Abstract
MicroRNAs (miRNAs) are small regulatory RNAs that individually regulate up to several hundred genes, and collectively may regulate as much as two-thirds of the transcriptome. Recent evidence supports a role for miRNA dysregulation in psychiatric and neurological disorders, including schizophrenia, bipolar disorder, and autism. Small changes in miRNA expression can fine-tune the expression of multiple genes within a biological network, suggesting that miRNA dysregulation may underlie many of the molecular changes observed in psychiatric disease, and that therapeutic regulation of miRNA levels may represent a novel treatment option.
Keywords: MicroRNA, schizophrenia, bipolar disorder, NMDA, miR-132
Introduction
Psychiatric disorders, including schizophrenia and bipolar disorder, are highly heritable, yet also highly resistant to genetic dissection. The results of recent genome-wide association studies indicate that most cases of psychiatric illness are the result of hundreds or thousands of common genetic variants acting in concert to produce a neuropsychiatric phenotype. Such heterogeneity will continue to confound the development of novel drug therapies in the absence of new approaches that address the complex gene networks dysregulated in disease. We suggest that studying microRNA function in psychiatric disease may represent a road forward: dysregulation of miRNA expression may account for some of the neurodevelopmental aspects of psychiatric disorders and the difficulty in identifying individual causative genes, while targeting a single miRNA for therapeutic purposes offers the ability to fine-tune the expression of entire gene networks. In the following review, we summarize the current state of the genetics of psychiatric disease and the basic biology of miRNAs and miRNA function in the nervous system. We then focus on evidence suggestive of miRNA dysregulation in psychiatric disease.
Genetics of psychiatric disorders: the “missing heritability” problem
Psychiatric disorders, including schizophrenia, bipolar disorder, and depression, are a major burden on both patients and society. On an annual basis, approximately 25% of adults in the US will suffer from a major psychiatric illness; of these, schizophrenia and bipolar disorder, the most psychologically and economically disabling of the disorders, account for 1.1 and 2.5% of cases, respectively (Kessler et al., 2005b; WHO, 2004). Both disorders are associated with a poor quality of life and reduced life-span, yet both lack effective treatments. Although lithium and certain anticonvulsants help many bipolar patients, the treatments cause significant side effects that often lead to patient non-compliance (Thase, 2007). Schizophrenia is even more difficult to treat: neither the efficacy nor the side effect profile of pharmaceutical treatments for schizophrenia has improved substantially since the introduction of antipsychotics in the 1950s (Lieberman et al., 2005; Miller et al., 2008). While drug therapy can suppress the positive symptoms associated with schizophrenia, the less florid, but equally disabling, negative symptoms are often treatment-resistant (Kapur and Remington, 2001; Tandon et al., 2008).
For both schizophrenia and bipolar disorder, development of new drug therapies is hindered by a lack of understanding of the biological underpinnings of the illnesses. Despite heritability estimates as high as 80% (Cardno and Gottesman, 2000; McGuffin et al., 2003), complex psychiatric diseases have been resistant to genetic dissection (Williams et al., 2009). Traditional genetic approaches, such as quantitative trait locus (QTL) studies, have linked more than 7,000 genetic polymorphisms to schizophrenia (Allen et al., 2008), but the majority of these studies have not been replicated in separate populations, or have produced loci that are too large for candidate gene identification. The loci that have been most clearly linked to psychiatric disorders, including 1q21, 8p, and 13q, have yet to yield candidate genes that have been definitively associated with schizophrenia (Bertram, 2008; Craddock et al., 2005; Porteous, 2008). Candidate gene analysis has likewise had limited success: there is clear molecular evidence supporting altered glutamate and dopamine (DA) signaling in schizophrenia-like symptoms, but genetic studies have often failed to associate mutations in key genes in these pathways with disease risk (Owen et al., 2004). For example, a recent meta-analysis of 118 schizophrenia QTL studies found that associations with the dopamine, glutamate, and GABA receptor genes did not meet genome-wide significance (Allen et al., 2008).
To date, the most striking advances in the genetics of schizophrenia have resulted from small family-based studies of rare diseases that include schizophrenia/psychosis as a phenotype. Family-based linkage studies attempt to identify risk loci by looking for co-segregation of a genomic locus with disease in multiple families or a single large pedigree. This was the method used to identify the Disrupted-in-Schizophrenia-1 (DISC1) locus, which was originally identified in a Scottish pedigree with a high rate of schizophrenia, bipolar disorder, and psychosis (St Clair et al., 1990). The Scottish family carries a balanced translocation (1;11) (q42.1; q14.3) that results in the truncation of the DISC1 gene. Subsequent work has shown that DISC1 plays a major role in human hippocampal structure and function, cerebral cortex development, and fetal and adult neurogenesis (Duan et al., 2007; Kamiya et al., 2005; Schurov et al., 2004). Binding partners of DISC1, including NDEL1 and PDE4B, have also been shown to be altered in some schizophrenic patients, suggesting that the DISC1 pathway may play a central role in schizophrenia in a subset of the population (Tomppo et al., 2009). However, it is important to note that DISC1 mutations are not found in the majority of patients: therefore, although DISC1 is highly penetrant in a subset of patients and may provide important insight into the neurobiology of schizophrenia, it is not a general risk factor (Sanders et al., 2008).
Similarly, genome-wide analysis of copy number variation (CNV) has identified a subset of psychiatric disease cases linked to CNV burden (ISC, 2008; Need et al., 2009; Walsh et al., 2008; Xu et al., 2008; Zhang et al., 2009). Several groups have suggested that schizophrenia-associated CNVs preferentially disrupt genes that may be involved in nervous system function (Walsh et al., 2008; Wilson et al., 2006) Although the increased CNV burden in schizophrenia and other neuropsychiatric disorders has been replicated in multiple populations, CNVs are believed to account for only a small minority of cases. Furthermore, many of the CNVs represent de novo or rare (“private”) genomic rearrangements, and therefore cannot account for the significant heritability of schizophrenia (Conrad et al., 2009).
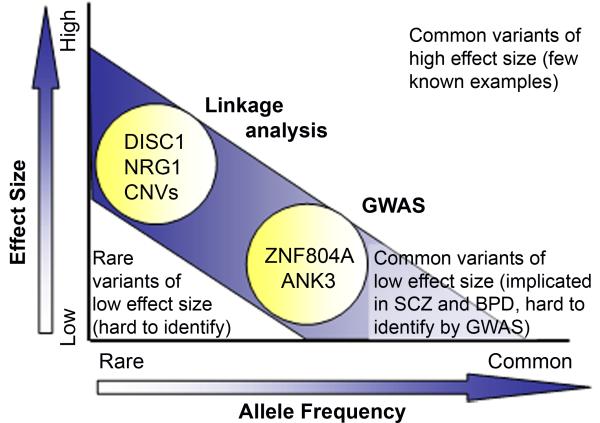
It is estimated that less than 10% of psychiatric illness are caused by rare, highly penetrant genes, such as DISC1, or by genomic rearrangements resulting in copy number variation (CNV), suggesting that the majority of cases are the result of the interaction of many genes, each with a small effect—the common variant/low penetrance model (Conrad et al., 2009; Purcell et al., 2009) (Figure 1). Genome-wide associations studies (GWAS), which use high-density single nucleotide polymorphism (SNP) genotyping to link phenotypes to underlying haplotypes, have been successfully applied to complex, presumably polygenic disorders such as breast cancer and obesity, but have had limited success when applied to psychiatric disorders. To date, approximately 10 GWAS studies have been directed at schizophrenia and/or bipolar disorder, the largest of which include 8,000 cases and 19,000 controls (Kirov et al., 2008; Need et al., 2009; O’Donovan et al., 2008; Purcell et al., 2009; Shi et al., 2009; Shifman et al., 2008; Stefansson et al., 2009; Sullivan et al., 2008). Although several of these studies are of sufficient power to detect loci carrying a relative risk of approximately 1%, only two genes, ZNF804A (O’Donovan et al., 2008) and ANK3 (Ferreira et al., 2008), have been linked to schizophrenia or bipolar disorder at a significant level. Statistical analysis of the most recent GWAS results suggests that both schizophrenia and bipolar disorder are highly polygenic, with hundreds, or possibly thousands, of common SNPs contributing to a large percentage of disease liability (Purcell et al., 2009).
Figure 1.
Probability of identifying psychiatric risk alleles by genetic analysis. Rare risk alleles with high penetrance, such as DISC1 and certain CNVs, including 22q11.2 microdeletions, can be identified using standard linkage analysis. Common variants that have an odds ratio of ~1.0 or higher, such as ZNF804A and ANK3, can be identified by GWAS. However, recent studies suggest that most cases of psychiatric disorders may be the result of many common variants, each with a very small effect size. These variants are undetectable by current genetic methods.
The failure of genetic association studies to shed significant light on the genetics of psychiatric disease has been termed the problem of “missing heritability” (Manolio et al., 2009; Purcell et al., 2009). Several potential hurdles may ultimately limit the success of genetic studies, including genetic and phenotypic heterogeneity, epistatic gene interactions, and the role that the environment plays in the development and expression of psychiatric illness (Burmeister, 1999; Burmeister et al., 2008). On a practical basis, however, it is not necessary to identify all causative genes in order to develop effective treatments: in this case, the rare variant and CNV models of psychiatric disease may be most instructive. The recent associations between genomic structural variants with schizophrenia, bipolar disorder, and autism indicate that there may be many biological pathways that, when disrupted, lead to affective and cognitive disorders; in this sense, schizophrenia and bipolar disorder may not be individual diseases, but rather phenotypes of altered neuronal development (Guilmatre et al., 2009).
This hypothesis is supported by a number of lines of evidence, particularly for schizophrenia. First, the genes that have been most clearly associated with schizophrenia are genes involved in neuronal development. DISC1 and its binding partners regulate hippocampal gray matter volume, neurite outgrowth, dendritic arborization, and neuronal migration and maturation (Callicott et al., 2005; Millar et al., 2007). Two other risk genes, NRG1 and ERBB4, interact to regulate neuronal migration, axon myelination, and synapse formation (Buonanno et al., 2008; Mei and Xiong, 2008). Second, the symptoms of psychiatric illness have a developmental trajectory that parallels the maturation of the brain. The timing of peak disease risk for all psychiatric disorders overlaps with the substantial cortical dendritic pruning that occurs during adolescence (Feinberg, 1982; Kessler et al., 2005a), and, although there are clear prodromal signs for some disorders, outright symptoms such as psychosis are rare during childhood (Borgmann-Winter et al., 2006; Paus et al., 2008). Finally, a number of studies have found significant structural differences between schizophrenic and control brains, including decreased cortical neuron spine density, enlarged lateral ventricle size, and decreased hippocampal and cortical volume (Fatemi and Folsom, 2009; Schultz and Andreasen, 1999).
The association of CNVs and genes involved in neurodevelopment with schizophrenia also has important implications for therapeutic treatments. If there are multiple biological pathways that can lead to psychiatric disease, with any single cause being relatively rare in the population, useful therapies must have a broad effect. Currently available antipsychotics reduce psychosis via activity at the D2 receptor, but also act non-specifically on almost all the catecholaminergic and monoaminergic systems of the brain (Carpenter and Koenig, 2008). These drugs not only have poor efficacy and serious side effects, but also fail to address what may be the underlying genetic and molecular causes of psychiatric disease (Gogos, 2007). In contrast, microRNAs, which have the ability to regulate a large network of protein coding targets, may prove a more promising therapeutic avenue.
MicroRNA biogenesis and function
MicroRNAs, first identified less than two decades ago, are one of several classes of small RNAs found only in eukaryotes (Lai, 2002). Initially, the pri-miR sequence is transcribed in a PolII-dependent manner into a double-stranded RNA hairpin with a stem-loop structure (Kim, 2005). As miRs are often found clustered in the same genomic locus, a pri-miR may contain sequences for several different miRNAs (Bartel, 2004). Following transcription, the RNase III endonuclease Drosha, in conjunction with DGCR8 (in mammals) or pasha (in D. melanogaster), excises the stem-loop, resulting in the pre-miR, which is exported from the nucleus by Exportin 5 (Carthew and Sontheimer, 2009). In the cytoplasm, the proteins Dicer and TRBP cleave the terminal loop, producing a ~22 nt single-stranded mature miRNA (Denli et al., 2004). This process produces both a major (abundant) miRNA and a minor (less abundant) form; the major form is the guide strand used to specify the interaction between the target mRNA and the silencing complex, while the minor form, known as the miRNA* form, is generally discarded and degraded (Bartel, 2004; Carthew and Sontheimer, 2009).
RNA silencing occurs within the RNA-induced silencing complex (RISC), a ribonucleoprotein complex composed of the miRNA, the target mRNA, Dicer, and the Argonaute proteins (AGO1-4 in mammals) (Carthew and Sontheimer, 2009). RISCs may also include proteins such as FMRP, the protein encoded by the FMR1 gene, and GW182, a component of cytoplasmic structures known as P bodies (Eulalio et al., 2008). The sequence of the miRNA specifies the target mRNA sequence via perfect Watson-Crick base-pairing between nucleotides 2-8 (the seed region) of the miRNA and a complementary site in the 3′ UTR of the target mRNA. The degree of complementarity between miRNA and mRNA dictates the method of RNA inhibition: perfect complementarity results in AGO-mediated cleavage of the target mRNA, while mismatches result in translational repression, a reversible process that may allow cells to store mRNAs in preparation for rapid translation (Bartel, 2009). MiRNAs may also regulate pre-mRNA processing, mRNA structure, and mRNA-protein interactions (Filipowicz et al., 2008). As a single miRNA transcript can catalyze multiple rounds of RNA cleavage, miRNA levels must be tightly regulated (Flynt and Lai, 2008; Kosik, 2006).
MiRNA target prediction is still in its infancy, but several rules for prediction have been identified. First, there must be perfect complementarity between the miRNA seed region and a region within the mRNA 3′ UTR, and the miRNA-mRNA interaction must be thermodynamically permissive (Ameres et al., 2007; Lai, 2002; Lewis et al., 2003). As any 7-nucleotide sequence is likely to appear thousands of times within a genome, the most accurate prediction algorithms require evolutionary conservation of 3′ UTR sites across eukaryotic genomes. Following these rules, each miRNA is predicted to have an average of 400 protein-coding targets, suggesting that 50-60% of the genome is subject to RNA inhibition (Brennecke et al., 2005; Friedman et al., 2009). However, there are multiple factors that can affect which mRNAs are subject to regulation, and the strength of the inhibition. Chief among these is selective avoidance: mRNAs that contain non-conserved 3′ UTR binding sites are generally not expressed in the same cell or tissue as their cognate miRNA. The UTR context of the miRNA binding site, the number of miRNA binding sites within a single 3′ UTR, mRNA polyadenylation, and alternative splicing also regulate miRNA-mRNA interactions (Bartel, 2009).
Several studies have measured proteome-wide changes following miRNA manipulation. Selbach and colleagues found that, as predicted by the number of conserved 3′ UTR binding sites, overexpression of a single miRNA results in the repression of hundreds of proteins, a change that can, for the majority of targets, also be measured at the RNA level by microarray. However, the repressive effect was generally 30-40%, and almost never exceeded 4-fold downregulation (Selbach et al., 2008). Similarly, Baek and colleagues found that overexpression or knockdown of a single miRNA downregulated or upregulated, respectively, hundreds of genes at both the mRNA and protein level (Baek et al., 2008).
Function of miRNAs in the nervous system
As might be expected from the proteome-wide studies, a single microRNA can have widespread effects on the molecular identity of an individual cell. Co-expression of specific miRNAs thus results in cell type-specific regulation of a battery of protein-coding genes in a manner that allows cells to establish and maintain tissue identity. This was clearly demonstrated by Lim and colleagues, who over-expressed either miR-1, a heart- and skeletal muscle-enriched miRNA, or miR-124, a brain-enriched miR, in HeLa cells (Lim et al., 2005). Upregulation of miR-1 resulted in the down-regulation of 96 genes that are normally expressed at a very low level in mature heart tissue, while upregulation of miR-124 repressed the expression of almost 200 genes that are normally expressed at low levels in the brain versus all other tissues (Lim et al., 2005). Within the nervous system, miR-124a and miR-9 regulate neuronal precursor fate by regulating glial versus neuronal patterns of gene expression and alternative splicing (Krichevsky et al., 2006). The expression of miR-124 is restricted to differentiating and mature neurons by the transcriptional repressor RE1 silencing transcription factor (REST); in the absence of REST, miR-124 inhibits a large number of non-neuronal genes, including the splicing factor PTBP1, the absence of which leads to the establishment of alternative splicing patterns specific to neurons (Conaco et al., 2006; Makeyev et al., 2007; Visvanathan et al., 2007).
RNAi has been shown to play a crucial role in neural development and patterning. Disruption of dicer during zebrafish development results in abnormal neurulation (Giraldez et al., 2005); although Dicer mouse mutants arrest prior to neurulation (Bernstein et al., 2003), selective deletion of Dicer in the developing mouse neocortex results in increased neuronal apoptosis and severe cortical hypotrophy (De Pietri Tonelli et al., 2008). In C. elegans, the left-right asymmetry of chemosensory ASE neurons is established by two miRNAs, lsy-6 and miR-273. In the left ASE, the transcription factor die-1 induces expression of lsy-6, which represses the homeobox gene cog-1; in the right ASE, miR-273 targets die-1 for inhibition, effectively repressing lsy-6 expression (Chang et al., 2004; Johnston et al., 2005).
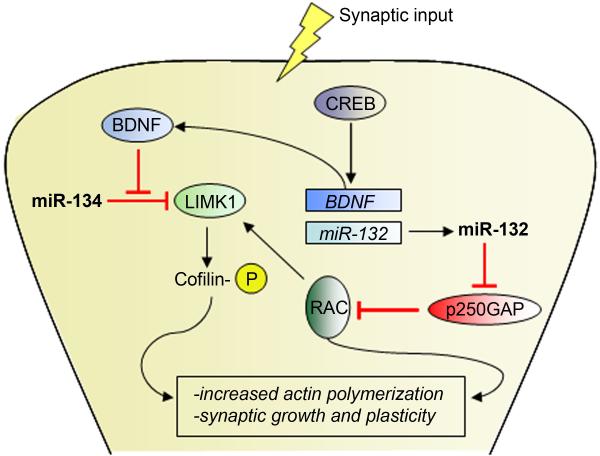
MiRNAs continue to play a major role in nervous system function during adulthood. A number of studies have linked miRNAs to both basic biology, such as synaptic plasticity and circadian regulation, and to disease-associated pathologies including Parkinson’s disease and Tourette’s Syndrome. Perhaps best-known is the role of miR-134, a regulator of activity-dependent RNA translation in dendritic spines. Local protein synthesis in dendrites was first identified in the 1980s, and later shown to be necessary for BDNF-induced changes in dendritic plasticity (Sutton and Schuman, 2006). Under static conditions, miR-134 binds to and inhibits the translation of LIMK1, a regulator of synaptic morphogenesis; BDNF signaling rapidly inhibits the effect of miR-134, allowing LIMK1 to be translated (Schratt et al., 2006). The brain-enriched miR-132 also regulates dendritic outgrowth via interactions with Rho family GTPase-activating protein p250GAP. MiR-132 transcription is induced by activity-dependent CREB signaling, resulting in translational inhibition of p250GAP, de-inhibition of Rac family GTPases, and a subsequent increase in dendritic size and branching (Wayman et al., 2008) (Figure 2).
Figure 2.
Function of microRNAs at the synapse. Synaptic input co-regulates miR-132 and miR-134, producing changes in synaptic plasticity. MiR-132, transcription of which is induced by CREB signaling, targets p250GAP for translational inhibition, resulting in an increase in RAC signaling and subsequent increase in LIMK1 activity. MiR-134 function is inhibited by activity-induced BDNF signaling, resulting in de-inhibition of LIMK1.
MiR-132 also plays a role in circadian rhythms, along with the brain-enriched miR-219. Both miRNAs are expressed in the suprachiasmatic nucleus (SCN), where they mediate separate aspects of the circadian pacemaker. MiR-219 is transcriptionally induced by the core circadian proteins CLOCK and BMAL and regulates phase length, whereas miR-132 transcription is induced by light-dependent activation of CREB and regulates light-induced phase-shifting (Cheng et al., 2007). In addition, miR-132 regulates neuronal excitability by potentiating the depolarizing effects of glutamate and NMDA, while miR-219 has the opposite effect (Cheng et al., 2007), suggesting that these miRNAs have a broader, as yet unknown role in regulating neuronal activity.
The widespread expression and activity of miRNAs in the brain suggest that they may be major players in neurological disorders. Indeed, two separate groups have associated global depletion of brain miRNA via selective deletion of Dicer with dopaminergic behaviors and Parkinson’s-like symptoms. Deletion of Dicer in dopamine neurons results in the progressive loss of dopamine neurons and expression of Parkinson’s-like behaviors, including reduced locomotion and increased immobility (Kim et al., 2007). Loss of Dicer in dopamine-receptive neurons also produces both behavioral changes, such as ataxia, and neuronal defects, such as reduced neuronal size and astrogliosis (Cuellar et al., 2008). As at least one miRNA, miR-133b, is significantly downregulated in midbrain tissue from Parkinson’s disease patient samples, it is possible that miRNA dysregulation regulates some aspects of Parkinson’s disease (Kim et al., 2007).
MicroRNAs have been suggested to play a role in several other neurological disorders. FMR1, the gene responsible for Fragile X mental retardation, encodes the protein FMRP, which is present in synapses and is part of the RISC complex that includes Dicer, miRNAs, and target mRNAs. Loss of FMRP impairs the function of RISC-mediated gene silencing, resulting in altered synaptic development (Jin et al., 2004). Some cases of Tourette’s syndrome may also be caused by miRNA dysregulation: mutations in the miR-189 binding site in the 3′ UTR of SLITRK1, a protein involved in neurite outgrowth, have been associated with Tourette’s syndrome in a small number of patients (Abelson et al., 2005). Finally, multiple studies have also suggested a link between miRNA dysregulation, including miR-29a/b-1, miR-106, and miR-9, and Alzheimer’s Disease, although the specific molecular mechanisms remain unknown (Hebert and De Strooper, 2009).
MiRNAs in psychiatric disease
The ability of miRNAs to have a broad effect on gene expression and functional pathways has important implications for psychiatric disease. Schizophrenia and bipolar disorder, which are now believed to represent different aspects of a clinical continuum, are characterized by dysregulation of multiple signaling pathways. For example, all currently available antipsychotics preferentially block neurotransmission at the dopamine D2 receptor (D2R), but may also affect signaling at as many as 10 other types of receptors, including the serotonergic, adrenergic, muscarinic, and histaminergic receptors (Meltzer and Huang, 2008). The action of antipsychotics at the D2R has led to the hypothesis that dopaminergic signaling is dysregulated in the schizophrenic brain, with the prefrontal areas exhibiting DA hypoactivity and the subcortical areas exhibiting DA hyperactivity (Alves Fda et al., 2008; Stone et al., 2007). This hypothesis is supported by a number of human and animal studies that have focused on the role of dopamine transmission (Schultz and Andreasen, 1999; Zhuang et al., 2001). However, dopamine antagonists do not treat negative symptoms of schizophrenia, nor do dopamine agonists fully recapitulate the spectrum of schizophrenic symptoms, suggesting that altered dopamine function represents only one aspect of schizophrenia.
Glutamatergic signaling, primarily via the NMDA receptor (NMDAR), has also been implicated in schizophrenia. The NMDAR antagonists phencyclidine (PCP) and MK-801 induce psychosis and cognitive impairment in normal human subjects, and NMDA receptor levels are reduced in schizophrenic patients (Pilowsky et al., 2006). Mouse models with hypoactive NMDAR function similarly exhibit hyperlocomotion, stereotypic movements, and deficits in cognitive and neurosensory function and social interaction (Enomoto et al., 2007). Many of these mouse models also exhibit reduced inhibitory GABAergic transmission. Importantly, NMDAR hypofunction recapitulates aspects of both positive and negative symptoms of schizophrenia. Recent experiments indicate that bipolar disorder shares many of the same genetic risk factors as schizophrenia, and is therefore likely to share many of the same molecular mechanisms (Potash and Bienvenu, 2009; Purcell et al., 2009).
The involvement of multiple signaling pathways in psychiatric disease complicates both the investigation of the underlying biological causes and efforts to develop effective therapies. Furthermore, it is likely that drugs that target a single receptor or signaling pathway are likely to be unsuccessful. Focusing on the role of miRNAs in psychiatric disease may both explain dysregulation of multiple pathways and offer a path to novel therapies that can target entire gene networks.
Several studies have directly looked at miRNA dysregulation in schizophrenia, although the results are somewhat conflicting (Table 1). Perkins and colleagues examined the expression of 264 miRNAs in prefrontal cortex (Brodmann’s Area 9) from a group of 15 schizophrenic or schizoaffective patients and 21 control samples, and identified 16 differentially regulated miRNAs, 15 of which were down-regulated in schizophrenia (Perkins et al., 2007). Of these, miR-26b, miR-30b, miR-29b, and miR-106b showed the greatest fold change, although all fold changes were less than 2-fold. Interestingly, for several of the differentially-expressed miRNAs, the ratio of mature miRNA to pri-miRNA was lower in schizophrenia, suggesting a disruption in miRNA biogenesis in schizophrenia.
Table 1.
MicroRNAs associated with psychiatric disease
| miRNA | Validated target/Function | Reference |
|---|---|---|
| 16 miRs dysregulated |
Changed in DL-PFC from SCZ patients (microarray) |
(Perkins et al., 2007) |
| let-7g, miR-181b | Changed in temporal gyrus from SCZ patients (microarray) |
(Beveridge et al., 2008) |
| 59 misR dysregulated |
Changed in temporal gyrus from SCZ patients (microarray) |
(Beveridge et al., 2009) |
| 26 miRs dysregulated |
Changed in DL-PFC from SCZ patients (microarray) |
(Beveridge et al., 2009) |
| miR-124, 383, 320, 596, 597, 598 |
Located within chromosome 8 QTL for SCZ, autism |
(Tabares-Seisdedos and Rubenstein, 2009) |
| 5 X-linked miRs | Private mutations in SCZ patients | (Feng et al., 2009) |
| miR-346 | Located in GRID2 intron, reduced in SCZ | (Zhu et al., 2009) |
| miR-34a | GRM7/reduced by both lithium and valproate treatment |
(Zhou et al., 2009) |
| miR-221, 152, 152, 494 |
Changed in BPD lymphocytes after lithium treatment |
(Chen et al., 2009) |
| miR-199a, 128a/b | Upregulated by haloperidol treatment in rats | (Perkins et al., 2007) |
| miR-195, miR-30a | BDNF regulation | (Mellios et al., 2008) |
| miR-96 | HTR1B/aggressive behavior in humans | (Jensen et al., 2009) |
| miR-219 | CAMK2G/reduced by MK-801 treatment in mice | (Kocerha et al., 2009) |
A second group has also observed miRNA dysregulation and altered miRNA biogenesis in schizophrenic brain tissue. In 2 separate studies evaluating the expression of 262 (Beveridge et al., 2008) or 322 (Beveridge et al., 2009) miRNAs in superior temporal gyrus (STG) and dorsolateral prefrontal cortex (DL-PFC) from schizophrenia and control samples, Beveridge and colleagues observed schizophrenia-associated upregulation of a very large number of miRNAs: 21% of expressed miRNAs in the STG and 9.5% of expressed miRs in the DL-PFC. Upregulated miRNAs included miR-181b, miR-219, and members of the miR-15 family; surprisingly, of the 81 dysregulated miRNAs, only 4 were upregulated in both the STG and DL-PFC (miR-128a, miR-16, miR-20a, and miR-338). Four miRNAs, including miR-24, miR-26b, miR-29c, and miR-7, overlapped with the set of significantly changed miRNAs from the Perkins study; in the Perkins study, all four miRNAs were downregulated, while in the Beveridge study all were upregulated. Changes in miRNA biogenesis were also contrary to those in the Perkins study: Beveridge and colleagues found that the pool of ~22-nt RNAs and levels of the miRNA processing enzymes Dicer and DGCR8 were significantly upregulated in schizophrenia, suggestive of a global increase in miRNA biogenesis.
Further studies in larger, separate populations will be necessary to reconcile the contradictory human data. However, there is strong experimental support for altered miRNA biogenesis in at least one subset of schizophrenic patients. Hemizygous deletions of the 22q11.2 locus in humans result in deficits in attention, learning, executive function, and emotional behavior, and account for up to 2% of all cases of schizophrenia (Karayiorgou et al., 1995). Stark and colleagues produced a mouse model hemizygous for a deletion of a 1.3 Mb region syntenic to 22q11.2 and observed a number of schizophrenia-like phenotypes (increased hyperactivity, poor pre-pulse inhibition, and reduced dendritic spine density), upregulation of pri-miRNA levels, and downregulation of mature miRNA levels in the brain (Stark et al., 2008). Contained within both the 22q11.2 region and the deleted murine locus is DGCR8, the absence of which results in a bottleneck in the processing of pre-miRNAs to mature miRNAs. Deletion of Dgcr8 alone was also sufficient to produce a number schizophrenia-like behaviors in mice.
Several groups have produced data associating specific miRNAs with schizophrenia. Hansen and colleagues genotyped SNPs near or within 28 brain-expressed miRNAs in three case/control populations of European ancestry, and found that minor alleles in miR-206 and miR-198 were over- or under-represented, respectively, in schizophrenia (Hansen et al., 2007). Feng and colleagues sequenced 59 X-linked SNPs in a small case/control population and found an increase in private, ultra-rare mutations of the pri- or mature miRNA sequences in schizophrenia (Feng et al., 2009). Zhu and colleagues used bioinformatics to identify the miRNAs that target the largest number of schizophrenia-associated genes, and found that two miRNAs, miR-566 and miR-346, target more schizophrenia-associated genes than would be expected by chance. Furthermore, miR-346 is located within an intron of GRID1, a glutamate receptor subunit gene that is downregulated in schizophrenia (Zhu et al., 2009). Finally, 6 miRNAs, including miR-124 and miR-383, are located with the 8p21-23 locus, a CNV “hot spot” linked to schizophrenia and autism (Tabares-Seisdedos and Rubenstein, 2009).
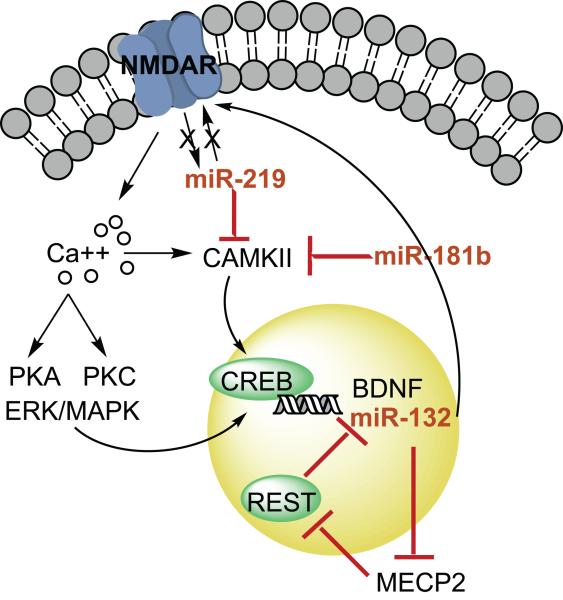
The specific molecular mechanisms through which altered miRNA activity may cause psychiatric phenotypes are still poorly understood. Recently, our group found that miR-219 mediates the behavioral effects of MK-801 treatment in mice (Kocerha et al., 2009). Acute, but not chronic, MK-801 treatment decreased levels of miR-219 in the prefrontal cortex, and inhibition of miR-219 prevented MK-801-induced hyperlocomotion and stereotypies. One of the targets of miR-219 is CAMK2G, a member of the calcium/calmodulin-dependent protein kinase family involved in NMDA signaling. When combined with data showing that miR-219 attenuates NMDA-induced neuronal depolarization (Cheng et al., 2007), these results indicate that miR-219 may inhibit NMDA signaling at the level of both the receptor and second messenger signaling. Upregulation of miR-219 in DL-PFC from schizophrenic patients, as observed by Beveridge and colleagues (Beveridge et al., 2009), is therefore consistent with the NMDAR hypoactivity hypothesis of schizophrenia.
BDNF is another major player in schizophrenia, bipolar disorder, and depression. In addition to inhibiting the effects of miR-134 on Limk1, described above, BDNF is itself the target of several miRNAs. Mellios and colleagues identified 2 miRNAs, miR-30a and miR-195, that are expressed in human prefrontal cortex, directly target the BDNF 3′ UTR, and reduce BDNF expression (Mellios et al., 2008). BDNF is indirectly regulated by miR-132: CREB-induced transcription of miR-132 results in a decrease of MECP2, the protein involved in Rett syndrome, and a subsequent decrease in BDNF due to de-repression of REST (Abuhatzira et al., 2007; Klein et al., 2007). CREB expression has been shown to be reduced in schizophrenia, suggesting that miR-132 expression may also be reduced (Yuan et al., 2009). Our lab has recently observed a significant reduction in miR-132 levels in prefrontal cortex from schizophrenic and bipolar patients (unpublished data); because miR-132 potentiates NMDAR depolarization (Cheng et al., 2007), reduced expression of miR-132 in schizophrenia, like up-regulation of miR-219, would also be consistent with NMDAR hypofunction in schizophrenia (Figure 3). The mechanism of the interaction between the NMDAR and miRNAs is still unknown: it is not clear whether miRNAs directly regulate receptor subunit activity and availability, or instead act on key members of the NMDAR signaling pathway.
Figure 3.
Possible roles of miRNAs in signaling pathways associated with psychiatric disorders. MiR-132, miR-219, and miR-181b all regulate NMDA-induced calcium signaling pathways. NMDA signaling inhibits miR-219, resulting in disinhibition of CAMK2G, which is also a target of miR-181b. Both miR-219 and miR-181b are upregulated in schizophrenic tissue. NMDA signaling increases miR-132 levels; miR-132 potentiates NMDAR activity, but inhibits BDNF transcription by targeting MECP2, which targets the transcriptional repressor REST. Reduced NMDA function, as observed in schizophrenia, would result in reduced miR-132 levels in schizophrenia tissue.
MiRNAs may also mediate some of the effects of psychiatric drug therapies. Zhou and colleagues found that in vitro lithium and valproic acid treatment differentially regulated 37 and 31 miRNAs, respectively, with 8 miRNAs in common (Zhou et al., 2009). Several of the miRNA target genes, including GRM7, DPP10, and THRB, are potential genetic risk factors for bipolar disorder. Similarly Chen and colleagues examined miRNA expression in lymphoblastoid cell lines derived from bipolar disorder patients or unaffected siblings, and identified alterations in expression of several miRNAs following lithium treatment (Chen et al., 2009). In rats, treatment with the antipsychotic haloperidol upregulates 3 miRNAs, miR-199a, miR-128a, and miR-128b (Perkins et al., 2007). All three miRNAs have been shown to be upregulated in brain tissue from schizophrenia patients (Beveridge et al., 2009), although it is unclear whether the upregulation is central to schizophrenia or a result of antipsychotic treatment.
Conclusion
Individual miRNAs regulate several hundred proteins in tandem, with a few targets downregulated greatly and many other targets downregulated to a lesser extent. The ability of miRNAs to fine-tune the activity of entire biological pathways may underlie some of the difficulties associated with linking psychiatric disorders to specific causative genes. A more complete picture of the miRNAs that are dysregulated in psychiatric illness may improve our understanding of the molecular mechanisms underlying neuropsychiatric phenotypes, and, due to their tuning effect on large numbers of proteins, miRNAs may ultimately represent a new therapeutic target for psychiatric disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, Davis NR, Ercan-Sencicek AG, Guez DH, Spertus JA, Leckman JF, Dure L.S.t., Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics. 2007;2:214–22. doi: 10.4161/epi.2.4.5212. [DOI] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Alves Fda S, Figee M, Vamelsvoort T, Veltman D, de Haan L. The revised dopamine hypothesis of schizophrenia: evidence from pharmacological MRI studies with atypical antipsychotic medication. Psychopharmacol Bull. 2008;41:121–32. [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–12. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bertram L. Genetic research in schizophrenia: new tools and future perspectives. Schizophr Bull. 2008;34:806–12. doi: 10.1093/schbul/sbn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–68. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgmann-Winter K, Calkins ME, Kniele K, Gur RE. Assessment of adolescents at risk for psychosis. Curr Psychiatry Rep. 2006;8:313–21. doi: 10.1007/s11920-006-0068-1. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A, Kwon OB, Yan L, Gonzalez C, Longart M, Hoffman D, Vullhorst D. Neuregulins and neuronal plasticity: possible relevance in schizophrenia. Novartis Found Symp. 2008;289:165–77. doi: 10.1002/9780470751251.ch13. discussion 177-9, 193-5. [DOI] [PubMed] [Google Scholar]
- Burmeister M. Basic concepts in the study of diseases with complex genetics. Biol Psychiatry. 1999;45:522–32. doi: 10.1016/s0006-3223(98)00316-3. [DOI] [PubMed] [Google Scholar]
- Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–40. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–32. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–7. [PubMed] [Google Scholar]
- Carpenter WT, Koenig JI. The evolution of drug development in schizophrenia: past issues and future opportunities. Neuropsychopharmacology. 2008;33:2061–79. doi: 10.1038/sj.npp.1301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr., Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–9. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang N, Burmeister M, McInnis MG. MicroRNA expression changes in lymphoblastoid cell lines in response to lithium treatment. Int J Neuropsychopharmacol. 2009:1–7. doi: 10.1017/S1461145709000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, O’Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–9. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–21. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–58. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto T, Noda Y, Nabeshima T. Phencyclidine and genetic animal models of schizophrenia developed in relation to the glutamate hypothesis. Methods Find Exp Clin Pharmacol. 2007;29:291–301. doi: 10.1358/mf.2007.29.4.1075358. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–53. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35:528–48. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res. 1982;17:319–34. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PLoS One. 2009;4:e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, MacLean AW, St CD, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–42. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gogos JA. Schizophrenia susceptibility genes: in search of a molecular logic and novel drug targets for a devastating disorder. Int Rev Neurobiol. 2007;78:397–422. doi: 10.1016/S0074-7742(06)78013-8. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Veber P. Saugier, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–56. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I, Hall H, Timm S, Wang AG, Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- ISC Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr., Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005;102:12449–54. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–78. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kapur S, Remington G. Atypical antipsychotics: new directions and new challenges in the treatment of schizophrenia. Annu Rev Med. 2001;52:503–17. doi: 10.1146/annurev.med.52.1.503. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–6. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005a;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005b;62:617–27. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, Cichon S, Hillmer A, Toncheva D, Owen MJ, O’Donovan MC. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–4. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, Orum H, Kauppinen S, Kenny PJ, Wahlestedt C. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–12. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–23. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–52. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–42. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer HY, Huang M. In vivo actions of atypical antipsychotic drug on serotonergic and dopaminergic systems. Prog Brain Res. 2008;172:177–97. doi: 10.1016/S0079-6123(08)00909-6. [DOI] [PubMed] [Google Scholar]
- Millar JK, Mackie S, Clapcote SJ, Murdoch H, Pickard BS, Christie S, Muir WJ, Blackwood DH, Roder JC, Houslay MD, Porteous DJ. Disrupted in schizophrenia 1 and phosphodiesterase 4B: towards an understanding of psychiatric illness. J Physiol. 2007;584:401–5. doi: 10.1113/jphysiol.2007.140210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, Caroff SN, Davis SM, Rosenheck RA, McEvoy JP, Saltz BL, Riggio S, Chakos MH, Swartz MS, Keefe RS, Stroup TS, Lieberman JA. Extrapyramidal side-effects of antipsychotics in a randomised trial. Br J Psychiatry. 2008;193:279–88. doi: 10.1192/bjp.bp.108.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciute D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Moller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C, Rujescu D, Meltzer HY, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–5. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Williams NM, O’Donovan MC. The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry. 2004;9:14–27. doi: 10.1038/sj.mp.4001444. [DOI] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci. 2008;9:947–57. doi: 10.1038/nrn2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilowsky LS, Bressan RA, Stone JM, Erlandsson K, Mulligan RS, Krystal JH, Ell PJ. First in vivo evidence of an NMDA receptor deficit in medication-free schizophrenic patients. Mol Psychiatry. 2006;11:118–9. doi: 10.1038/sj.mp.4001751. [DOI] [PubMed] [Google Scholar]
- Porteous D. Genetic causality in schizophrenia and bipolar disorder: out with the old and in with the new. Curr Opin Genet Dev. 2008;18:229–34. doi: 10.1016/j.gde.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Potash JB, Bienvenu OJ. Neuropsychiatric disorders: Shared genetics of bipolar disorder and schizophrenia. Nat Rev Neurol. 2009;5:299–300. doi: 10.1038/nrneurol.2009.71. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schultz SK, Andreasen NC. Schizophrenia. Lancet. 1999;353:1425–30. doi: 10.1016/s0140-6736(98)07549-7. [DOI] [PubMed] [Google Scholar]
- Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–10. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–7. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, Kendler KS, Li T, O’Donovan M, O’Neill FA, Owen MJ, Walsh D, Weinberger DR, Sun C, Flint J, Darvasi A. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–6. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–60. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–7. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Morrison PD, Pilowsky LS. Glutamate and dopamine dysregulation in schizophrenia--a synthesis and selective review. J Psychopharmacol. 2007;21:440–52. doi: 10.1177/0269881106073126. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–84. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–89. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- Tandon R, Belmaker RH, Gattaz WF, Lopez-Ibor JJ, Jr., Okasha A, Singh B, Stein DJ, Olie JP, Fleischhacker WW, Moeller HJ. World Psychiatric Association Pharmacopsychiatry Section statement on comparative effectiveness of antipsychotics in the treatment of schizophrenia. Schizophr Res. 2008;100:20–38. doi: 10.1016/j.schres.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Thase ME. STEP-BD and bipolar depression: what have we learned? Curr Psychiatry Rep. 2007;9:497–503. doi: 10.1007/s11920-007-0068-9. [DOI] [PubMed] [Google Scholar]
- Tomppo L, Hennah W, Lahermo P, Loukola A, Tuulio-Henriksson A, Suvisaari J, Partonen T, Ekelund J, Lonnqvist J, Peltonen L. Association Between Genes of Disrupted in Schizophrenia 1 (DISC1) Interactors and Schizophrenia Supports the Role of the DISC1 Pathway in the Etiology of Major Mental Illnesses. Biol Psychiatry. 2009 doi: 10.1016/j.biopsych.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–9. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–8. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . The global burden of disease: 2004 update. Geneva: 2004. [Google Scholar]
- Williams HJ, Owen MJ, O’Donovan MC. New findings from genetic association studies of schizophrenia. J Hum Genet. 2009;54:9–14. doi: 10.1038/jhg.2008.7. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet. 2006;15:743–9. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yuan P, Zhou R, Wang Y, Li X, Li J, Chen G, Guitart X, Manji HK. Altered levels of extracellular signal-regulated kinase signaling proteins in postmortem frontal cortex of individuals with mood disorders and schizophrenia. J Affect Disord. 2009 doi: 10.1016/j.jad.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Qian Y, Alliey-Rodriguez N, Kelsoe JR, Greenwood T, Nievergelt C, Barrett TB, McKinney R, Schork N, Smith EN, Bloss C, Nurnberger J, Edenberg HJ, Foroud T, Sheftner W, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon F, Schulze TG, Berrettini W, Potash JB, Belmonte PL, Zandi PP, McInnis MG, Zollner S, Craig D, Szelinger S, Koller D, Christian SL, Liu C, Gershon ES. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry. 2009;14:376–80. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Kalbfleisch T, Brennan MD, Li Y. A MicroRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res. 2009;109:86–9. doi: 10.1016/j.schres.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Oosting RS, Jones SR, Gainetdinov RR, Miller GW, Caron MG, Hen R. Hyperactivity and impaired response habituation in hyperdopaminergic mice. Proc Natl Acad Sci U S A. 2001;98:1982–7. doi: 10.1073/pnas.98.4.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]