Abstract
Parallel biosensors for proteins are becoming more essential for the thorough and systematic investigation of complex biological processes. These tools also enable improved clinical diagnoses relative to single protein analyses due to their greater information content. If implemented correctly, affinity-based techniques can provide unique advantages in terms of sensitivity and flexibility. Aptamers are increasingly being used as the affinity reagents of choice for protein biosensing applications. Here, we describe the development and characterization of an aptamer-based method for parallel protein analyses that relies on recognition of the target protein by two unique aptamers targeting different epitopes on the protein. Our results show that the technique achieved simultaneous and quantitative detection of thrombin and platelet derived growth factor-BB (PDGF-BB) with high specificity both in buffered solutions and in serum samples.
Keywords: Biosensor, multiplex, aptamer, thrombin, PDGF-BB, quantitative PCR
1. Introduction
Improvements in clinical, laboratory, and biodefense applications rely on the continuing development of biosensing techniques for various biomolecules. In particular, considerable current work is focused on novel tools for parallel biosensors. While nucleic acid detection tools are relatively mature at this point, further evolution in the area of protein biosensing technologies, especially in a multiplex fashion, is required.
While classical parallel protein analytical techniques, such as two-dimensional gel electrophoresis followed by mass spectroscopy (2DE/MS) (O'Farrell 1975), will continue to be valuable tools for discovery-oriented protein measurements, affinity-based techniques have emerged as popular approaches for the detection of single and focused sets of proteins (MacBeath 2002). Antibody-based multiplex sensing assays have been developed in various planar and bead array formats and are becoming accepted methods for protein expression and interaction studies (MacBeath and Schreiber 2000; Schweitzer et al. 2002; Xie et al. 2009). However, the function of these arrays may be impaired by the high local concentrations of surface-bound antibodies and non-specific interactions between the substrate and the antibodies or analytes (Zichi et al. 2008). Additional limitations of antibody-based proteomics methods, in general, include batch-to-batch variation in antibody quality and the cost of antibody generation and purification (Gloriam et al. 2009). Therefore, novel approaches are being sought that can complement or expand existing antibody-based techniques.
Aptamers are gaining popularity as alternative affinity reagents for biosensing applications (Breaker 2002; Hianik and Wang 2009; Lai et al. 2007; Lee et al. 2009; Lee et al. 2008; Navani and Li 2006; Swensen et al. 2009; Xie and Walton 2009; Zuo et al. 2009). Aptamers are single-stranded nucleic acids that bind other molecules with high specificity and affinity (Ellington and Szostak 1990; Tuerk and Gold 1990) and may provide unique advantages for multiplex analytical applications (Bock et al. 2004; Collett et al. 2005; Zichi et al. 2008). Aptamers are selected in vitro and in theory can be selected to bind almost any target with automated and high throughput techniques for aptamer selection having been demonstrated (Cox et al. 2002). Furthermore, aptamers are nucleic acids rather than proteins and therefore can be easily synthesized commercially or enzymatically with high reproducibility and minimal batch-to-batch variation (Nimjee et al. 2005; Taussig et al. 2007), which is valuable for the development of reproducible biosensors.
The majority of current aptamer-based biosensor designs are specific for a single analyte (Bang et al. 2005; Fischer et al. 2008; Fredriksson et al. 2002; Gronewold et al. 2009; He et al. 2009; Kim et al. 2009; Lee et al., 2009; Peng et al. 2009; Yao et al. 2009). While chip-based aptamer arrays for multiplexed protein measurements are currently in development both by academic and commercial groups (Bock et al. 2004; Cho et al. 2006; Collett et al. 2005; Hesselberth et al. 2003; Kirby et al. 2004; Seetharaman et al. 2001; Stadtherr et al. 2005). Aptamer-based sensing combined with capillary electrophoresis has been used to measure four proteins simultaneously (Zhang et al. 2008). Nonetheless, the development of additional solution-phase and bead-based assays is still valuable, as they can potentially be customized for specific sets of target proteins and may have sensitivity advantages over current techniques.
In this work, we describe proof-of-concept development of a solution-phase biosensor for multiplex protein measurements that relies on dual-aptamer analyte recognition. We have shown previously that the detection dynamic range based on aptamer conformational rearrangement can be limited (Xie and Walton 2009). We hypothesized that altering the sensing design to use increased aptamer concentrations, two specific recognition events, and an affinity-based separation would have improved sensitivity, specificity, and dynamic range compared to the structure-switching aptamer approach. Applying our new technique to measure the concentrations of thrombin and platelet-derived growth factor (PDGF-BB), we found a reproducible, linear dynamic response to analyte concentrations over a three order-of-magnitude range of concentration. For both proteins, accurate and reproducible measurements were obtained using multiple readouts in both buffered solutions and in complex mixtures.
2. Experimental
2.1 Proteins and Aptamer Sequences
Thrombin was purchased from Haematologic Technologies (Essex Junction, VT). PDGF-BB was purchased from R & D Systems (Minneapolis, MN). Haptoglobin, fibrinogen, C-reactive protein and lysozyme were from Sigma Aldrich (St Louis, MO). The sequences for the thrombin sensing and capture aptamers were 5′-GATGTCCACGAGGTCTCTATTGCATTCGCACTTCCGATTTTTTCAGTCCGTGGTAGGGCAGGTTGGGGTGACTCGTACGCTGCAGGTCGAC-3′ (MB-T, 91 nt, italicized: MB sequence) and 5′-GTGACTACTGGTTGGTGAGGTTGGGTAGTCACAAATTTTTTTTTT-Bio-3′ (Bio-T), respectively. The sequences for the PDGF-BB sensing and capture aptamers were 5′-GATGTCCACGAGGTCTCTGCGGCATCAACAATCTCGAATTTTTTACTCAGGGCACTGCAAGCAATTGTGGTCCCAATGGGCTGAGTATCGTACGCTGCAGGTCGAC-3′ (MB-P, 106 nt, italicized: MB sequence) and 5′-TACTCAGGGCACTGCAAGCAATTGTGGTCCCAATGGGCTGAGTATTTTTT-Bio-3′ (Bio-P), respectively. Each of these aptamers was derived from sequences available in the literature (Fredriksson et al. 2007; Green et al. 1996; Macaya et al. 1995; Tasset et al. 1997). Since PDGF-BB is a homodimeric protein, the same sequence can be used for the aptamer domain of both the capture and sensing aptamers, as each PDGF-BB molecule has two epitopes for aptamer recognition. Aptamer-protein affinities were measured by filter binding assays. Capture aptamers were synthesized with 3′-biotins spaced from the aptamer sequence by oligo(dT) sequences. Primer sequences specific for MB-T were 5′-ATTGCATTCGCACTTCCGAT-3′ (T5′p) and 5′-AGTCACCCCAACCTGCCCTA-3′ (T3′p). Primer sequences specific for MB-P were 5′-GCGGCATCAACAATCTCGAA-3′ (P5′p) and 5′-ATACTCAGCCCATTGGGACC-3′ (P3′p). Aptamer specific primers were used for single-protein and multiplex qPCR readouts. The 5′ and 3′ primers were chosen to be specific to the MBs and to the aptamer 3′ ends, respectively (Aptamer specific primers are labeled as dashed arrows in Fig. 1). Using the hybridization server on mfold (Zuker 2003), both sets of template specific primers were confirmed not to cross-prime the other sensing aptamer template (data not shown). Universal primer sequences for both sensing aptamers were 5′-GATGTCCACGAGGTCTCT-3′ (U5′p) and 5′-GTCGACCTGCAGCGTACG-3′ (U3′p) (Universal primers are labeled as solid arrows in Fig.1). Universal primers were used for multiplex experiments using electrophoresis readouts. All oligonucleotides were synthesized and purified from Integrated DNA Technologies, Inc (Coralville, IA).
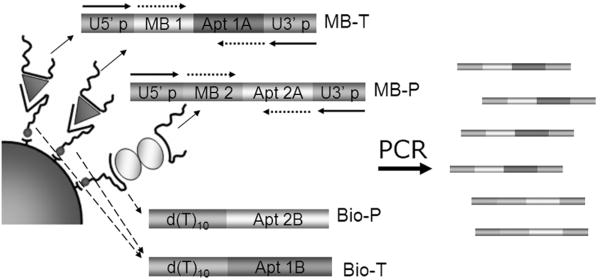
Fig. 1. Dual-aptamer detection schematic.
Thrombin and PDGF-BB are represented by triangles and double-ellipses, respectively. The sensing aptamers (MB-T, MB-P) containing thrombin aptamer (Apt 1A) and PDGF-BB aptamer (Apt 2A) are each labeled with an MB tag (MB1 and MB2) and flanked by a pair of universal primers (5′p and 3′p). Capture aptamers (Bio-T and Bio-P) containing thrombin aptamer (Apt 1B) and PDGF-BB aptamer (Apt 2B) are biotinylated to be attached to streptavidin-coated magnetic beads. Oligo(dT)s are used as the spacer between the aptamer sequences and the beads. After separation, eluted sensing aptamers are amplified by multiplex PCR and quantified.
2.2 Dual aptamer sensing assays
Prior to use, aptamers were annealed at 95°C for 3 min and cooled to room temperature over a period of 30 min. Proteins and annealed aptamers were mixed in binding buffer (50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 0.1% BSA, and 5 μg/ml poly(dI-dC)) in a total volume of 30 μL. Binding reactions were incubated at room temperature for 30 min followed by the addition of streptavidin coated magnetic beads (Dynal/Invitrogen, Oslo, Norway) for an additional 30 min at room temperature. A magnet was then applied to retain the beads, and the supernatant was removed. The beads were then washed three times with 100 μL of wash buffer (binding buffer plus 0.02% Tween 20). Retained sensing aptamers were eluted from the magnetic beads in 100 μL of water at 95°C for 5 min. Some or all of the recovered sensing aptamers were amplified by PCR.
Experiments determining the optimal concentration of thrombin or PDGF-BB sensing aptamer were performed in binding reactions with varying concentrations of sensing aptamer (5, 50, 250, 500, 1000 and 2500 nM) and 500 nM of capture aptamer in the presence and absence of 500 nM protein. After binding and separation as described above, sensing aptamers were eluted and amplified by qPCR. The metric for optimal sensing was ΔCt where ΔCt = (Ct value in the absence of protein) - (Ct value in the presence of protein). Higher ΔCt values indicate better recovery of sensing aptamers with minimal background.
2.3 PCR and qPCR
For electrophoresis readout experiments, eluted sensing aptamers were added with universal primers U5′p and U3′p at 500 nM each and amplified for 20 cycles with iQ Supermix (Bio-Rad, Hercules, CA) in 50 μL reactions. The PCR program was 94°C for 45 s, 54°C for 45 s and 72°C for 1 min. PCR products were analyzed by electrophoresis (5 μL mixed with 2 μL of loading buffer and electrophoresed at 100 V for 1 hr in 10 % acrylamide gels). Gels were stained with SYBR Gold nucleic acid gel stain (Invitrogen, Carlsbad, CA) and visualized in a Molecular Imager ChemiDoc XRS System (Bio-Rad, Hercules, CA).
For qPCR readout experiments, primers specific for sensing aptamers (e.g. T5′p and T3′p for MB-T; P5′p and P3′p for MB-P) were added at 500 nM each and amplified for 40 cycles with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in 50 μL reactions using the same PCR program as above. Threshold cycle numbers (Ct) for deriving sensing aptamer qPCR standard curves were generated from PCRs containing MB-T or MB-P template serially diluted from 3 × 108 to 3000 or 300 initial template molecules. In all standard curves and the correlation analysis, error bars represent at least duplicate experiments with each experiment assayed twice by PCR (four total PCRs). Signal-to-noise ratio was calculated as the fold difference in signal obtained from the maximal signal relative to the background. For qPCR data, this is 2 raised to the power (Ct value for background - Ct value for maximal sensitive concentration)
2.4 Detection in bovine and human sera
Binding reactions were performed as described above except in undiluted fetal bovine serum (FBS) (GIBCO, Invitrogen, Carlsbad, CA) or human serum (Sigma, St. Louis, MO) with thrombin and PDGF-BB spiked in. After separation and elution, the recovered aptamers were amplified with sensing aptamer specific primers to derive Ct values.
3. Results and Discussion
In developing aptamer-based biosensors, or any affinity-based biosensor, sensing of the analyte depends on the complex formation between the analyte and the detector molecule, in our case an aptamer. Because the formation of the 1-to-1 complex is an equilibrium process, the degree to which complex forms is controlled by the affinity (e.g., the KD) of the interaction and the concentrations of the analyte and aptamer. For significant complex formation to occur, either the analyte or aptamer concentration must be in the range of or above the KD of the interaction. Thus, a critical decision must be made regarding the detecting aptamer concentration (Fig. S1). Using an aptamer concentration below the analyte concentration (limiting aptamer), the dynamic range of the technique is limited, and the total signal recovered saturates at the limiting aptamer concentration (Fig. S1a and S1b). In contrast, with aptamer in excess relative to analyte, both the dynamic range and total signal are maximized over the entire analyte concentration range (Fig. S1a and S1b).
That said, in techniques where the aptamers are labeled for detection, for instance by fluorescence, the feasibility of using excess aptamer is usually reduced by the presence of a high concentration of unbound aptamer whose signal masks that of the analyte-aptamer complexes. The dynamic range and sensitivity advantages of the excess aptamer design must then be weighed against the need for a stringent separation, wherein the aptamer-analyte complexes (the signal) are potentially less than 1% of the total aptamers present (the background) (Fig. S1). As discussed above, the necessary separation can be achieved by methods such as microarrays or electrophoresis (Stadtherr et al. 2005; Zhang et al. 2008). Here, we describe a method where we have combined aptamer-based separation and aptamer-based detection, using excess aptamers, to design a parallel protein analytical technique (Fig. 1) with specificity for the target protein and the potential for excellent sensitivity and broad dynamic range.
In our technique, one aptamer, termed the capture aptamer, is biotinylated to immobilize the protein onto streptavidin-coated magnetic beads, while the other aptamer, termed the sensing aptamer, is used for quantification of the protein concentration by PCR, microarray, or any other nucleic acid readout. Detection of the sensing aptamer rather than the protein directly leverages the simplicity of multiplex PCR (Chamberlain et al. 1988; Zangenberg G 1999), through the incorporation of universal primer sequences on all sensing aptamers. The use of PCR also provides straightforward signal amplification, a unique advantage of nucleic acid analytical approaches (Schweitzer et al. 2002). In addition, we have incorporated molecular barcodes (MBs) into our sensing aptamers which permits facile parallel detection of the sensing aptamers by sequence alone, as has been shown using oligonucleotide microarrays (Giaever et al. 2002; Shoemaker et al. 1996; Walton et al. 2006). Readout of the concentration of the MB sequence then serves as an indirect readout of the concentration of the protein target. In this study, we demonstrate proof-of-concept use of our technique for multiplex detection of thrombin and PDGF-BB.
We first confirmed the fidelity of our sensing aptamers as qPCR templates using serial dilutions of sensing aptamers and template specific primers (similar results were obtained using universal primers) (Fig. S2). PCR for both templates showed the expected log-linear response over at least a five order-of-magnitude dynamic range (Fig. S2). Responses plateaued outside of the regions shown on the plot (data not shown). We then examined the quantitative response of sensing aptamer concentration relative to protein concentration. To identify the optimal conditions for protein sensing, ΔCt values (defined as the difference in Ct value in the presence and absence of 500 nM protein) were measured at various sensing aptamer conditions for both proteins (Fig. S3). Capture aptamer concentrations were fixed at 500 nM, which should bind 46% of the thrombin and 81% of PDGF-BB based on our measurement of the aptamer-protein affinities (Fig. S4 and Table S1). At varying sensing aptamer concentrations, binding reactions were set up, the beads were washed, and the retained sensing aptamers were eluted. Ct values were derived experimentally, and ΔCt values were calculated for each protein. A maximum ΔCt of 16.1 was observed for thrombin at 250 nM sensing aptamer (MB-T) concentration. Similarly, a maximum ΔCt of 7.5 was observed at 50 nM sensing aptamer (MB-P) concentrations for PDGF-BB. In each case, higher concentrations of sensing aptamer resulted in higher non-specific retention of sensing aptamers in the absence of protein (Table S2), while lower concentrations had reduced sensitivity in the presence of protein. The larger maximal ΔCt observed for thrombin versus PDGF-BB is a result of higher background using the PDGF-BB aptamers (Table S2).
The optimal concentrations derived above (as selected from the concentrations with the maximum ΔCt value) were then used to generate standard curves for single protein measurements (Fig. S5). Serial dilutions of thrombin and PDGF-BB were added to binding reactions containing capture and sensing aptamers. After separation, the eluted sensing aptamers were quantified by qPCR. The technique showed sensitivity down to 1 nM protein concentration for thrombin and 10 nM for PDGF-BB. Sensitive detection extended over at least a two order-of-magnitude dynamic range. A linear dose response was observed over the sensing dynamic range, resulting in a maximal signal-to-noise ratio of ∼ 23000 (2(25.4-10.9)) for thrombin and ∼84 (2(19.3-12.9)) for PDGF-BB, an improvement over single aptamer approaches (Hu et al. 2009; Shlyahovsky et al. 2007; Wang et al. 2009a; Zhang et al. 2009) as well as over a related surface-based aptamer approach (Lee et al. 2009). While maximizing dynamic range is valuable for any sensing technique, other factors, such as specificity for the analyte of interest, must be examined in technique development.
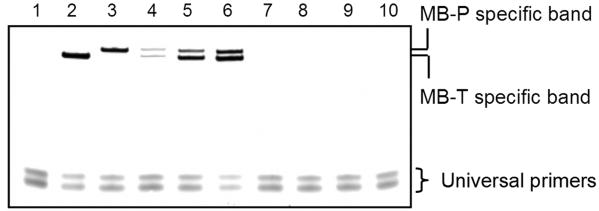
We then examined whether the concentration sensitivity would be maintained using the assay in multiplex format (Fig. 2). After addition of either or both proteins, binding, and washing, sensing aptamers were eluted and amplified by multiplex PCR using the universal primers. PCR products were separated by electrophoresis and visualized. When thrombin was added alone, a band corresponding to the 91 bp MB-T amplification product was observed (Fig. 2, lane 2). Likewise, when PDGF-BB was added alone, a band representing the 106 bp MB-P was observed (Fig. 2, lane 3). Upon addition of both proteins, both bands appeared and band intensity increased dose-dependently with increasing protein concentrations (Fig. 2, lanes 4-6). No signal was returned from either aptamer pair in the presence of haptoglobin, fibrinogen, C-reactive protein, or lysozyme, further demonstrating good specificity for detection of only the specific protein targets (Fig. 2, lanes 7-10).
Fig. 2. Multiplex detection of proteins using electrophoresis as the readout method.
Control or target protein samples were added to binding buffer containing 250 nM MB-T, 500 nM Bio-T, 50 nM MB-P and 500 nM Bio-P. After binding and separation, eluted sensing aptamers were amplified with universal primers for 20 cycles. PCR products were separated by acrylamide gel and detected by gel staining. Higher bands are the products from amplification of the PDGF-BB and thrombin sensing aptamers. Remaining primers run to the bottom of the gel. Lane 1: Buffer; Lane 2: 333 nM thrombin; Lane 3: 333 nM PDGF-BB; Lanes 4-6: both thrombin and PDGF-BB at 33, 100 and 333 nM from left to right; Lanes 7-10: haptoglobin, fibrinogen, C-reactive protein and lysozyme each at 333 nM. Image contrast was adjusted to reduce background.
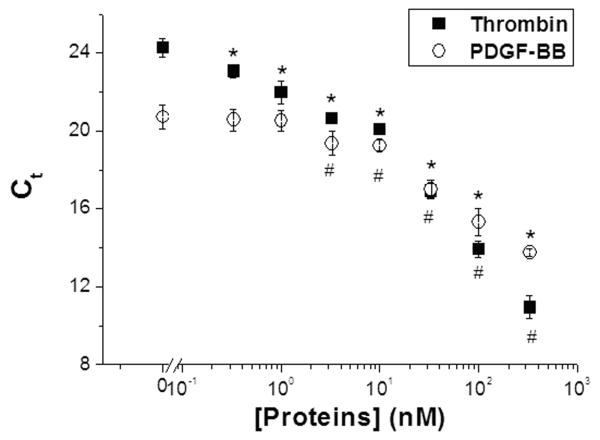
Dose-response curves for multiplex detection of both proteins were derived with serial dilutions of both proteins ranging from 333 pM to 333 nM (Fig. 3). Eluted sensing aptamers were quantified with qPCR using MB-T or MB-P specific primers. In the multiplex format, the detection limits were 333 pM for thrombin and 3.3 nM for PDGF-BB, respectively, essentially in agreement with the single protein measurements. In both cases, we expect technical improvements in binding, washing, and elution protocols could extend the dynamic range both higher and lower in concentration. That said, the current dynamic range exceeds that of a recently described fluorescence-based method by an order-of-magnitude (Wang et al. 2009b), while the sensitivity approaches that of a capillary electrophoresis based technique (Zhang et al. 2008).
Fig. 3. Multiplex detection using qPCR as the readout method.
Both sets of aptamer pairs were added with serial dilutions of protein mixtures containing both thrombin and PDGF-BB. After binding, separation, and elution, eluted sensing aptamers were amplified with qPCR using template specific primers. (* and # denote Ct values significantly lower than background for thrombin and PDGF-BB, respectively (p < 0.01)). Lower detection limits were 330 pM and 3.3 nM for thrombin and PDGF-BB, respectively.
Given that our initial multiplexing experiments were performed with both proteins at identical concentration in all tests, we examined the concentration sensitivity in the presence of high and low concentrations of the competing proteins (Figs. 4 and S6). Serial dilutions of one protein were measured in multiplex in the presence and absence of 333 nM of the other protein. After separation, eluted aptamers were amplified by PCR and assayed by electrophoresis (Fig. 4). In all cases and concentrations, the signal returned from each aptamer pair was essentially identical to that detected in the single-protein case (compare corresponding Ct values from Figs. S5 and S6).
Fig. 4. Specificity of detection in the presence of competing protein.
Both pairs of aptamers were added to serial dilutions of PDGF-BB in the absence or presence of thrombin (top) or to serial dilutions of thrombin in the absence or presence of PDGF-BB (bottom). After separation, eluted sensing aptamers were amplified with universal primers and read out using electrophoresis. All four aptamers were used at 500 nM concentration to assess how much cross reactivity occurred at high aptamer concentrations. Little cross-reactivity was seen in either case.
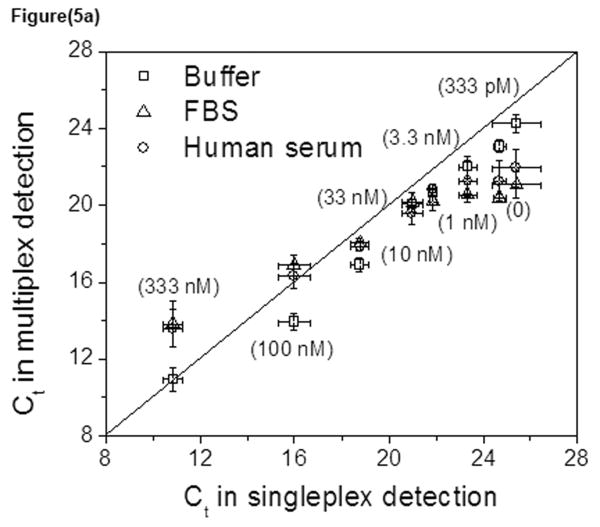
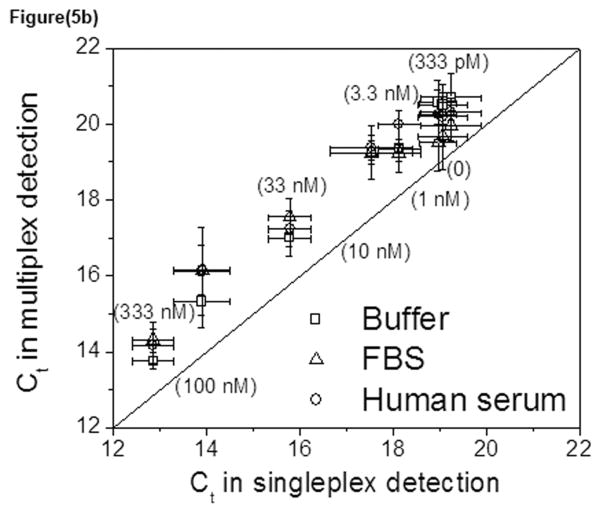
Standard curves were also derived in complex biological fluids, fetal bovine serum (FBS) and human serum (Fig. S7). Results showed that both proteins can still be detected in undiluted serum at low nanomolar concentrations. It was important to confirm that the results in the complex environment agreed with the standard curves derived for the individual proteins in buffered solution (Fig. 5). For thrombin, the multiplex data derived in buffer, FBS and human serum all agreed well with the singleplex data, showing that competition of the multiplex measurement did not impact the accuracy of our method (Fig. 5a). For PDGF-BB, the majority of the data derived from multiplex measurements underestimates the PDGF-BB concentration by about two-fold as compared to the singleplex measurement (Fig. 5b). This implies a greater effect of the binding environment on PDGF-BB detection as compared to thrombin, perhaps due to greater non-specific binding of the sensing aptamer. Nonetheless, the sensitivity and accuracy of concentration detection for the protein are largely maintained.
Fig. 5. Correlation of various analyses.
Multiplex Ct values derived in buffer or serum samples were plotted against the Ct value derived from singleplex experiments in buffer for thrombin (a) and PDGF-BB (b).
It could be argued that our method requires more processing as compared to other aptamer-based protein sensing methods (Zhang et al. 2008). Moreover, detection with our technique does not provide the sensitivity of more traditional methods such as ELISA (Kajizuka et al. 2010). However, we feel that our approach has potential utility that makes it worthwhile to continue optimization of the approach. In particular, our method is designed specifically for parallel analyses due to the requirement for multiple specific interactions, the affinity-based separation, and incorporation of the MBs. Thus, our technique avoids any issues that would arise using other protein analytical approaches, especially in analyzing proteins of similar size, charge, or hydrophilicity.
The advantages of using a nucleic acid based-readout for other molecular species has also been demonstrated in several antibody-based protein detection techniques, such as in immuno-PCR (Niemeyer et al. 2005; Sano et al. 1992), immuno-RCA (Schweitzer et al. 2000), and the proximity ligation assay (Gullberg et al. 2004). Multianalyte approaches using these techniques have also been demonstrated (Fredriksson et al. 2007; Hendrickson et al. 1995; Joerger et al. 1995; Schweitzer et al. 2002; Schweitzer et al. 2000). In each of these cases, the antibody must be conjugated with the DNA for use in the assay. This serves as an advantage of our approach, in which the affinity reagent can be amplified directly without the need for conjugation to another species. It is, however, important to note that our current detection limits do not reach those of these amplified, antibody-based approaches. We feel that this is a reflection of the affinities of the aptamers for their targets. The aptamers used in this work do not have the affinity for their targets of the antibodies used for protein recognition in other approaches. As it is known that aptamers with affinities comparable to antibodies have been generated with both natural and modified nucleotides (Hermann and Patel 2000; Keefe and Cload 2008), we expect to be able to overcome this limitation. With improved affinities, we anticipate being able to enhance the stringency of our binding, blocking, and washing conditions to improve our sensitivity, especially in complex backgrounds such as sera.
4. Conclusions
In this study, a novel technique for parallel protein measurement using dual-aptamer recognition was developed. Simultaneous and quantitative detection of thrombin and platelet derived growth factor (PDGF-BB) was achieved with good specificity and reproducibility in both buffered solutions and a serum background. We have demonstrated that the technique is flexible and has the potential for straightforward customization to sets of proteins, depending on the availability of aptamer pairs for each protein and their specificity and compatibility. The sensitivity and dynamic response of the approach warrants further selection and characterization of aptamer pairs and their use in the described approach.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Brad Hall, Dr. Na Li, and Dr. Andrew Ellington for helpful discussions. We also thank the members of the Cellular and Biomolecular Laboratory (http://www.egr.msu.edu/cbl/) for their advice and support. Financial support for this work was provided in part by Michigan State University, the National Science Foundation (#0425821), and the National Institutes of Health (#CA126136, #GM079688, #RR024439).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bang GS, Cho S, Kim BG. Biosensors & bioelectronics. 2005;21(6):863–870. doi: 10.1016/j.bios.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Bock C, Coleman M, Collins B, Davis J, Foulds G, Gold L, Greef C, Heil J, Heilig JS, Hicke B, Hurst MN, Husar GM, Miller D, Ostroff R, Petach H, Schneider D, Vant-Hull B, Waugh S, Weiss A, Wilcox SK, Zichi D. Proteomics. 2004;4(3):609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Curr Opin Biotechnol. 2002;13(1):31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Nucleic Acids Res. 1988;16(23):11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal Chim Acta. 2006;564(1):82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Collett JR, Cho EJ, Ellington AD. Methods. 2005;37(1):4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Cox JC, Hayhurst A, Hesselberth J, Bayer TS, Georgiou G, Ellington AD. Nucleic Acids Research. 2002;30(20) doi: 10.1093/nar/gnf107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fischer NO, Tarasow TM, Tok JBH. Anal Biochem. 2008;373(1):121–128. doi: 10.1016/j.ab.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Nature Methods. 2007;4(4):327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Nature biotechnology. 2002;20(5):473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gloriam DE, Orchard S, Bertinetti D, Bjorling E, Bongcam-Rudloff E, Bourbeillon J, Bradbury AR, de Daruvar A, Dubel S, Frank R, Gibson TJ, Haslam N, Herberg FW, Hiltke T, Hoheisel JD, Kerrien S, Koegl M, Konthur Z, Korn B, Landegren U, van der Maarel S, Montecchi-Palazzi L, Palcy S, Rodriguez H, Schweinsberg S, Sievert V, Stoevesandt O, Taussig MJ, Uhlen M, Wingren C, Gold L, Woollard P, Sherman DJ, Hermjakob H. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900185-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Biochemistry. 1996;35(45):14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- Gronewold TMA, Baumgartner A, Hierer J, Sierra S, Blind M, Schafer F, Blumer J, Tillmann T, Kiwitz A, Kaiser R, Zabe-Kuhn M, Quandt E, Famulok M. J Proteome Res. 2009;8(7):3568–3577. doi: 10.1021/pr900265r. [DOI] [PubMed] [Google Scholar]
- Gullberg M, Gustafsdottir SM, Schallmeiner E, Jarvius J, Bjarnegard M, Betsholtz C, Landegren U, Fredriksson S. Proc Natl Acad Sci U S A. 2004;101(22):8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JL, Wu ZS, Zhang SB, Shen GL, Yu RQ. Talanta. 2009;80(3):1264–1268. doi: 10.1016/j.talanta.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Hendrickson ER, Truby TM, Joerger RD, Majarian WR, Ebersole RC. Nucleic Acids Res. 1995;23(3):522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Science. 2000;287(5454):820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Robertson MP, Knudsen SM, Ellington AD. Anal Biochem. 2003;312(2):106–112. doi: 10.1016/s0003-2697(02)00441-4. [DOI] [PubMed] [Google Scholar]
- Hianik T, Wang J. Electroanalysis. 2009;21(11):1223–1235. [Google Scholar]
- Hu J, Zheng PC, Jiang JH, Shen GL, Yu RQ, Liu GK. Anal Chem. 2009;81(1):87–93. doi: 10.1021/ac801431m. [DOI] [PubMed] [Google Scholar]
- Joerger RD, Truby TM, Hendrickson ER, Young RM, Ebersole RC. Clin Chem. 1995;41(9):1371–1377. [PubMed] [Google Scholar]
- Kajizuka M, Miyachi T, Matsuzaki H, Iwata K, Shinmura C, Suzuki K, Suda S, Tsuchiya KJ, Matsumoto K, Iwata Y, Nakamura K, Tsujii M, Sugiyama T, Takei N, Mori N. Prog Neuro-Psychopharmacol Biol Psychiatry. 2010;34(1):154–158. doi: 10.1016/j.pnpbp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Keefe AD, Cload ST. Current opinion in chemical biology. 2008;12(4):448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Kim KS, Lee HS, Yang JA, Jo MH, Hahn SK. Nanotechnology. 2009;20(23):6. doi: 10.1088/0957-4484/20/23/235501. [DOI] [PubMed] [Google Scholar]
- Kirby R, Cho EJ, Gehrke B, Bayer T, Park YS, Neikirk DP, McDevitt JT, Ellington AD. Analytical Chemistry. 2004;76(14):4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- Lai RY, Plaxco KW, Heeger AJ. Analytical Chemistry. 2007;79(1):229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- Lee J, Icoz K, Roberts A, Ellington AD, Savran CA. Analytical Chemistry. 82(1):197–202. doi: 10.1021/ac901716d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Icoz K, Roberts A, Ellington AD, Savran CA. Anal Chem. 2009 doi: 10.1021/ac901716d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, So HM, Jeon EK, Chang H, Won K, Kim YH. Analytical and Bioanalytical Chemistry. 2008;390(4):1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaya RF, Waldron JA, Beutel BA, Gao HT, Joesten ME, Yang MH, Patel R, Bertelsen AH, Cook AF. Biochemistry. 1995;34(13):4478–4492. doi: 10.1021/bi00013a041. [DOI] [PubMed] [Google Scholar]
- MacBeath G. Nature Genetics. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Navani NK, Li Y. Current opinion in chemical biology. 2006;10(3):272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Niemeyer CM, Adler M, Wacker R. Trends Biotechnol. 2005;23(4):208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Annual Review of Medicine. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. Journal of Biological Chemistry. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Peng YG, Zhang DD, Li Y, Qi HL, Gao Q, Zhang CX. Biosensors & bioelectronics. 2009;25(1):94–99. doi: 10.1016/j.bios.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Sano T, Smith CL, Cantor CR. Science. 1992;258(5079):120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nature biotechnology. 2002;20(4):359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B, Wiltshire S, Lambert J, O'Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC. Proc Natl Acad Sci U S A. 2000;97(18):10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Nature biotechnology. 2001;19(4):336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- Shlyahovsky B, Li D, Katz E, Willner I. Biosensors & bioelectronics. 2007;22(11):2570–2576. doi: 10.1016/j.bios.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Nat Genet. 1996;14(4):450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- Stadtherr K, Wolf H, Lindner P. Anal Chem. 2005;77(11):3437–3443. doi: 10.1021/ac0483421. [DOI] [PubMed] [Google Scholar]
- Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Journal of the American Chemical Society. 2009;131(12):4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset DM, Kubik MF, Steiner W. Journal of Molecular Biology. 1997;272(5):688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- Taussig MJ, Stoevesandt O, Borrebaeck CAK, Bradbury AR, Cahill D, Cambillau C, de Daruvar A, Dubel S, Eichler J, Frank R, Gibson TJ, Gloriam D, Gold L, Herberg FW, Hermjakob H, Hoheisel JD, Joos TO, Kallioniemi O, Koegll M, Konthur Z, Korn B, Kremmer E, Krobitsch S, Landegren U, van der Maarel S, McCafferty J, Muyldermans S, Nygren PA, Palcy S, Pluckthun A, Polic B, Przybylski M, Saviranta P, Sawyer A, Sherman DJ, Skerra A, Templin M, Ueffing M, Uhlen M. Nature Methods. 2007;4(1):13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Walton SP, Mindrinos MN, Davis RW. Biochem Biophys Res Commun. 2006;348(2):689–696. doi: 10.1016/j.bbrc.2006.07.108. [DOI] [PubMed] [Google Scholar]
- Wang J, Meng W, Zheng X, Liu S, Li G. Biosensors & bioelectronics. 2009a;24(6):1598–1602. doi: 10.1016/j.bios.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Wang LQ, Li LY, Xu Y, Cheng GF, He PA, Fang YZ. Talanta. 2009b;79(3):557–561. doi: 10.1016/j.talanta.2009.05.034. [DOI] [PubMed] [Google Scholar]
- Xie S, Moya C, Bilgin B, Jayaraman A, Walton SP. Expert Rev Proteomics. 2009;6(5):573–583. doi: 10.1586/epr.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Walton SP. Anal Chim Acta. 2009;638(2):213–219. doi: 10.1016/j.aca.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CY, Qi YZ, Zhao YH, Xiang Y, Chen QH, Fu WL. Biosensors & bioelectronics. 2009;24(8):2499–2503. doi: 10.1016/j.bios.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Zangenberg G, S R, Reynolds R. PCR Applications: Protocols for functional Genomics. Academic Press; 1999. Multiplexed PCR: Optimization Guidelines; pp. 73–94. [Google Scholar]
- Zhang HQ, Li XF, Le XC. Journal of the American Chemical Society. 2008;130(1):34–35. doi: 10.1021/ja0778747. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang W, Wang J, Yang C, Yang F, Yang X. Talanta. 2009;78(4-5):1240–1245. doi: 10.1016/j.talanta.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Zichi D, Eaton B, Singer B, Gold L. Current opinion in chemical biology. 2008;12(1):78–85. doi: 10.1016/j.cbpa.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Zuker M. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XL, Xiao Y, Plaxco KW. Journal of the American Chemical Society. 2009;131(20):6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.