stabilization of p53. In two feedback loops, p53 positively regulates MDM2 transcription, and negatively regulates ARF transcription.
B) In the p53-ARF axis in autophagy, p53 and ARF positively regulate autophagy in stressed cells. In contrast, in the unstressed cell, p53 negatively regulates ARF via transcriptional repression, and it also negatively regulates autophagy.
Autophagy is a key cell survival pathway
Autophagy is an evolutionarily-conserved homeostatic process where cytosolic components are targeted for removal or turnover in membrane-bound compartments (autophagosomes) that fuse with the lysosome (Figure 2; see also Box 1). Types of autophagy include macro-autophagy, chaperone-mediated autophagy, and micro-autophagy. Macro-autophagy refers to the rather non-selective sequestration of cytoplasmic substrates. In contrast, chaperone-mediated autophagy involves recognition of specific substrates by the heat shock cognate 70 (Hsc70) machinery. Micro-autophagy involves lysosomal invagination and ingestion of minor portions of the cytosol. For this review, the focus is on macro-autophagy. As only a few key components of the molecular machinery of macro-autophagy are referenced, readers are referred to two excellent recent reviews for a more detailed overview of this pathway [5, 6].
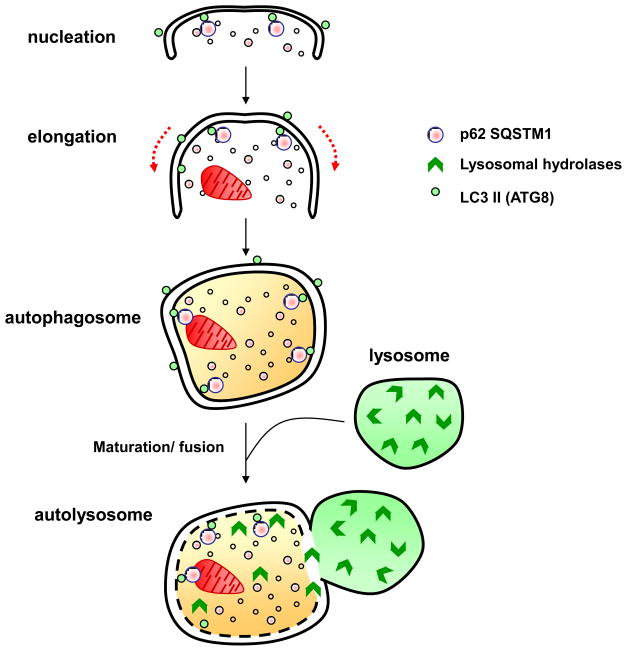
Figure 2. Major cellular events during autophagy.
i) Nucleation: appearance of double layered, isolation membrane, the phagophore, ii) Elongation: extension of the phagophore, iii) autophagosome formation, encapsulation of cellular components and iv) Maturation: fusion of the autophagosome with the lysosome, resulting in formation of the autophagolysosome. Ultimately, the components in the autophagolysosome are degraded by lysosomal hydrolyses and the breakdown products are recycled in cellular bio-synthetic pathways.
Box 1. Macroautophagy.
Macroautophagy, often referred to simply as autophagy, is a relatively non-specific degradation process that delivers organelles and long-lived proteins to the lysosome via a double membrane vesicle termed the autophagosome (Figure 1). This process targets defective organelles, misfolded proteins and proteins too large to be delivered to the proteasome for degradation. Autophagy ultimately yields amino acids and fatty acids for use in catabolic breakdown or re-cycling in biosynthetic pathways, therefore maintaining cellular homeostasis during periods of stress. Autophagy can at times be selective for particular organelles, and based on the organelles involved, macroautophagy can be further sub-classified into mitophagy (mitochondria), reticulophagy (endoplasmic reticulum) and pexophagy (peroxisomes). At present the regulatory pathways governing the selective autophagy of organelles are only now becoming elucidated.
Macro-autophagy, hereafter referred to as autophagy, regulates the turnover of long-lived proteins and damaged or excess organelles. This process occurs constitutively at low levels, and is greatly induced during periods of metabolic stress. During such periods, lysosome-mediated digestion of proteins serves to release free amino acids that can be used to fuel ATP generation, thereby promoting survival of the cell. The pro-survival function of autophagy is evolutionarily conserved from yeast to mammals and is best characterized during nutrient deficiency. Knockout mice for the autophagy genes Atg5 and Atg7 develop normally in utero, but these mice die shortly after birth, as they do not survive the fasting process that occurs prior to nursing [7, 8]. Therefore, in response to nutrient limitation, autophagy is required to promote cell survival. There is some support for the premise that autophagy also constitutes a form of cell death. However, at present it is unclear if autophagy is truly a form of cell death, or merely represents an exhausted attempt to preserve cell viability. Additionally, direct evidence that autophagy induces rather than simply accompanies cell death in a physiological context are lacking. Therefore, the most compelling evidence supports a role for autophagy in cell survival. Not surprisingly, this process is believed to be harnessed by malignant cells to increase their likelihood of survival.
Autophagy and cancer
That autophagy has an important role in tumor development and progression is quite clear. However, the exact nature of this role is different for tumor initiation compared to tumor progression. Genetic evidence firmly indicates that autophagy is suppressive to tumor initiation. A key regulatory protein in autophagy is Beclin-1, which binds and activates the class III PI3-kinase Vps34, which is required for the induction of autophagy. The Beclin-1 heterozygous knockout mouse is predisposed to multiple tumor types, including lymphoma and liver cancer; in tumors from these mice, the wild type allele of Beclin-1 is not lost, so Beclin-1 is a haplo-insufficient tumor suppressor gene [9]. Consistent with this premise, Beclin-1 is mono-allelically deleted in certain cancers [10]. Two Beclin-1 binding proteins that both play important roles in the induction of autophagy are UVRAG and Bif-1; knockout mice for these proteins are predisposed to multiple spontaneous cancers [11, 12], and like Beclin-1, UVRAG is mono-allelically mutated in cancer [12].
There are several mechanisms where suppression of autophagy leads to tumor initiation. White and colleagues have found that compromised autophagy, conferred by mono-allelic loss of Beclin-1 or deficiency of the key autophagy gene Atg5, leads to impaired survival following metabolic stress, but is also associated with increased reactive oxygen species and DNA damage; this leads to the appearance of cells containing gene amplification and aneuploidy [13–15]. The genetic instability induced by autophagy inhibition is believed to promote tumorigenesis by increasing mutation rate. As an added and possibly unrelated mechanism, autophagy is induced during, and facilitates the process of, senescence [16], suggesting that autophagy inhibition might facilitate transformation by oncogenes that typically induce senescence, such as Ras. Additionally, in some instances autophagy facilitates the completion of apoptosis (see Box 2) [17]. The combined data suggest that autophagy constitutes a multi-faceted tumor suppressive pathway.
Box 2. Links between autophagy and apoptosis or senescence.
Interesting links between autophagy and apoptosis, and autophagy and p53-medated senescence, have been uncovered. Specifically, individual depletion of either DRAM or the critical autophagy gene ATG5 inhibits p53-mediated apoptosis, suggesting that autophagy may facilitate apoptosis in some instances [17]. In support of this premise, the p53 target genes PUMA and Bax can induce mitophagy (autophagy of mitochondria), and inhibition of PUMA- or Bax-induced mitophagy dampens the apoptotic response [50]. Instead of apoptosis, in other cell types the p53 tumor suppressor protein can induce senescence. Recently, an interesting association between autophagy and senescence has been uncovered. Cellular senescence is a permanent cell cycle arrest induced by oncogene activation, DNA damage or replicative stress. Recently, Narita and colleagues found that a subset of autophagy genes are up-regulated during oncogene-induced senescence [16, 67]. Moreover, these researchers found that inhibition of autophagy delayed the onset of senescence [16, 67]. How autophagy facilitates senescence is not currently understood.
There are compelling data that established (as opposed to initiating) tumor cells use autophagy as a critical survival pathway. Autophagy allows cells to recoup ATP and essential building blocks for biosynthesis when they are starved of nutrients or exposed to hypoxia, and tumor cells can be exposed to such conditions regularly. Several groups have shown that the autophagy inhibitors chloroquine and 3-methyladenine are effective anti-tumor drugs for Burkitt s lymphoma and chronic myelogenous leukemia [18–20]. These data support the emerging premise that autophagy promotes the survival of established tumors. The general consensus in the field is that inhibiting autophagy can lead to tumor formation because it normally removes damaged mitochondria and misfolded proteins, which would otherwise lead to increased reactive oxygen species and mutagenic damage. In contrast, established tumors encounter increasing environmental stress, and autophagy is critical for tumors to survive such episodes. It is interesting to note that autophagy genes have not been found to be bi-allelically deleted or mutated in cancer, suggesting that a balance between the pro-tumorigenic and pro-survival functions of autophagy may be actively sought by the tumor cell.
p53 regulates stress-induced autophagy
The p53 tumor suppressor protein is a latent and inactive transcription factor in unstressed cells. Following genotoxic stress or oncogene activation, this protein becomes stabilized and activated as a transcription factor, capable of inducing a transient cell cycle arrest, senescence or apoptosis. The groups of Levine and Jin along with Thompson first showed that, like genotoxic stress, metabolic stress (nutrient deprivation) also activates p53 [21, 22]. As early as 20 minutes after glucose deprivation, p53 becomes phosphorylated on serine 15, a hallmark of p53 activation [21, 22]. This phosphorylation is mediated by the kinase AMPK, which serves as a fuel sensor in the cell by assessing the ratio of AMP to ATP. The activation of p53 by metabolic stress and AMPK leads to cell cycle arrest, thereby allowing cells to survive periods of nutrient deprivation. Some facets of this pathway remain unclear. First, it is not clear whether p53 is a direct or indirect target of AMPK, and it seems likely that an intermediate kinase is involved. Second, the downstream transcriptional pathway of p53 following nutrient stress has not been elucidated, and it is unclear if there are transcriptional targets of p53, or post-translational modifications, that are specific to this pathway.
In addition to nutrient deprivation, genotoxic stress also induces p53-dependent autophagy. Induction of p53 by DNA damaging agents in mouse embryo fibroblasts leads to inhibition of mammalian Target of Rapamycin (mTOR) and induction of autophagy, as assessed by autophagosome formation and LC3 lipidation and cleavage [21]. There were two mechanisms proposed for how p53 inhibits mTOR, which is a master regulator of autophagy: first, in a p53-dependent manner, genotoxic stress induces the activation of AMPK, which inhibits mTOR. Inhibition of AMPK by Compound C, a known AMPK inhibitor, completely abolishes the p53-dependent inhibition of mTOR [21]. Additionally, p53 transcriptionally activates negative regulators of mTOR: the β subunits of AMPK (AMPK β1 and AMPK β2) as well as TSC-2 and PTEN. Interestingly, the latter regulation is tissue specific; p53 up-regulates TSC-2 and PTEN only in tissues highly dependent on insulin-mediated glucose utilization during energy metabolism [23]. More recently Budanov and Karin showed that the products of two p53 target genes, Sestrin1 and Sestrin2, can bind and activate AMPK, causing it to phosphorylate TSC2 and thereby inhibit mTOR [24]. Nutrient starvation significantly increases the expression of Sestrin2, and loss of Sestrin2 markedly reduces the levels of p53-mediated autophagy [24]. The combined data indicate that p53 uses multiple avenues in order to inhibit mTOR and subsequently induce autophagy (Figure 3).
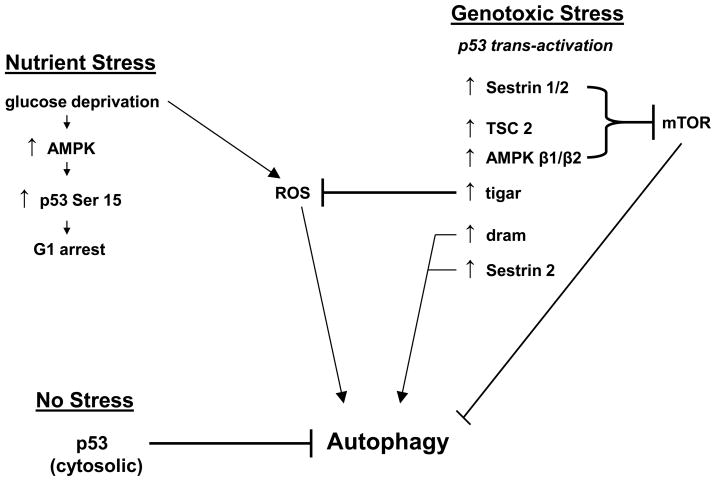
Figure 3. Pathways upstream and downstream from p53-mediated autophagy.
p53 is downstream of both nutrient stress and genotoxic stress. In nutrient-deprived cells, such as following glucose deprivation, p53 is phosphorylated and induces G1 arrest, thereby allowing for cell survival. In response to genotoxic stress, p53 likewise becomes active as a transcription factor, and transactivates a number of genes with direct roles in autophagy, such as DRAM, or that encode inhibitors of mTOR activity, including Sestrin 1 and 2, TSC2 and the β1 and -2 subunits of AMPK. In the unstressed cell, silencing or inhibiting p53 induces autophagy. Therefore, p53 is intimately tied to autophagy as both a positive and negative regulator, depending on the presence of stress.
Another mechanism whereby p53 induces autophagy emerged with the discovery by Ryan and colleagues that p53 directly transactivates the gene encoding DRAM (damage-regulated autophagy modulator). DRAM encodes a highly evolutionarily-conserved protein with six putative transmembrane domains that co-localizes with cathepsin D in the lysosome [17]. p53 binds directly to a consensus p53 binding site in the DRAM promoter following genotoxic stress and transactivates this gene [17]. Interestingly, silencing DRAM markedly inhibits both p53-mediated autophagy and apoptosis, suggesting these two outcomes are mechanistically linked (see Box 2). Along these lines, induction of DRAM is essential for the ability of some anti-tumoral compounds to induce p53-mediated cell death and autophagy [25]. More recently the p53 family member p73 has also been found to induce autophagy. p73 also transactivates DRAM, but this gene is not essential for the ability of p73 to induce autophagy [26]. Rather, it appears that p73 directly regulates an entire program of autophagy genes, including the well known autophagy genes Atg5 and Atg7 [27].
The emerging role of p53 in nutrient deprivation and autophagy prompted the group of Thompson to test the hypothesis that tumors without functional p53 might be more sensitive to nutrient deprivation, due to their decreased ability to respond to such stress with growth arrest and autophagy. Using two compounds that activate AMPK, the compound AICAR as well as the diabetes drug metformin, this group showed that p53-null tumor cells were markedly growth inhibited by these compounds, while neither compound affected the growth of p53 +/+ isogenic tumor cells [28]. These data offer promise that novel metabolic therapies may be developed for cancer. They may also shed light on the unexplained finding that individuals treated with metformin have reduced risk for cancer [29].
p53 regulates basal autophagy
Seemingly in contrast to the data that p53 induces autophagy in stressed cells, Kroemer and colleagues found that p53 inhibits basal autophagy in unstressed cells [30–32]. This group showed that silencing of p53, or pharmacological inhibition of this protein using the compound pifithrin-α, led to an increase in the steady state level of basal autophagy in the G1 and S phase of the cell cycle, in both mammalian cells and in C. elegans [30–32]. This activity required cytosolic p53, and indeed degradation of cytosolic p53 was shown to be required for autophagy induction in response to endoplasmic reticulum (ER) stress and other agents [31]. p53 has been implicated in the negative regulation of organismal aging (see [33] for review), and Kroemer s group found that silencing p53 in C. elegans induces autophagy, and this may explain an increase in lifespan of this organism [34]. Interestingly, the Kroemer group found that some tumor-derived point mutants of p53 can still inhibit autophagy; specifically, mutant forms of p53 that consistently localize to the cytosol are highly efficient autophagy inhibitors [35]. These findings suggest that wild type cytosolic p53 may inhibit autophagy via a protein-protein interaction that is retained by mutant p53; as such, analyses of tumor-derived mutant forms of p53 should help elucidate the nature of this cytosolic activity.
The ARF tumor suppressor induces autophagy
The p14ARF tumor suppressor (p19ARF in mouse, and hereafter referred to as ARF) is best known as a positive regulator of the p53 tumor suppressor protein that is negligibly expressed in normal cells but is up-regulated in tumors or following silencing of p53. It was first reported in 2006 that both human and mouse ARF can be translated from an internal methionine (Met48 in human, Met45 in mouse) to generate a short version of the protein denoted smARF (short mitochondrial ARF) [36]. smARF lacks the N-terminus of ARF, which encodes much of ARF s tumor suppressor functions. Immuno-localization and biochemical fractionation studies determined that, in contrast to full-length ARF, smARF predominantly localized to mitochondria [36]. The mitochondrial localization of smARF is supported by reports that both human and mouse smARF interact with the protein p32, which is mitochondrial [37, 38], though one group reports that smARF also localizes to the nucleus and cytoplasm [39].
Full-length ARF and smARF are generated in equal proportions using in vitro translation reactions; however, smARF has a very short half-life in vivo, as it is rapidly eliminated by proteasome-mediated degradation. Consequently, smARF is present at very low steady-state levels (less than 5% of total ARF protein) [36]. Kimchi and colleagues found that enforced expression of smARF resulted in abnormalities in mitochondrial membrane potential, which were caused by increased autophagy. This group showed that acute overexpression of smARF by transient transfection eventually leads to cell death accompanied by indicators of autophagy, and that this occurred in a p53- and caspase-independent manner [36].
Conflicting reports exist regarding the potential role of full-length ARF, versus smARF, in autophagy. Abida and Gu engineered a version of ARF in which the internal methionine was mutated, and showed that this form could still induce autophagy in transfected cells [40]. In contrast, Reef and Kimchi published findings that full length (nucleolar) ARF is incapable of inducing autophagy except when ARF was expressed at non-physiological levels, at which time ARF was observed in extra-nuclear compartments [41]. At present the contribution of full-length ARF to autophagy remains unresolved. Given that smARF is tightly regulated by proteasomal degradation, and that there are links between autophagy and proteasome-mediated degradation, it is tempting to speculate that smARF may be preferentially stabilized by metabolic stress, and that this form of ARF could play the major role in autophagy induced by such stress.
ARF-mediated autophagy can enhance cell survival and promote tumor progression
Recently our group reported that the Arf gene product (encoding both full-length ARF and smARF) has a pro-survival role in autophagy. For this study we took advantage of the fact that p53 transcriptionally represses the Arf gene, and that p53-null cells express high levels of endogenous ARF [3, 4]. Building upon reports that ARF is frequently overexpressed in B cell tumors [2], we silenced this gene in B cell lymphomas using two different short hairpins for ARF, and found that silencing ARF impaired autophagy, and also inhibited the ability of these cells to establish as tumors in vivo [42]. These data support a cytoprotective role for ARF and autophagy. Consistent with this premise, we found that silencing ARF reduced the survival of lymphoma cells and mouse embryo fibroblasts exposed to nutrient deprivation [42]. These data may explain why a large percent of human tumors with mutant p53 retain high levels of ARF expression [2, 43].
Other evidence that ARF confers a selective advantage to some tumors is emerging. In most cancer-prone animal models, deletion of ARF accelerates tumorigenesis, in part through its ability to positively regulate p53 (see [44] for review). Recently however, deletion of ARF was shown to impede the development of prostate cancer in a conditional PTEN knockout mouse [45]. Deletion of the PTEN tumor suppressor gene, which occurs frequently in cancer, leads to increased AKT activity; the latter is known to inhibit autophagy, in part by its ability to activate mTOR, a negative regulator of autophagy. In mice with deletion of PTEN in the prostate, tumors eventually develop, but in mice with both PTEN and ARF deleted, cancer development was inhibited [45]. Therefore in this cancer model ARF confers a positive growth advantage. One could hypothesize that the impaired autophagy induced by PTEN loss was exacerbated by the further loss of ARF, and the level of autophagy in these tumors was too low for survival. This remains speculative however, as the authors did not analyze tumors for the level of autophagy.
One wonders why the tumor-promoting role of ARF was not seen until only recently. One possibility is that the cytoprotective role of ARF and autophagy may be restricted to certain tumor or tissue types. Along these lines, we found that silencing ARF in three different primary lymphoma cell lines reduced survival and progression in vivo [42]. In contrast, silencing ARF in a p53-null sarcoma cell line enhanced the progression of this tumor in a xenograft assay, despite clearly inhibiting autophagy [46]. Additionally, silencing of Beclin-1 restricts the growth of some tumor lines, but enhances others [46]. These data indicate that autophagy may be cytoprotective only for certain tumors.
Mechanisms of ARF-induced autophagy
Using the technique of 2-Dimensional In-gel Electrophoresis, ARF-interacting mitochondrial proteins have recently been identified. This study revealed that at the mitochondria ARF physically interacts with the Bcl2 family member Bcl-xl [47]. Bcl-xl plays a known role in autophagy; it interacts with Beclin-1 and negatively regulates the kinase activity of the Vps34/Beclin-1 complex, which is a key mediator of autophagy [48, 49]. Notably, ARF was shown to inhibit the ability of Bcl-xl to associate with Beclin-1 complexes. Interestingly, the Beclin-1/Bcl-xl complex is notoriously difficult to detect by co-immunoprecipitation; indeed, in cells with high levels of ARF, the Beclin-1/Bcl-xl complex is undetectable. However, in these same cells, when ARF is silenced, the complex is readily detectable by immunoprecipitation followed by imunoblotting [47]. While regulation of Beclin-1/Bcl-xl complex formation is one mechanism how ARF induces autophagy, the fraction of ARF that interacts with Bcl-xl in cells is low, and there are undoubtedly other mechanisms remaining to be identified. Additionally it remains unclear whether another mitochondrial ARF-interacting protein, p32, which plays a clear role in apoptosis, likewise plays a role in autophagy [37].
Conclusions and Future Directions
The pathways in which p53 and ARF function in autophagy, in the stressed and unstressed cell, are depicted in Figure 4. At first glance, the roles of p53 in autophagy appear contradictory. Specifically, stress-activated p53 induces autophagy, while unstressed p53 represses basal levels of autophagy. One possibility is that genotoxic stress-induced p53, particularly the p53-induced genes PUMA and Bax, may selectively induce mitochondrial autophagy (mitophagy), thus allowing for more effective release of mitochondrial inducers of apoptosis such as cytochrome c, and more efficient apoptosis [50]. In contrast Kroemer s group found that silencing IREα, which controls one arm of the endoplasmic reticulum (ER) stress pathway, effectively inhibits the ability of p53 to suppress autophagy [31]; in other words, silencing of p53 may predominantly signal ER stress and concomitant autophagy. As such, careful delineation of the types of autophagy being induced (that is, mitophagy versus reticulophagy), along with the upstream signaling pathway (such as nutrient deprivation, ER stress or genotoxic stress) may clarify some of these apparent discrepancies. Other apparent inconsistencies need to be put into some context: for example, p53 transactivates at least one negative regulator of autophagy, TIGAR (TP53-induced glycolysis and apoptosis regulator). During nutrient and metabolic stress TIGAR reduces reactive oxygen species (ROS) levels and significantly down-regulates autophagy [51]. Where TIGAR fits into the p53-autophagy pathway is not known. Additionally, the rules that govern whether a tumor is inhibited, or enhanced, by autophagy inhibition need to be determined.
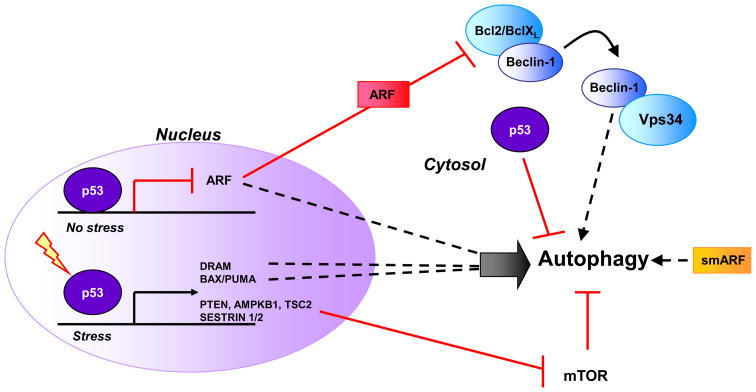
Figure 4. Regulation of autophagy by p53 and ARF.
Based on cellular localization and type of stress p53 can be a positive or negative regulator of autophagy. In the nucleus p53 transcriptionally regulates a number of autophagy modulators. Cytosolic p53 inhibits autophagy in unstressed cells. ARF positively regulates autophagy by disrupting the Bcl-xl/Beclin 1 complex, releasing Beclin-1 to induce autophagy. A truncated form of ARF, smARF directly localizes to the mitochondria to induce autophagy.
The ability to inhibit autophagy and sensitize tumors to metabolic stress is a promising avenue for cancer therapy. As indicated above, efforts to use chemical inhibitors of autophagy, such as chloroquine and metformin, have been successful in pre-clinical settings (see Table 1). Specifically, pre-clinical studies with chloroquine, which inhibits lysosome function and hence autophagy, showed considerable efficacy when combined with a genotoxic agent in a mouse model of B cell lymphoma [18]. Similarly, chloroquine was found to synergize with the chromatin-modifying agent SAHA in the killing of imatinib-resistant chronic myelogenous leukemia [19]. A fraction of the heat shock protein 70 (HSP70) is known to function in a cancer-specific manner to stabilize lysosomes [52, 53]. Along these lines recently an inhibitor of Hsp70 (2-Phenylethynesulfonamide) has been described that impairs lysosome function and inhibits autophagy; notably, this compound has shown success in a mouse model of B cell lymphoma [54]. These compounds should lead the way in our efforts to use autophagy modulators for cancer therapy. Caution should be exercised regarding this route of cancer therapy, however. Specifically, most cancer cells are believed to be impaired in the pathways of apoptosis. Autophagy inhibition in this setting has been shown to induce necrotic cell death, which can be detrimental to tumor progression. However, White and colleagues found that inhibition of autophagy in apoptosis-defective tumor cells leads to necrosis; in their model system necrosis stimulates inflammation and cytokine production, and led to increased tumor development [55]. Therefore, enthusiasm for autophagy inhibition as a novel pathway for cancer therapy should be tempered with caution until more studies can show that the necrosis induced by them does not promote tumor progression.
Table 1.
| Compound: | Mechanism of Action | References: |
|---|---|---|
| ABT 737 | Binds and inhibits Bcl-xl | [56, 57] |
| Chloroquine | A lysomotropic drug that inhibits autophagy; also used to treat malaria and rheumatoid arthritis | [58] |
| 3-methyladenine | A PI3 kinase inhibitor that inhibits autophagy, via the class III PI3 kinase Vps34 | [59–62] |
| Metformin | A diabetes drug that leads to activation of AMPK and induction of autophagy | [28] |
| Rapamycin | mTOR inhibitor that induces autophagy | [63] |
| RAD001 (Everolimus) | mTOR inhibitor that induces autophagy | [64, 65] |
| Temsirolimus (CCI-779) | mTOR inhibitor that induces autophagy | [66] |
| 2-Phenylethynesulfonamide (PES); Also known as pifithrin-μ | An Hsp70 inhibitor that impairs lysosome function and inhibits autophagy. | [54] |
| LY294002 | PI3Kinase inhibitor | [60] |
| Wortmannin | PI3Kinase inhibitor | [60] |
Acknowledgments
The authors would like to thank Marika K. Bergenstock for aid with figures, members of the Murphy lab, Donna George and Julia I-Ju Leu for critical reading of the manuscript. The authors acknowledge funding from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Eischen CM, et al. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stott FJ, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esclatine A, et al. Macroautophagy signaling and regulation. Curr Top Microbiol Immunol. 2009;335:33–70. doi: 10.1007/978-3-642-00302-8_2. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 9.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 10.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 11.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 18.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maclean KH, et al. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 24.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorin S, et al. c-Jun NH2-terminal kinase activation is essential for DRAM-dependent induction of autophagy and apoptosis in 2-methoxyestradiol-treated Ewing sarcoma cells. Cancer Res. 2009;69:6924–6931. doi: 10.1158/0008-5472.CAN-09-1270. [DOI] [PubMed] [Google Scholar]
- 26.Crighton D, et al. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ. 2007;14:1071–1079. doi: 10.1038/sj.cdd.4402108. [DOI] [PubMed] [Google Scholar]
- 27.Rosenbluth JM, et al. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buzzai M, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 29.Evans JM, et al. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tasdemir E, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 31.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasdemir E, et al. p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle. 2008;7:3006–3011. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
- 33.Donehower LA. Does p53 affect organismal aging? J Cell Physiol. 2002;192:23–33. doi: 10.1002/jcp.10104. [DOI] [PubMed] [Google Scholar]
- 34.Tavernarakis N, et al. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 35.Morselli E, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 36.Reef S, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reef S, et al. The autophagic inducer smARF interacts with and is stabilized by the mitochondrial p32 protein. Oncogene. 2007;26:6677–6683. doi: 10.1038/sj.onc.1210485. [DOI] [PubMed] [Google Scholar]
- 39.Ueda Y, et al. Small mitochondrial ARF (smARF) is located in both the nucleus and cytoplasm, induces cell death, and activates p53 in mouse fibroblasts. FEBS Lett. 2008;582:1459–1464. doi: 10.1016/j.febslet.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Abida WM, Gu W. p53-Dependent and p53-independent activation of autophagy by ARF. Cancer Res. 2008;68:352–357. doi: 10.1158/0008-5472.CAN-07-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reef S, Kimchi A. Nucleolar p19ARF, unlike mitochondrial smARF, is incapable of inducing p53-independent autophagy. Autophagy. 2008;4:866–869. doi: 10.4161/auto.6691. [DOI] [PubMed] [Google Scholar]
- 42.Humbey O, et al. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res. 2008;68:9608–9613. doi: 10.1158/0008-5472.CAN-08-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geradts J, et al. Immunohistochemical [corrected] detection of the alternate INK4a-encoded tumor suppressor protein p14(ARF) in archival human cancers and cell lines using commercial antibodies: correlation with p16(INK4a) expression. Mod Pathol. 2001;14:1162–1168. doi: 10.1038/modpathol.3880452. [DOI] [PubMed] [Google Scholar]
- 44.Sherr CJ, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, et al. Differential p53-independent outcomes of p19(Arf) loss in oncogenesis. Sci Signal. 2009;2:ra44. doi: 10.1126/scisignal.2000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pimkina J, Murphy ME. ARF, autophagy and tumor suppression. Autophagy. 2009;5:397–399. doi: 10.4161/auto.5.3.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pimkina J, et al. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maiuri MC, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Yee KS, et al. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bensaad K, et al. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirkegaard T, et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 463:549–553. doi: 10.1038/nature08710. [DOI] [PubMed] [Google Scholar]
- 53.Nylandsted J, et al. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leu JI, et al. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tasdemir E, et al. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle. 2007;6:2263–2267. doi: 10.4161/cc.6.18.4681. [DOI] [PubMed] [Google Scholar]
- 57.Levine B, et al. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 59.Belanger M, et al. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy. 2006;2:165–170. doi: 10.4161/auto.2828. [DOI] [PubMed] [Google Scholar]
- 60.Blommaart EF, et al. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 61.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abedin MJ, et al. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 63.Hartford CM, Ratain MJ. Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin Pharmacol Ther. 2007;82:381–388. doi: 10.1038/sj.clpt.6100317. [DOI] [PubMed] [Google Scholar]
- 64.Martinet W, et al. Everolimus-induced mTOR inhibition selectively depletes macrophages in atherosclerotic plaques by autophagy. Autophagy. 2007;3:241–244. doi: 10.4161/auto.3711. [DOI] [PubMed] [Google Scholar]
- 65.Cao C, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 2006;66:10040–10047. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]
- 66.Yazbeck VY, et al. Temsirolimus downregulates p21 without altering cyclin D1 expression and induces autophagy and synergizes with vorinostat in mantle cell lymphoma. Exp Hematol. 2008;36:443–450. doi: 10.1016/j.exphem.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Narita M, Young AR. Autophagy facilitates oncogene-induced senescence. Autophagy. 2009;5:1046–1047. doi: 10.4161/auto.5.7.9444. [DOI] [PubMed] [Google Scholar]