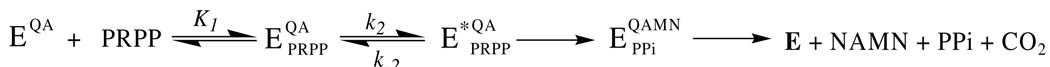Abstract
Quinolinate phosphoribosyltransferase (QAPRTase, EC 2.4.2.19) catalyzes an unusual phosphoribosyl transfer that is linked to a decarboxylation reaction to form the NAD precursor nicotinate mononucleotide, carbon dioxide, and pyrophosphate from quinolinic acid (QA) and 5-phosphoribosyl 1-pyrophosphate (PRPP). Structural studies and sequence similarities with other PRTases have implicated Glu214, Asp235, Lys153 and Lys284 in contributing to catalysis through direct interaction with PRPP. The four residues were substituted by site-directed mutagenesis. A nadC deletant form of BL21DE3 was created to eliminate trace contamination by chromosomal QAPRTase. The mutant enzymes were readily purified and retained their dimeric aggregation state on gel filtration. Substitution of Lys153 with Ala resulted in an inactive enzyme, indicating its essential nature. Mutation of Glu214 to Ala or Asp caused at least a 4000-fold reduction in kcat, with 10-fold increases in Km and KD values for PRPP. However, mutation of Glu214 to Gln had only modest effects on ligand binding and catalysis. pH profiles indicated that the deprotonated form of a residue with pKa of 6.9 is essential for catalysis. The WT-like pH-profile of the E214Q mutant indicated that Glu214 is not that residue. Mutation of Asp235 to Ala did not affect ligand binding or catalysis. Mutation of Lys284 to Ala decreased kcat by 30-fold and increased Km and KD values for PRPP by 80-fold and at least 20-fold, respectively. The study suggests that Lys153 is necessary for catalysis and important for PRPP binding, Glu214 provides a hydrogen bond necessary for catalysis but does not act as a base or electrostatically to stabilize the transition state, Lys284 is involved in PRPP binding and Asp235 is not essential.
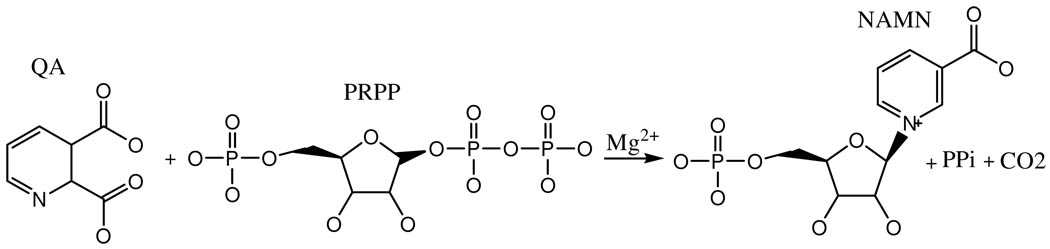
Quinolinate phosphoribosyltransferase (QAPRTase, EC 2.4.2.19) is a key enzyme in the metabolism of quinolinic acid (QA), an intermediate in the de novo pathway of NAD biosynthesis (1). The enzyme catalyzes the transfer of the phosphoribosyl group from 5-phosphoribosyl-1-pyrophosphate (PRPP) to the nitrogen atom of QA to form nicotinate mononucleotide (NAMN), carbon dioxide (CO2), and pyrophosphate (PPi) in the presence of Mg2+ (Scheme 1). Although QAPRTase has been regarded as a drug target for Mycobacterium tuberculosis, the finding that the NAD salvage pathway is active in that organism under aerobic conditions has made that contention more controversial (2,3). In addition, QA is a known neurotoxin (4), for which QAPRTase provides the sole metabolic fate (2).
Scheme 1.
The reaction catalyzed by QAPRTase
QAPRTase is a member of the phosphoribosyltransferase (PRTase) group of enzymes, which catalyze the formation of nucleotides from nitrogenous bases and their common substrate PRPP (5, 6). PRTases have been recently classified into four subclasses (type I, II, III, and IV) based on their structural architecture (7–12). The type II class is represented by QAPRTase, NAPRTase and NMPRTase (8–10). Crystallographic studies of QAPRTase from Salmonella typhimurium revealed a homodimeric protein whose 297-residue subunits (13) are folded into two structural domains (8). The deep active site is formed by the C-terminal α/β barrel domain, and is capped by the N-terminal open-faced sandwich domain provided by the adjacent subunit (8). The more highly resolved structures of various complexes of the hexameric M. tuberculosis QAPRTase (2) further document details of this interfacial active site. A recent set of structures of the yeast enzyme has added detail to this picture (14).
Previously, our lab determined that the QAPRTase reaction proceeds via an ordered-sequential mechanism, in which QA binds first, followed by PRPP, and NAMN is released prior to PPi (15). This mechanism is compatible with the S. typhimurium QAPRTase structure. Comparison of the native and substrate bound structures show that binding of QA induces a structural rearrangement of residues in the active site proposed to facilitate PRPP binding and the subsequent reaction (2). Binding studies employing the nonproductive carbocyclic QA analog phthalic acid (benzene-1,2-dicarboxylic acid, PA), also revealed a tighter binding of PA and PRPP in the QAPRTase•PA•PRPP ternary complex than in their respective binary complexes. Kinetic studies showed that a nonproductive QAPRTase•PRPP complex can also form and cause substrate inhibition (15).
Although the kinetic mechanism of QAPRTase has been established, its catalytic mechanism has not. In one proposal, the pyrophosphate group of PRPP is protonated and expelled to leave an oxycarbonium ribosyl phosphate intermediate which is then attacked by N1 of QA (2, 8). Tighter coupling of expulsion and attack could result in an oxycarbonium-like transition state (2). The crystal structure of M. tuberculosis QAPRTase in complex with the substrate analogs 5-phosphoribosyl-1-(β-methylene)pyrophosphate (PRPCP) and PA show the C2 carboxylate of PA and the α-phosphoryl group of PRPCP as neighbors to the C1 of PRPCP (2), potentially stabilizing an oxycarbonium intermediate or transition state. The proposal of an oxycarbonium-like transition state also receives support from similar transition state structures characterized by kinetic isotope effects for OPRTase, a type I PRTase, and a variety of chemically analogous ribosyltransferases, glycosyltransferases and hydrolases (16, 17). The product of the phosphoribosyl transfer reaction is the putative quinolinic acid mononucleotide (QAMN) intermediate. Subsequent decarboxylation of the unstable QAMN intermediate leads to the formation of NAMN (2, 8).
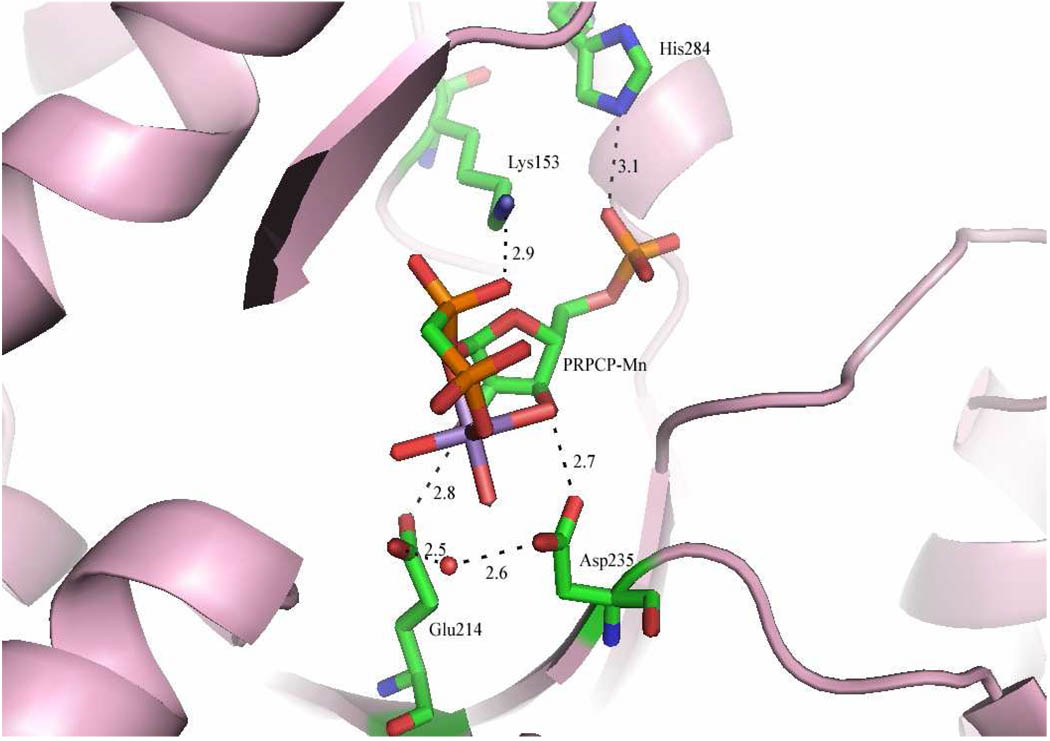
QAPRTase might use geometric, chemical and electrostatic means to stabilize the positively charged oxycarbonium-like transition state and assist catalysis of phosphoribosyl transfer. In the structure of the M. tuberculosis QAPRTase•PA•PRPCP•2Mg2+ complex, Glu214 and Asp235 (residue numbering is based on the mature S. typhimurium protein sequence, starting with Pro1) are seen to contact the 2- and 3-hydroxyl groups of PRPCP (Figure 1) and are likely enzymic candidates for the stabilization of the transition state (2, 8). One oxygen of the carboxylate sidechain of Glu214 forms a hydrogen bond (2.8 Å) with the 2-hydroxyl group of PRPCP, while the second carboxylate oxygen is 2.5 Å from a water molecule that coordinates the dihydrated Mg2+ required for PRPP binding and catalysis (Figure 1). In the product bound QAPRTase•NAMN complex, Glu214 is 4.2 Å from the 2-hydroxyl of NAMN. The shorter distance (2.8 Å) of Glu214 from the 2-hydroxyl group of PRPCP in the ternary complex suggests that Glu214 might geometrically orient PRPP in catalysis. Alternatively, the negative charge of the carboxylates may serve to stabilize the oxycarbonium-like transition state. The position of Glu214 in QAPRTase is also appropriate for Oppenheimer’s proposed mechanism for the chemically analogous NAD glycohydrolase, in which an active site glutamate carboxylate deprotonates the 2-hydroxyl of NAD in order to increase the electron density near C1, thereby stabilizing an oxycarbonium transition (18). At the 3-hydroxyl group of PRPCP, Asp235 is seen to form a hydrogen bond (2.7 Å) with the 3-hydroxyl oxygen, and the fixed water molecule that binds Glu214 and coordinates the dihydrated Mg2+ (Figure 1). In their three-dimensional locations, the two acidic residues Glu214 and Asp235 are analogs of the highly conserved but sequence-adjacent acidic residues present in the PRPP binding motif of most type I PRTases that also interact with the 2- and 3-hydroxyl groups of PRPP (2, 19). The residues are also apparent analogs of the acidic residues of other glycosyltransferases that are believed to take part in stabilizing an oxycarbonium ion (20).
Figure 1.
Active site of M. tuberculosis QAPRTase•PA•PRPCP•2Mn2+ Michaelis complex. The image shows the interactions of Lys153, Glu214, Asp235 and His284 (corresponding to Lys284 of S. typhimurium) with PRPCP-Mn2+ complex. Hydrogen bonds are shown as dashed lines and the water molecule involved in coordinating one of the Mn2+ is represented by a red sphere. Carbon atoms are colored green, nitrogen are blue, oxygen are red and phosphorous in orange and Mn2+ in purple. PA and the second Mn2+ coordinated by two oxygen of the pyrophosphate group of PRPCP are not shown. All distances shown in black are given in Å. The image was generated using PyMOL (Delano Scientific) and PDB entry 1QPR.
The non-bridging 5-phosphate oxygen atoms of PRPCP are seen to interact with the backbone amide nitrogens of Asn260 and Gly280 in the QAPRTase•PA•PRPCP•2Mg2+ complex. The sidechain nitrogen of Lys284 is within hydrogen-bonding distance (3.1 Å) to the 5-phosphate oxygen (Figure 1). These interactions were suggested to anchor the 5-phosphate on the protein so that C1 of PRPP can swing towards QA during catalysis (2). This proposal is also supported by Schramm’s suggestion that ribosyltransferases assist in catalysis through geometric distortion from the ribosyl-5-phosphate moeity to form the transition state (20). As shown in Figure 1, a non-bridging α-phosphoryl oxygen of PRPCP hydrogen bonds (2.9Å) to the side chain of Lys153. This observation led to the hypothesis that Lys153 might protonate the leaving pyrophosphate of PRPP in the proposed catalytic mechanism of the QAPRTase reaction to form the oxycarbonium-like transition state (8).
In this paper, detailed kinetic analysis of site-directed mutants and corroboration with previously determined structural and kinetic data resolved the functions of the active site residues Lys153, Glu214, Asp235 and Lys284 in catalysis. The effects of the mutations on the physical, steady-state, pre-steady state kinetics, binding of substrates and pH-profiles are described with respect to WT QAPRTase.
Materials and Methods
Materials
Oligonucleotide primers were obtained from IDT, Inc. Rapid PCR, DNA purification and DNA extraction kits were purchased from Qiagen. Restriction endonucleases, enzymes and substrates used in PCR reactions were supplied by Roche Applied Biosciences. The QuikChange mutagenesis kit was purchased from Stratagene. The host strain E. coli DY330 [W3110 ΔlacU169 gal490 pglΔ8 λc1857 λ(cro-bioA)] (21) was obtained from Donald Court. BL21DE3 cells were from Invitrogen. FPLC equipment used for protein chromatography was supplied by Amersham Biosciences. DEAE-Toyo Pearl M650 resin was purchased from Tosoh Biosciences. Gel filtration molecular weight standards were obtained from Sigma. [3H]QA and [3H]PA were purchased from Moravek Biochemicals, Inc. (Brea, CA). Chromatography paper was Whatman 3MM. NAMN and PRPP were from Sigma. [14C]PRPP and [14C]NAMN were enzymatically prepared as described (15).
Sequence Analysis
The EC number of QAPRTase was used to search the protein database at the European Bioinformatics Institute website. The database search initially identified 352 sequences; 3 sequences that were not defined as QAPRTase were discarded. As suggested (22), in order to exclude redundant entries, multiple-sequence alignments were performed on sequence entries from the same organism using ClustalW (23). Analyses of those showing more than 80% identity allowed exclusion of the redundant entries. The remaining 230 sequences were again aligned with ClustalW and viewed with BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Construction of ZB100
A nadC deleted strain, ZB100, was constructed in two steps by replacing the nadC of E. coli with a spectinomycin resistance gene (SpcR), using the recombineering technology of Yu et al. (21). This method uses an inducible λ bacteriophage recombination system (RED), to replace the targeted genomic sequence with a drug-resistance cassette. In the first step, the DY330 E. coli strain that harbors a λ prophage encoding the λ, exo, bet, and cro genes regulated by a temperature sensitive λ-phage cI-repressor (24) was used to carry out the exchange. PCR was used to construct a SpcR cassette with flanking sequences that targeted the upstream and downstream regions of E. coli nadC. The following pair of primers were used: forward primer, 5'AGG ATC TCC TTA TAT GTG GTG CTA ATA CCC GGT TCA GAG TAG CAT GTT TCT ATG AGG AGG CAG ATT GCC TTG, and reverse primer, 5'AAC CGC GTT GCT TGT CCA TCT GTT TCG CTC CTT GAT TTG GTT GGC GCT ACC TGT GGA TAA CCG TAT TAC CGC C with a previously amplified SpcR sequence from pFW11 (a gift from Bettina Buttaro, Temple University School of Medicine) as a template. The underlined sequences are complementary to the SpcR template, while the non-underlined sequences target the E. coli nadC sequence. The gel-purified cassette was subsequently electroporated into DY330 cells that had been induced for λ recombination functions according to Yu et al. (21). After electroporation, the cells were incubated with shaking at 30°C for 1 hr prior to selection of the spectinomycin resistant colonies. To obtain a host cell line suitable for over-expression of mutant plasmids, a second step was carried out in which generalized P1 transduction (25) was used to transfer the replaced nadC locus of the spectinomycin resistant recombinant DY330 strain into E. coli BL21DE3. Selection of spectinomycin-resistant colonies confirmed that the resulting strain BL21(DE3)SpcR ΔnadC (ZB100) contained the SpcR sequence. Replacement of the chromosomal nadC was confirmed by PCR analysis and gene sequencing. The absence of nadC in ZB100 was also confirmed by a nicotinic acid requirement (5 µg/mL) on M9 minimal media.
Over-expression and Purification of QAPRTase
Using the nadC coding sequence of S. typhimurium previously cloned into the pRSETC expression vector (8) as a template, K153A, E214A, E214D, E214Q, D235A and K284A mutations were generated using the QuikChange mutagenesis procedure (Stratagene) with the following primers: K153A (forward CAG TTG CTC GAC ACG CGT GCG ACG CTG CCG GGT CTG CGC; reverse GCG CAG ACC CGG CAG CGT CGC ACG CGT GTC GAG CAA CTG), E214A (forward, G CCG GTA GAA GTC GCG GTC GAA AAT CTG G; reverse, C CAG ATT TTC GAC CGC GAC TTC TAC CGG C), E214D (forward, G CCG GTA GAA GTC GAC GTC GAA AAT CTG G; reverse, CCA GAT TTT CGA CGT CGA CTT CTA CCG GC), E214Q (forward, G CCG GTA GAA GTC CAG GTC GAA AAT CTG G; reverse, C CAG ATT TTC GAC CTG GAC TTC TAC CGG C), D235A (forward, GCG GAT ATT ATC ATG CTG GCG AAT TTC AAC ACC GAC CAG; reverse, CTG GTC GGT GTT GAA ATT CGC CAG CAT GAT AAT ATC CGC) and K284A (forward, TCC GTT GGC GCG CTG ACC GCG CAC GTC CGC GCG CTC G; reverse, C GAG CGC GCG GAC GTG CGC GGT CAG CGC GCC AAC GGA) with the position of the mismatch indicated in bold. The plasmids were subsequently transformed into competent ZB100 by standard procedure (26). The presence of the mutation and absence of other alterations in the mutated nadC sequence were confirmed by sequencing the entire gene with T7 primers. Mutant proteins were over-expressed and purified as reported for WT by Eads et al. (8), with the use of a larger (7 × 90 cm) DEAE column in the purification of E214A. This modification overcame poor resolution and yield during the first ion exchange chromatography step. During the purification procedure, precautions were taken to assure that there was no batch-to-batch contamination of mutant enzymes by WT. All columns were washed with elution buffer, followed by a wash with 1 M NaOH, prior to equilibration of the column with the loading buffer. SDS PAGE was used to determine the purity of the protein. Enzymes were stored in 65% saturated ammonium sulfate at 4°C. Prior to all kinetic and binding assays, WT and mutant enzymes were desalted using the centrifuge column technique (27) in 1 mL syringes containing Sephadex G-50 pre-equilibrated in buffer QA (50 mM KH2PO4 at pH 7.2, 6 mM MgCl2).
Oligomerization state of QAPRTases
Approximately 0.5 mL samples of 5 µM or 50 µM of the desalted enzymes dissolved in buffer QA were separately applied to a (1.6 × 60 cm) Superdex HR-200 column (Amersham Biosciences) pre-equilibrated with buffer QA. The enzymes were eluted at a flow rate of 1 ml/min, and absorbance was monitored at 280 nm. Protein standards chromatographed under similar conditions included cytochrome c (12.4 kDa), carbonic anyhdrase (29 kDa), ovalbumin (44 kDa), bovine serum albumin (67 kDa), yeast alcohol dehydrogenase (150 kDa) and β-amylase (200 kDa). Molecular masses of QAPRTases were estimated from a calibration curve of log molecular mass versus retention time generated with the standards. The experiments for WT and mutant enzymes were performed individually in buffer QA and buffer QA that contained 2 mM DTT.
Enzymatic and Equilibrium Binding Assays for QAPRTase
A spectrophotometric assay that measures the increase in absorbance at 266 nm as QA is converted to NAMN at 30°C (15) was used to quantitate QAPRTase activity. For mutant enzymes with severely reduced activities, for which the observable rate was below the detection limit of the spectrophotometric assay, a more sensitive radiolabel transfer assay modified from Cao et al. was used to measure the conversion of QA to NAMN. The system contained [3H]QA, PRPP and MgCl2 in a final volume of 100 µL in buffer QA. The amount of MgCl2 was adjusted to maintain a 5 mM excess of Mg2+ over PRPP concentration. The amount of WT enzyme in the reaction mixtures ranged from 3–5 µg, whereas amounts of mutant enzymes (E214A, E214D, Lys153 and K284A) ranged from 200–600 µg. All reactions were incubated at 30°C for 1–3 h, and 10 µL of the reaction mixture was applied to a 3MM Whatman paper that had been pre-spotted with 3 N HCl, QA and NAMN. Chromatograms were developed in 1 M ammonium acetate (pH 7.5): 95% ethanol (4:6; v:v). The [3H]QA and [3H]NAMN spots, located under 254 nm UV light, were cut out from the chromatogram, eluted in 0.3 mL of 0.1 N HCl, and radioactivity was determined by liquid scintillation. The higher sensitivity of the radiolabel transfer assay allowed the low activities of E214A and E214D to be measured reliably. All kinetic data were fit to the hyperbolic form of the Michaelis-Menten equation (28) using Prism (GraphPad Software, San Diego, CA). Protein concentration was measured spectrophotometrically based on the extinction coefficient of QAPRTase (E280nm (1 mg/mL) = 0.82). Enzyme molarity was calculated on the basis of the dimer molecular weight of 64 856 Da (13).
Equilibrium binding of [14C]PRPP and [3H]QA to WT and mutant QAPRTase forms was measured using the centrifugal dialysis method with Microcon YM-10 apparatus (Amicon) as detailed by Cao et al. (15). Data obtained were fit to a single site binding model using Prism (GraphPad), and binding constants are reported with their standard errors. Scatchard analysis (29) was used to display the binding data.
Chemical Quench Experiments
Rapid quench experiments were carried out at 25°C in buffer QA using RQF-3 Chemical-Quench-Flow apparatus (KinTek Instruments). Reactions were initiated when 15 µl enzyme (50 µM) pre-equilibrated with 500 µM [3H]QA was mixed with 15 µl of PRPP (2 mM) in buffer QA, with reaction times ranging from 3 ms to 10 s. The reactions were quenched with 1.5 M HCl and collected in 1.5 ml eppendorf tubes, which were centrifuged at 3000 × g for 2 min to remove denatured protein. The quenched mixtures (10 µL) were chromatographed on 3MM Whatman paper with a 4:6 mixture of 1M ammonium acetate: 95% ethanol. [3H]QA and [3H]NAMN were measured as described for the radiolabel transfer assay of mutant enzymes. Control experiments in the absence of enzyme in which the reaction was quenched with HCl determined the small amount of 3H in [3H]QA that chromatographed as [3H]NAMN.
Stopped-Flow Fluorescence Measurements
Stopped-flow measurements were performed with KinTek SF-2001 (KinTek Instruments) stopped-flow fluorescence/absorbance apparatus. This apparatus has a thermostatted observation cell that was maintained at 25°C. Fluorescence was excited at 290 nm and detected after passing through a 340 nm cut-off filter. One syringe contained 3 µM QAPRTase pre-incubated with 300 µM QA in buffer QA, and the second syringe contained PRPP at indicated levels. The reaction was initiated by rapidly mixing equal volumes of PRPP with pre-incubated QAPRTase•QA binary complex. The time dependent decrease of intrinsic tryptophan fluorescence (S. typhimurium QAPRTase contains 3 tryptophan residues per subunit) was measured at various PRPP concentrations over 0.15–3 s, with 1000 points collected in each trace. Typically, 5–8 traces were averaged for each set of concentrations. The averaged time dependent decrease in fluorescence was fit to a single exponential decay function (eq I) using the KinTek analysis program to obtain the observed rate constant kobs.
| (I) |
In eq I, F = fluorescence observed, at time, t, Fo = amplitude of fluorescence change, kobs = observed rate constant and C = fluorescence intensity at t = 0. The values of kobs were plotted against the various PRPP concentrations and fitted to eq II (30) using Prism.
| (II) |
Data analysis by eq II allowed the calculation of the binding constant of PRPP using eq III.
| (III) |
pH-rate Profiles
Values of kcat were determined for NAMN formation by WT and mutant enzymes (E214A, E214D and E214Q) over the pH range of 6.0 – 8.5 using saturating concentrations (10 times Km) of QA and PRPP. The buffer system contained 50 mM Bis-Tris, 50 mM Tris, 50 mM KH2PO4 and 6 mM MgCl2, with pH adjusted with 1 M KOH or 1 M HCl as needed. Control reactions performed at 20 mM and 50 mM buffer concentrations at pH 7.2, ensured that the buffer system was neither inhibiting nor accelerating the QAPRTase reaction. The spectrophotometric assay was used to determine kcat for WT and E214Q, while the radiolabel transfer assay was used to determine kcat for E214A and E214D. The kcat values were normalized and plotted as log kcat against pH. Data were fitted with the program HABELL (28) using eq III, in which kcat is the value of kcat at a particular pH, C is the pH-independent value of the parameter, [H] represents the hydrogen ion concentration and Ka is the acid dissociation constant for a functional group on QAPRTase required in a given deprotonated state.
| (IV) |
Results
Amino Acid Sequence Analysis
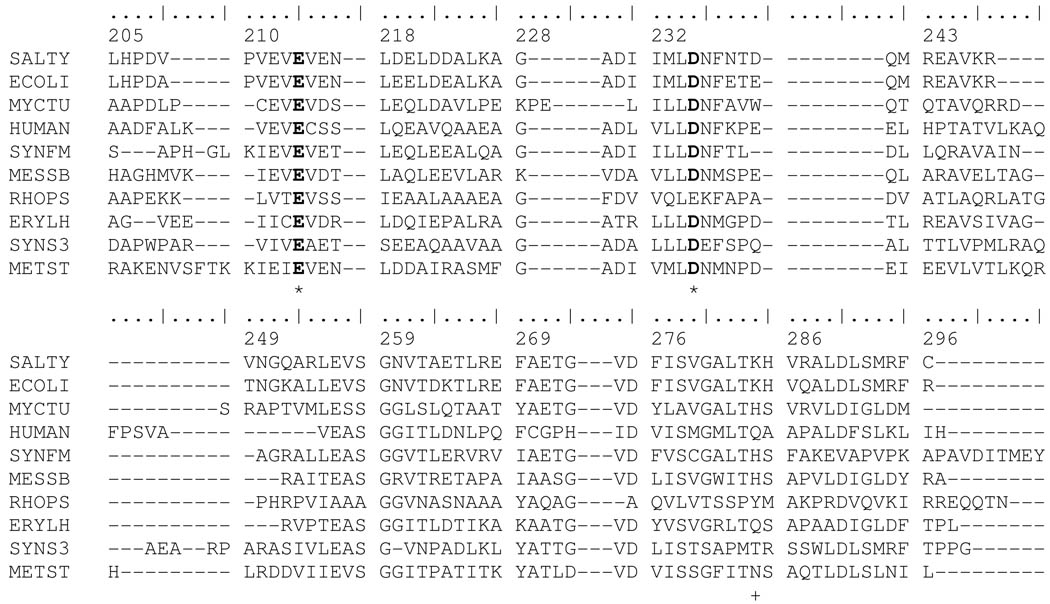
The degree of amino acid sequence variation among the 230 available distinct QAPRTases was determined by multiple sequence alignment. The sequence database was comprehensive as of April 2007. Figure 2 shows a portion of the amino acid alignment, revealing that Lys153 and Glu214 are absolutely conserved. Asp235 is 97% conserved, being conservatively replaced by glutamic acid in the distantly related gram negative Rhodopseudomonas palustris, Rhodobacter sphaeroides, Magnetospirillum magneticum and the archaebacteria Pyrobaculum islandicum. The high conservation of the residues together with their active site locations suggests an important role in PRPP interaction. In contrast, a modest conservation was observed for Lys284 (27%) with substitutions by His (53%), Gln (7%), Thr (6%), Tyr, Asn, Met, Phe, Arg and Leu.
Figure 2.
Partial sequence alignment of QAPRTase from various organisms. 10 of the 230 sequences aligned as described in Methods are displayed. Sequences are: SALTY, Salmonella typhimurium LT2; ECOLI, Escherichia coli K-12; MYCTU, Mycobacterium tuberculosis H37Rv; HUMAN, Homo sapiens; SYNFM, Syntrophobacter fumaroxidans; MESSB, Mesorhizobium sp. BNC1; RHOPS, Rhodopseudomonas palustris BisB5; ERYLH, Erythrobacter litoralis; SYNS3, Synechococcus sp. CC9311; METST, Methanosphaera stadtmanae. Residue numbering at top is from the mature S. typhimurium protein sequence. Highly conserved Glu214 and Asp235 residues are indicated in bold and marked with asterisks. Moderately conserved Lys284 is marked with a plus sign. A full display and sequence sources is found in Supporting Information.
Purification and Aggregation State of Mutant QAPRTases
Glu214 was individually replaced with Ala, Asp and Gln, whereas Ala was substituted for Asp235, Lys153 and Lys284. In order to avoid contamination by the presence of WT QAPRTase from the E. coli BL21 host strain during over-expression of mutant QAPRTase, a nadC deleted host strain, ZB100, was successfully created for this study by replacing its nadC with a spectinomycin resistance gene using the recombineering method of Yu et al., as described in Materials and Methods. The viable spectinomycin-resistant recombinants allowed for the over-expression of the pure S. typhimurium QAPRTase and the mutant forms. The mutant enzymes were purified to homogeneity in 250 mg yield from a 6L culture, as determined by SDS-PAGE analysis. Purification of E214A benefited from the use of a larger column as described in Materials and Methods. The similarity in expression and purification properties of WT with the other mutant enzymes indicated that there were no global structural changes due to the mutations.
Size exclusion chromatography used to investigate the oligomerization state of the mutants showed that they eluted as single symmetrical peaks with retention times and peak-widths at half height similar to the WT enzyme (data not shown). The peaks eluted at volumes corresponding to apparent molecular masses of 63 – 65 kDa, close to the calculated dimeric molecular mass of 65 kDa for WT. The elution profiles for WT and mutant enzymes were similar at sample concentrations of 5 µM or 50 µM, confirming that the mutations did not affect the global structure or aggregation state of the enzymes.
Steady-State Kinetic Studies
Table 1 shows the steady-state kinetic parameters of the WT and mutant enzymes. K153A mutant enzyme had no detectable activity, indicating that the amino acid change is not tolerated by the enzyme. The kcat value for this mutant must have decreased by more than 4000-fold, the detectable level of activity measured for other extremely sluggish mutant enzymes in this study. The steady-state kinetic parameters were dramatically impacted by mutation at Glu214. The catalytic efficiency (kcat/Km) decreased by 19500-fold for E214A with regards to QA. This reduction was distributed between a 4000-fold decrease in kcat, QA and a 5-fold increase in Km,QA. The 4300-fold decrease in kcat,PRPP and 11-fold increase in Km,PRPP observed for E214A resulted in a 45000-fold reduction in kcat/Km,PRPP. Likewise, approximately 4000-fold decreases in kcat,QA and kcat,PRPP were observed for the E214D mutant. Its Km,PRPP increased by 10-fold and Km,QA increased by 5-fold, producing a 19500-fold decrease in kcat/Km,QA and a 37000-fold reduction in kcat/Km,PRPP. The severe effects on kcat in the E214A and E214D mutants indicated an essential role for Glu214 in catalysis. In contrast, replacement of Glu214 by glutamine only modestly affected the steady-state parameters. The Km values of E214Q for QA and PRPP increased by only 2-fold, while kcat/Km for QA and PRPP decreased by 3-fold and 5-fold respectively.
Table 1.
Kinetic parameters and Equilibrium Binding Constants for Wild-type and Mutant QAPRTases
| kinetic constant |
wild-type | E214A | E214D | E214Q | D235A | K284A |
|---|---|---|---|---|---|---|
| kcat, (s−1) | 1.0 ± 0.1 | 2.4 × 10−4 | 2.5 × 10−4 | 0.5 ± 0.03 | 1.6 ± 0.1 | 0.033 ± 0.007 |
| KM, QA (µM) | 27 ± 6 | 121 ± 47 | 120 ± 20 | 41 ± 8 | 31 ± 5 | 1463 ± 182 |
|
kcat/KM, QA (µM−1 s−1) |
3.7 × 10−2 | 1.9 × 10−6 | 1.9 × 10−6 | 1.2 × 10−2 | 5.2 × 10−2 | 2.3 × 10−5 |
|
KM, PRPP (µM) |
57 ± 7 | 604 ± 94 | 550 ± 117 | 128 ± 25 | 138 ± 12 | 4120 ± 1000 |
|
kcat/KM, PRPP (µM−1 s−1) |
1.8 × 10−2 | 4.0 × 10−7 | 4.9 × 10−7 | 3.9 × 10−3 | 1.2 × 10−2 | 7.0 × 10−6 |
| KD, QA (µM) | 25 ± 7 n = 2.0 ± 0.1 |
45 ± 7 n = 2.1 ± 0.2 |
20 ± 4 n = 1.9 ± 0.1 |
50 ± 10 n = 2.0 ± 0.2 |
25 ± 5 n = 2.2 ± 0.3 |
25 ± 10 n = 2.0 ± 0.2 |
| KD,PRPP (µM) | 53 ± 11 n = 2.0 ± 0.1 |
770 ± 200 n = 2.1 ± 0.3 |
417 ± 76 n = 2.0 ± 0.1 |
113 ± 24 n = 2.2 ± 0.4 |
127 ± 25 n = 1.7 ± 0.3 |
~ 1000 + |
|
KD,PRPP (with PA) (µM) |
22 ± 3 n = 2.1 ± 0.1 |
219 ± 33 n = 2.3 ± 0.2 |
124 ± 16 n = 2.5 ± 0.2 |
68 ± 8 n = 1.5 ± 0.1 |
ND | ND |
| K1 (µM−1) * stopped-flow |
0.002 | 0.0017 | ||||
|
k2 (s−1) stopped-flow |
10.3 | 10.6 | ||||
|
k−2 (s−1) stopped-flow |
1.1 | 0.78 |
ND, not detected
single point measurement, see Results
Surprisingly, substitution of alanine for the highly conserved Asp235 had virtually no effect on the steady-state kinetic parameters of the mutant enzyme. The kcat value for D235A was increased slightly, Km for QA was approximately equal to that of WT, and that for PRPP increased by about 2-fold, resulting in kcat/Km values for both substrates that were similar to WT.
Mutation of the modestly conserved Lys284 to alanine lowered kcat by 30-fold. The values of Km,PRPP and Km,QA for K284A increased by 80-fold and 50-fold, respectively, producing a 2570-fold decrease in kcat/Km,PRPP and 1600-fold decrease in kcat/Km,QA, suggesting a role for Lys284 in catalysis.
Substrate Binding by Mutant Enzymes
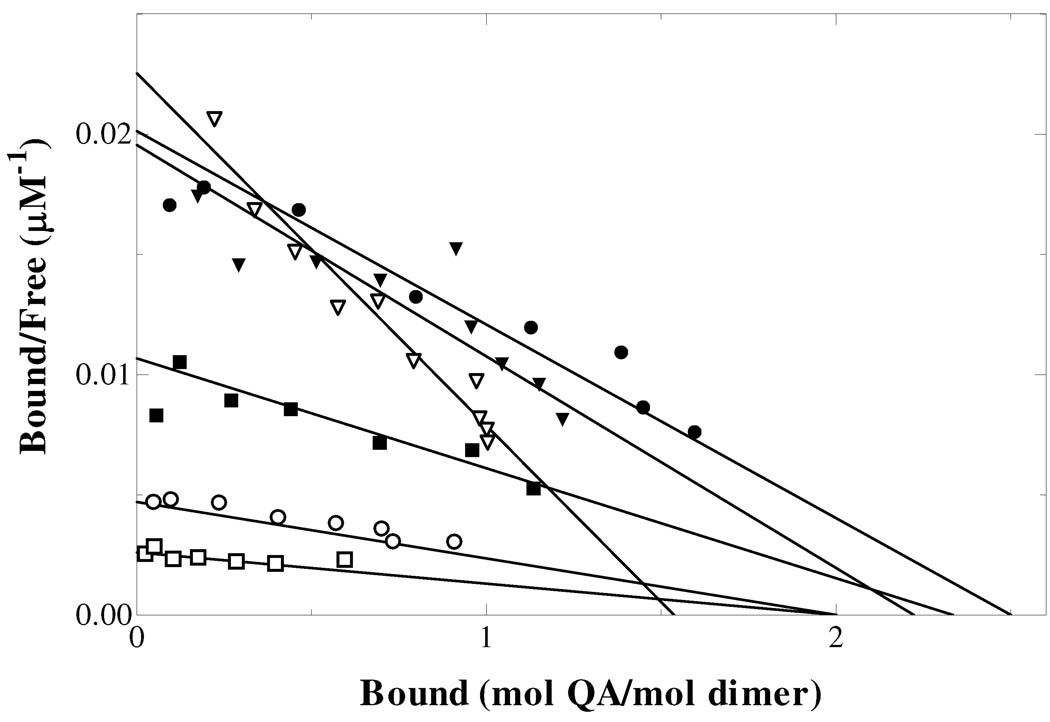
The ordered mechanism of QAPRTase allows for the formation of a nonproductive QAPRTase•PRPP binary complex as well as the QAPRTase•QA complex. To determine the effect of the mutations on binding to [3H]QA and [14C]PRPP, a centrifugal filtration technique was used and results were compared to those of WT previously determined by Cao et al. (15). Scatchard plot analysis of [3H]QA binding in binary complex with the mutant enzymes (Figure 3, Table 1) showed modest effects on QA binding. Despite the enzymatic inactivity observed for K153A, the mutant was able to bind QA with a KD value only 2-fold higher than WT. Compared to WT, KD,QA for E214A and E214Q increased by ~2-fold, and those for E214D, D235A and K284A were similar to WT.
Figure 3.
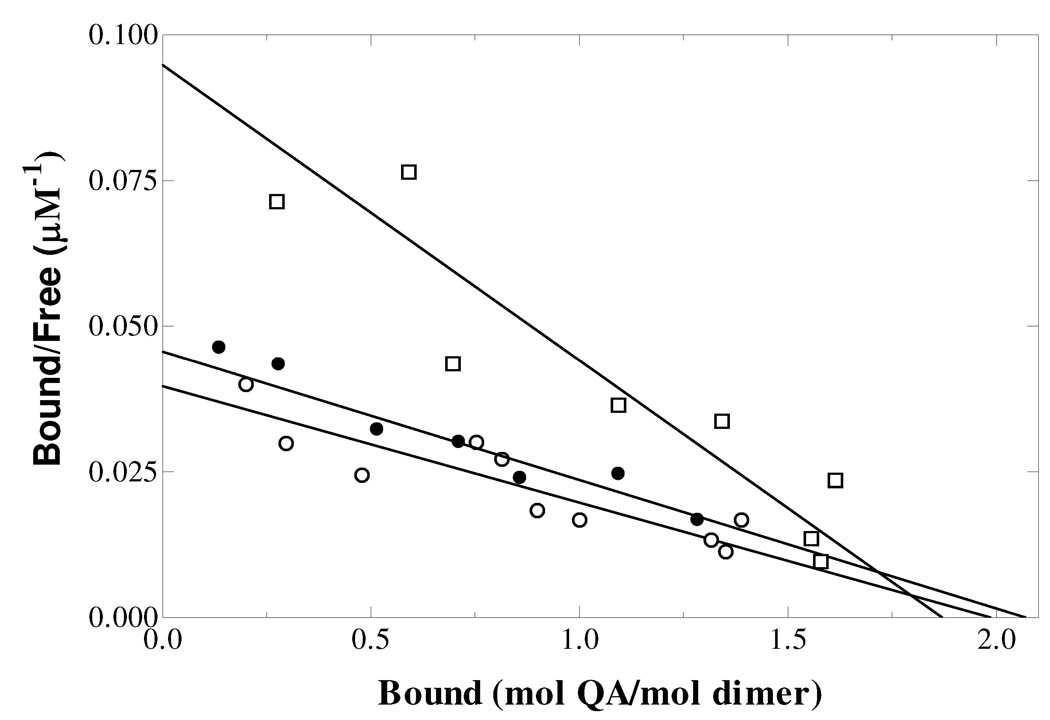
Scatchard plot for the binding of [3H]QA to mutant enzymes. E214A (●), E214D (□) and E214Q (○). The values of KD were 45 µM, 20 µM and 50 µM respectively.
Binding of K153A to PRPP was not detected by the centrifugal filtration technique. Based on our own observed limits for measurable binding by this technique, KD,PRPP for K153A was estimated to be greater than 1mM. Further kinetic analysis was not performed for K153A. Consistent with the modest changes in KM,PRPP, the KD,PRPP for E214A, E214D and E214Q increased by 15-fold, 10-fold and 2-fold respectively. The binding constant of PRPP to D235A also increased by 2-fold. Due to the low binding affinity of PRPP for K284A, a full analysis was not performed. A single point measurement for K284A binding to PRPP was consistent with a KD,PRPP of ~ 1 mM, about 20-fold higher than WT.
Previously, Cao et al. observed that WT enzyme binds PRPP 2.4 times tighter in the ternary complexes with the unproductive QA analog PA (15). The orientation of residues in the active site of the apoenzyme is different from that of QAPRTase•QA complex, suggesting that the tightening represents the structural rearrangement of active site residues induced by the binding of QAPRTase to QA (2, 8). Similarly, [14C]PRPP binding tightened in the presence of PA in E214A, E214D and E214Q as shown in Figure 4. The presence of PA resulted in 6-fold tightening of PRPP binding to E214D, with 3.4-fold and 2-fold tightening effects displayed by E214A and E214Q respectively. The result indicates that the rearrangement of the active site residues caused by QA binding can still occur in the three mutants of E214.
Figure 4.
Scatchard plot for the binding of [14C]PRPP to mutant apo-enzymes and QAPRTase•PA binary complex. [14C]PRPP binding by E214A (□), E214D (○), E214Q (▽) and to the E214A•PA (■), E214D•PA (●), and E214Q•PA (▼) complexes. The KD values are reported in Table 1.
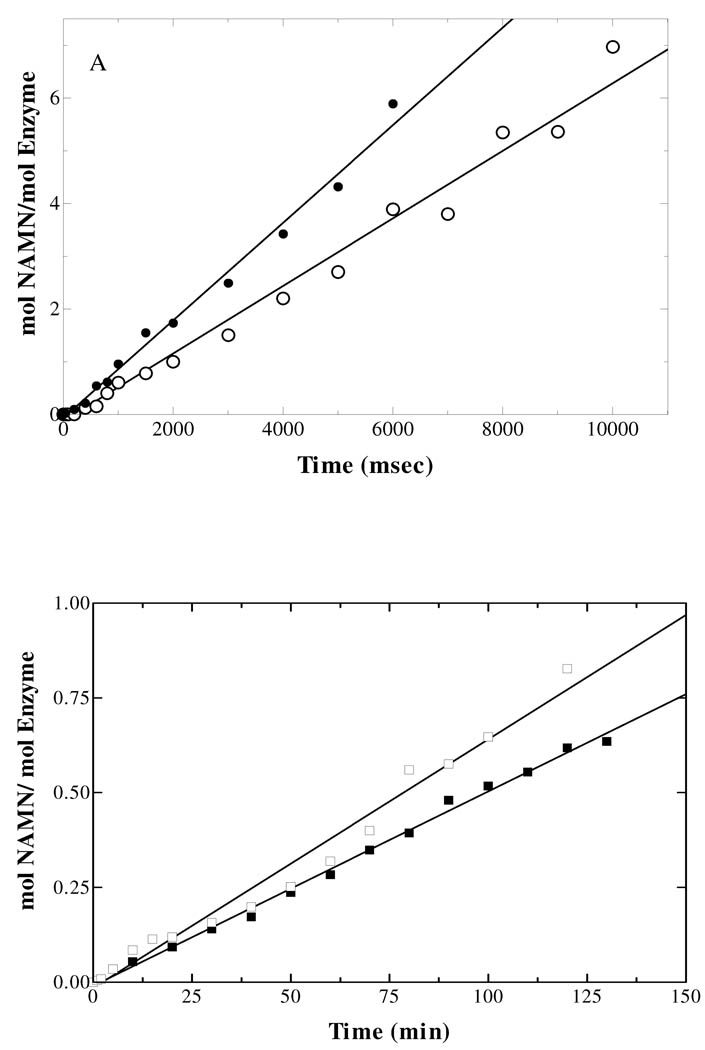
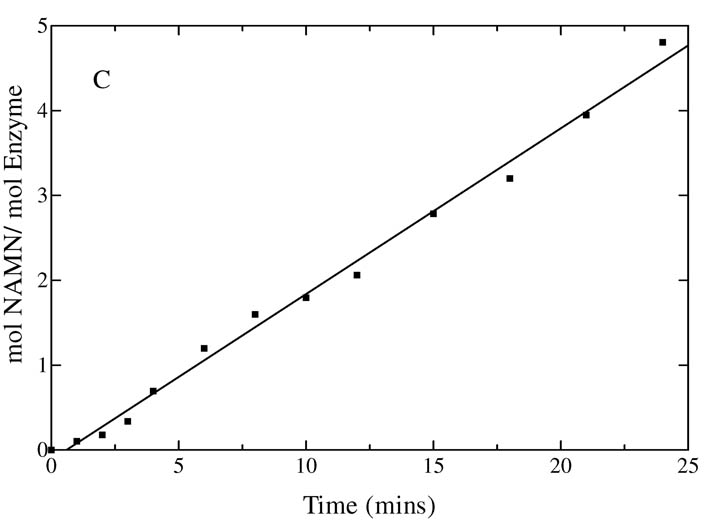
Pre-steady State
The lack of a burst of NAMN formation in the pre-steady state observed for QAPRTase indicated that the chemistry of the reaction or a step preceding it is rate limiting (15). As observed by Cao et al. (15), chemical quench experiments used to measure the rate of the on-enzyme phosphoribosyl group transfer for WT enzyme at 23°C showed a linear rate of NAMN production with a kcat value of 0.6 s−1 (Figure 5A), a value comparable to the kcat value under steady-state at 30°C. Like the WT enzyme, E214Q also lacks a pre-steady state burst (Figure 5A) with an initial rate of NAMN formation of 0.5 s−1, consistent with the steady-state value and similar to the behavior of WT. The pre-steady states of NAMN formation by E214A and E214D were slow enough to be measured by the manually quenched radiolabel transfer assay as their rates were approximately 0.0125 min−1. The initial rate of NAMN formation was linear and no burst was observed (Figure 5B). The pre-steady state kcat for E214A and E214D was 1.6 × 10−5 s−1, a 4000-fold decrease compared to WT. Both values are in close agreement with the kcat values recorded under steady-state conditions. For K284A as well, the initial rate of NAMN formation was linear with a rate of 0.03 s−1 in close agreement with the steady-state rate, and no burst phase was observed (Figure 5C). The absence of a burst of product formation in all the mutants implies that the step accounting for the slow overall rate of NAMN formation occurred after the formation of QAPRTase•QA•PRPP ternary complex or during the phosphoribosyl transfer step.
Figure 5.
Pre-steady state kinetics of [3H]NAMN formation by WT and mutant enzymes. Rapid mixing was performed as described in Methods. (A) The kcat for WT(●) reaction was 0.6 s−1 and that for E214Q (○) was 0.5 s−1. (B) The kcat values for E214A(■), E214D (□) were 1.6×10−5 s−1. (C) kcat for K284A was 0.03 s−1.
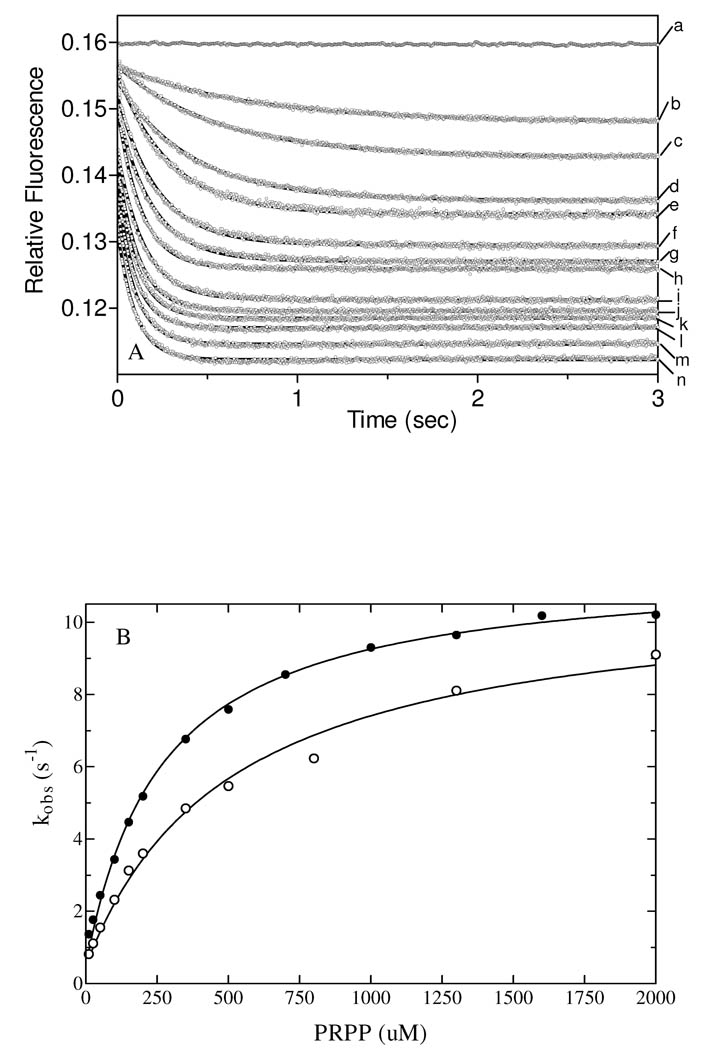
Kinetics of PRPP binding
In the ordered sequential mechanism of QAPRTase, the QAPRTase•QA•PRPP ternary complex must form and reach its correct conformation before chemistry takes place. The fluorescence of the 3 tryptophan residues of QAPRTase was used to follow the potential isomerization of the productive ternary complex in the stopped-flow apparatus. When varied concentrations of PRPP were mixed with a fixed amount of enzyme pre-incubated with saturating QA, a 20% decrease in fluorescence was observed. Fluorescence traces with WT and E214Q fitted well to single exponentials (eq I) at all the PRPP concentrations investigated. The fluorescence trace for PRPP binding to WT•QA complex is shown in Figure 6A. The observed rate constants (kobs) increased hyperbolically with increasing PRPP concentration for WT and E214Q (Figure 6B). Values of kobs were considerably faster than the rate limiting chemistry step kcat. This faster rate indicates a mechanistic step that represents a process leading to the productive Michaelis complex. The hyperbolic dependence of kobs on PRPP concentration for WT and E214Q fitted well to eq II, a model accounting for a two-step binding process (30) that can be described as illustrated in the first two steps of Scheme 2, where E* represents the ternary complex with altered fluorescence. The PRPP binding process includes an equilibrium association/dissociation process (K1) followed by a slower conformational change (k2 and k−2). The best-fit K1 value for WT was 0.002 µM −1and k2 and k−2 were 10 s−1 and 1.1 s −1, respectively. The 53 µM KD value for PRPP binding to WT calculated from the stopped-flow data analysis using eq III is identical to that measured directly (Table 1), consistent with the two-step mechanism of Scheme 2.
Figure 6.
Stopped-flow studies with WT and E214Q QAPRTases. (A) Reaction traces obtained from Stopped-flow fluorescence analysis. Time dependence decay of intrinsic WT QAPRTase fluorescence upon mixing the QAPRTase•QA complex (6 µM QAPRTase incubated with 300 µM QA) with various concentrations of PRPP at pH 7.2, 25 °C (λex = 290 nm, λem = 340 nm). Final PRPP concentrations were (a) 0, (b) 10, (c) 25, (d) 50, (e) 100, (f) 150, (g) 200, (h) 500, (i) 350, (j) 700, (k) 1000, (l) 1300, (m) 1600 and (n) 2000 µM. The traces were fit to a single exponential decay function (eq I). (B) Dependence of the observed rate constants of data from Figure A on PRPP concentration for WT (●) and E214Q (○).
Scheme 2.
Kinetics of PRPP binding
A similar stopped-flow analysis was also performed for binding of WT to NAMN, in order to investigate any conformational change associated with NAMN binding. Various concentrations of NAMN were mixed with fixed amount of WT enzyme pre-incubated with saturating PPi. A 25% decrease in fluorescence was observed and was fitted to the single exponential equation. In contrast to binding of WT to PRPP, the kobs were linearly dependent on the concentration of NAMN (data not shown). This behavior suggests a one-step binding rapid equilibrium process (30) of NAMN to WT enzyme, which includes the process that alters fluorescence. The kinetics of NAMN binding were not further investigated here.
As followed by fluorescence, PRPP binding kinetics of E214Q were indistinguishable from WT, with a K1 value of approximately 0.002 µM −1and k2 and k−2 of 10 s−1 and 1 s −1. The 43 µM KD value of PRPP for E214Q calculated from the data analysis is similar to the value measured directly (Table 1). When stopped-flow studies were performed with E214A and E214D to determine whether their ability to induce a pre-catalytic conformation in the ternary complex had been compromised, no significant change in fluorescence was observed for either mutant over a 60 min period. Incubation of the reaction mixture for more than 30 mins revealed a very slow and apparently zero-order decrease in fluorescence. However, this long-term change may have arisen from the slow precipitation of magnesium phosphate that resulted from the presence of excess MgCl2 and PRPP in the reaction mixture. The lack of a first-order exponential fluorescence decay in the mutant enzymes might indicate that isomerization to form the productive Michealis complex (Scheme 2, step 2) had been jeopardized. It was not possible to reliably measure the kinetics of PRPP binding to K284A because of the precipitation of magnesium phosphate resulting from the high PRPP and Mg2+ concentrations required.
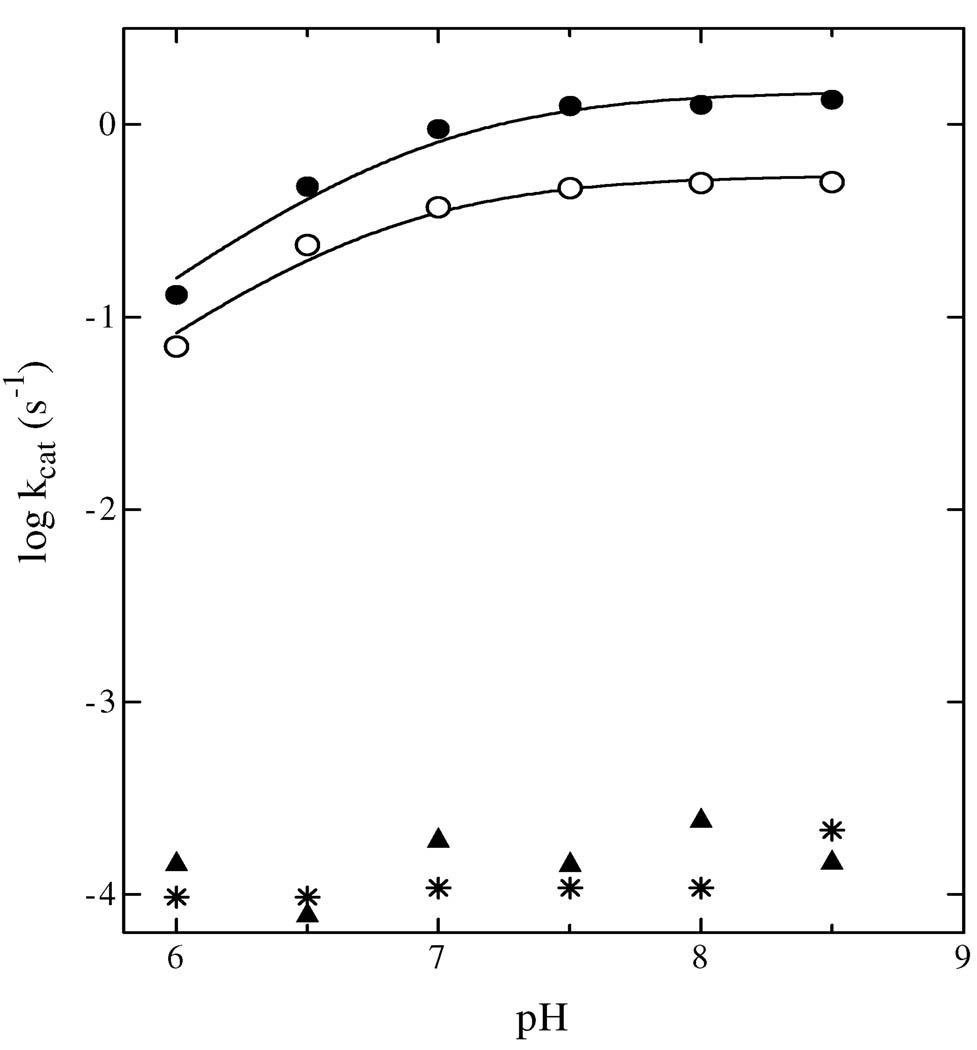
Effects of pH on kcat of WT and Glu214 Mutants
To determine whether the strictly conserved Glu214 was required in its deprotonated form to abstract a proton from the 2-hydroxyl group of PRPP, the pH dependence of kcat for WT was determined by measuring kcat within the pH range of 6.0 – 8.5, at saturating concentrations of substrates. A plot of log kcat versus pH for NAMN formation catalyzed by WT revealed that a residue with pKa = 6.92 ± 0.08 (Figure 7) was required in its deprotonated form for the QAPRTase reaction. The pH-rate profiles for E214A and E214D mutants remained flat over the pH range examined (Figure 7). The pH-rate profile for E214Q showed a similar requirement like WT for a residue with pKa = 6.75 ± 0.08 in its deprotonated form. Since the amide pKa for Gln is about 15 (31), these results indicate that ionization of the carboxylate group of Glu214 is not responsible for the pH dependence on catalysis.
Figure 7.
pH Dependence of log kcat with WT (●), E214Q (○), E214A (*) and E214D (▲). Lines represent fits of the data to eq IV with pKa = 6.92 (WT) and 6.75 (E214Q).
Discussion
This mutagenesis study of QAPRTase indicated that Lys153, Glu214 and Lys284, all residues interacting with PRPP, are important for the QAPRTase reaction. The absolutely conserved Lys153 is essential, and, with the moderately conserved Lys284, they both contribute to catalysis through their roles in PRPP binding. In contrast to the hypothesis that Glu214 functions as a catalytic base or provides electrostatic stabilization for the reaction, it appears more simply to provide a hydrogen bond necessary for geometric catalysis and contributes to PRPP binding. The highly conserved Asp235 is surprisingly not essential.
The interaction of the positively charged Lys153 with the phosphoryl group of PRPP suggests that the residue might participate in protonation of the pyrophosphate leaving group during catalysis. The inability of K153A to bind PRPP suggests a role for Lys153 in PRPP binding. However, confirming the potential role of Lys153 in protonation of the pyrophosphate leaving group from an inactive enzyme was not possible. The bulk of this discussion focuses on the Glu214 mutants.
Glu214 was examined on the basis of its absolute conservation among QAPRTases, its observed location in forming an apparent hydrogen bond to oxygen at position 2 of PRPP, and its proposed role in stabilizing the putative oxycarbonium-like transition state. The three proposed modes of catalysis are (i) stabilization of the transition state by electrostatic effect provided by Glu214, (ii) geometric stabilization of the ribosyl group to facilitate conformations required for catalysis, and (iii) deprotonation of the 2-hydroxyl group of PRPP to increase the electron density at C1 of PRPP. The existence of geometrically analogous carboxylate residues among other PRTases makes this examination additionally attractive. The greatly diminished catalytic rate of E214A demonstrated the essential role of Glu214 in catalysis. Substitution of Glu214 by Asp might conserve the proposed electrostatic and deprotonation means of catalysis. Remarkably, E214D experienced a drastic decrease in catalytic rate similar to that of E214A mutant, suggesting that the exact geometry of Glu214 is critical.
In dissecting the role of Glu214, it was first noted that the poor activity of the mutants was not due to gross changes in the overall structure of the enzymes. Importantly, the lack of pre-steady state burst for NAMN formation by E214A and E214D indicated that an event after binding of PRPP or during the chemistry step had been disrupted. We showed that the ability of E214A and E214D mutants to tighten PRPP binding to the non-catalytic QAPRTase•PA binary complex was unaffected, even though binding affinity of PRPP for the mutant apoenzymes was reduced by 10-fold. This tightening corresponds to the structural rearrangement of active site residues noted in the binary QAPRTase•QA and the catalytically unproductive QAPRTase•PA•PRPCP complexes (2, 8).
Stopped-flow experiments used to characterize the binding of WT QAPRTase•QA binary complex to PRPP displayed a hyperbolic dependence on PRPP, consistent with a two-step binding process as illustrated in Scheme 2. The rate of the conformational change k2, for the rate constant of forward QAPRTase catalysis is 10 s−1. However, both steady-state and pre-steady state kcat values (1 s−1 and 0.6 s−1) are at least 10-fold smaller than kobs from the stopped-flow analysis. Therefore, the previously determined kinetic mechanism by Cao et. al (15) is expanded to include the additional conformational step (Scheme 2) that occurred after initial binding of PRPP to the QAPRTase•QA binary complex. This two-step mechanism is consistent with the comparable values of KD,PRPP derived from the stopped-flow experiments to those measured by centrifugal dialysis method. The additional conformational change observed must lead to the catalytically productive Michealis complex, and is distinct from the tightening of PRPP binding, or the structural rearrangement observed in the non-catalytic QAPRTase•PA•PRPCP complex. Because fluorescence kinetic transients were not observed with E214A and E214D in the stopped-flow experiment, it was concluded that the conformational change leading to a productive Michealis complex has been compromised in the mutants.
It is tempting to speculate on the relationship between the structures of complexes described earlier (2, 8), and the PRPP binding and fluorescence changes reported here. In the functional data, three different states seem to be represented: an initial state that occurs in the QAPRTase•QA complex, a more tightly bound state reported by the 2.4-fold tightening of binding in the ternary complex and a third state reported here by the decrease in tryptophan fluorescence that occurs subsequent to formation of the ternary complex. Sharma et al. observed structural changes occurring on formation of the QAPRTase•QA complex from the apoenzyme, but no further changes when PRPP was present (2). Additionally no direct hydrogen bonding between the PA and PRPCP molecules were observed in the ternary complex. This finding suggests that the previously reported ternary structure is not that of the final E*•QA•PRPP complex (Scheme 2), but may represent the tightly bound E•QA•PRPP complex, whose further movement toward productive catalysis is disfavored by the substitution of the carbocyclic PA as opposed to the native QA substrate. This finding is supported by the fact that Glu214 is seen to form structurally favorable interactions with the 2-hydroxyl of PRPP in the ternary structure. The measured equilibrium between the two fluorescence states is only 10:1, in favor of the less fluorescent, but catalytically poised state. Sharma et al. point out that in the QAPRTase•NAMN complex, whose structure is apoenzyme-like, there has been a 2.6 Å translation of C12, which includes a lengthening (to 4.2 Å) of the hydrogen bond from 2-hydroxyl of Glu214. One could speculate that as the ribose moiety travels the 2.6 Å required, the Glu214 interaction initially becomes better as the complex reaches a pre-catalytic state that might resemble the type of “Near Attack Conformer” described by Bruice et al. (32). It is this conformation that facilitates the formation of a chemical transition state.
The mutation of Glu214 to either Ala or Asp affected both PRPP binding and a pre-catalytic conformational change, thus favoring the hypothesis that Glu214 might stabilize the transition state geometrically. The nearly full activity observed with the electrically neutral carboxamide of E214Q destroys all support for any purely electrostatic role for Glu214. The pH-profile of WT indicated that a general base with a pKa of 6.9 is involved in the catalytic mechanism. The retention of the residue required in its deprotonated form by the E214Q mutant confirmed that Glu214 does not function to abstract a proton from the 2-hydroxyl group of PRPP. The carboxamide functional group of Gln in E214Q can hydrogen bond with PRPP and with the water molecule involved in coordinating the dihydrated Mg2+. The stopped-flow experiments, steady and pre-steady state kinetic parameters of E214Q were identical to WT, indicating that the hydrogen bond ability rather than the general base properties of Glu214 is essential for the QAPRTase reaction.
The low residual activity measured for the E214A and E214D mutants raised questions regarding whether the activity observed was due to artifacts of the assay procedure or contaminants. We observed that the reaction catalyzed by the mutants showed a direct dependence on enzyme concentration, gave the correct product on chromatography, and was inhibited by PA, a specific inhibitor of QAPRTase. Additionally, to avoid possible contamination by E. coli host QAPRTase, mutant enzymes were over-expressed in the nadC deletant E.coli strain ZB100, which lacks QAPRTase. It is conceivable that the low activity observed with E214A and E214D results from WT QAPRTase that is present because of natural errors of misincorporation giving back mistranslated protein molecules with the WT glutamate in position 214. The average error frequency of translation (for any particular amino acid at any particular position) has been estimated as 4 × 10−4 for normally growing cells (33), but depends on factors such as type of codon used to specify the substituted amino acid, strain and type of microorganism (34, 35). The remaining activity with these mutants (1 part in 4 × 103 compared to WT) would correspond to a mistranslation rate of 2.5 × 10−4, which is close to these published values. We also took great care to reduce the possibility that any enzyme from WT purifications did not contaminate subsequent mutant preparations performed on the same equipment. Nevertheless the minute amount of contamination required to produce the observed activity is impressive. However, the greatly reduced kcat values for the mutants were accompanied by reproducibly altered KM values for the substrates. A contamination by an active enzyme would give a constant value for KM that is equal to WT regardless of the amount of the contaminant (36). The observed KM values roughly corresponded to KD values, which are themselves assignable only to the bulk of the enzyme preparation, demonstrably the mutant form. In addition, the undetectable activity of K153A indicates the absence of contamination. We thus conclude that although mistranslation or contamination may account for some fraction of enzyme activity in E214A or E214D, it is highly unlikely that this accounts for the overall observed behavior.
Astonishingly, Asp235 is dispensable for catalysis despite its high conservation and its interaction with PRPP in the crystal structures of the complexes. Substitution of Asp235 by Ala did not perturb catalysis or substrate binding. Contribution of Asp235 in catalysis might have been masked by other interactions with residues in the active site that might have rearranged as a consequence of the removal of Asp235.
Surprisingly, despite the similarity in structural fold among the type II PRTases, interactions of residues with the ribosyl hydroxyl groups of PRPP are not highly conserved. Structure-based sequence comparison of M. tuberculosis QAPRTase and Thermoplasma acidophilum NAPRTase revealed that Asp237 of NAPRTase, seen to interact with the 3-hydroxyl group of PRPP, is equivalent to Asp235 in QAPRTase (37, 38). On the other hand, Glu214 of QAPRTase is not conserved in NAPRTase, and is replaced by Leu213 (38). Instead, a highly conserved histidine residue, shown to be the site of phosphorylation that increases the catalytic efficiency of NAPRTase, was revealed to be structurally analogous to Glu214 (9). Chappie et al. suggested assistance of the phosphohistidine residue in catalysis by NAPRTase by electrostatic stabilization of the positively charged oxycarbonium-ion-like transition state (9). It is likely that this residue makes important hydrogen bond interactions necessary for catalysis as determined for Glu214 in QAPRTase.
Comparison of the M. tuberculosis QAPRTase and human NMPRTase showed that Glu214 and Asp235 are replaced by Val277 and Asp313 respectively, in NMPRTase (38). But, unlike Asp235 of QAPRTase which forms a hydrogen bond with 3-hydroxyl of the ribose group, Asp313 of NMPRTase is seen to interact with the 2-hydroxyl group of the ribose moiety of NMN like Glu214 of QAPRTase. This interaction might also function to stabilize the transition state in NMPRTase.
The reduced binding of PRPP to K284A suggests a role for Lys284 in the ground state binding of PRPP. Asn260 and Gly280, which also interact with the 5-phosphate of PRPP, might share a role in retaining the position of the 5-phosphate on the enzyme during catalysis. As confirmed from this work, Glu214 provides a hydrogen bond that leads to the formation of a productive Michealis complex necessary for catalysis, while Lys153 and Lys284 are required for PRPP binding.
Supplementary Material
ACKNOWLEDGMENTS
We thank Giao Le for insightful discussion.
Abbreviations
- QA
quinolinic acid
- NAD
nicotinamide adenine dinucleotide
- PRPP
5-phosphoribosyl-1-pyrophosphate
- NAMN
nicotinate mononucleotide
- PPi
pyrophosphate
- NAPRTase
nicotinate phosphoribosyltransferase
- NMPRTase
nicotinamide phosphoribosyltransferase
- QAMN
quinolinate mononucleotide
- PRPCP
5-phosphoribosyl-1-(β-methylene)pyrophosphate
- PA
phthalic acid.
Footnotes
Supported by NIH grant GM48623 to CG.
SUPPORTING INFORMATION
Multiple sequence alignment of the QAPRTase residues from 230 organisms (Figure S1) is available online at http://pubs.acs.org.
REFERENCES
- 1.Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, Begley TP. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 2003;10:1195–1204. doi: 10.1016/j.chembiol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Sharma V, Grubmeyer C, Sacchettini JC. Crystal structure of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis: a potential TB drug target. Structure. 1998;6:1587–1599. doi: 10.1016/s0969-2126(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 3.Boshoff HI, Xu X, Tahlan K, Dowd CS, Pethe K, Camacho LR, Park T, Yun C, Schnappinger D, Ehrt S, Williams KJ, Barry CE., III Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J. Biol. Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarcz R, Whetsell WO, Jr, Mangano RM. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science. 1983;219:316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- 5.Musick WDL. Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. CRC Crit. Rev. Biochem. 1981;11:1–34. doi: 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- 6.Schramm VL, Grubmeyer C. Phosphoribosyltransferase mechanisms and roles in nucleic acid metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:261–304. doi: 10.1016/S0079-6603(04)78007-1. [DOI] [PubMed] [Google Scholar]
- 7.Sinha SC, Smith JL. The PRT protein family. Curr. Opin. Struct. Biol. 2001;11:733–739. doi: 10.1016/s0959-440x(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 8.Eads JC, Ozturk D, Wexler TB, Grubmeyer C, Sacchettini JC. A new function for a common fold: The crystal structure of quinolinic acid phosphoribosyltransferase. Structure. 1997;5:47–58. doi: 10.1016/s0969-2126(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 9.Chappie JS, Canaves JM, Han GW, Rife CL, Xu CP, Stevens RC. The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure. 2005;13:1385–1396. doi: 10.1016/j.str.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Zhang X, Bheda P, Revollo JR, Imai SI, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme. Nat. Struct. Mol. Biol. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 11.Kim C, Xuong NH, Edwards S, Madhusudan, Yee MC, Spraggon G, Mills SE. The crystal structure of anthranilate phosphoribosyltransferase from the enterobacterium Pectobacterium carotovorum. FEBS Lett. 2002;523:239–246. doi: 10.1016/s0014-5793(02)02905-8. [DOI] [PubMed] [Google Scholar]
- 12.Champagne KS, Piscitelli E, Francklyn CS. Substrate recognition by the hetero-octameric ATP phosphoribosyltransferase from Lactococcus lactis. Biochemistry. 2006;45:14933–14943. doi: 10.1021/bi061802v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes KT, Dessen A, Gray JP, Grubmeyer C. The Salmonella typhimurium nadC gene: sequence determination by use of Mud-P22 and purification of quinolinate phosphoribosyltransferase. J. Bacteriol. 1993;175:479–486. doi: 10.1128/jb.175.2.479-486.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.di Luccio E, Wilson DK. Comprehensive X-ray structural studies of the quinolinate phosphoribosyl transferase (BNA6) from Saccharomyces cerevisiae. Biochemistry. 2008;47:4039–4050. doi: 10.1021/bi7020475. [DOI] [PubMed] [Google Scholar]
- 15.Cao H, Pietrak BL, Grubmeyer C. Quinolinate phosphoribosyltransferase: kinetic mechanism for a type II PRTase. Biochemistry. 2002;41:3520–3528. doi: 10.1021/bi012148g. [DOI] [PubMed] [Google Scholar]
- 16.Tao W, Grubmeyer C, Blanchard JS. Transition state structure of Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1996;35:14–21. doi: 10.1021/bi951898l. [DOI] [PubMed] [Google Scholar]
- 17.Schramm VL. Enzymatic transition states and transition state analog design. Ann. Rev. Biochem. 1998;67:693–720. doi: 10.1146/annurev.biochem.67.1.693. [DOI] [PubMed] [Google Scholar]
- 18.Oppenheimer NJ. NAD hydrolysis: chemical and enzymatic mechanisms. Mol. Cell Biochem. 1994;138:245–251. doi: 10.1007/BF00928468. [DOI] [PubMed] [Google Scholar]
- 19.Arent S, Kadziola A, Larsen S, Neuhard J, Jensen KF. The extraordinary specificity of xanthine phosphoribosyltransferase from Bacillus subtilis elucidated by reaction kinetics, ligand binding, and crystallography. Biochemistry. 2006;45:6615–6627. doi: 10.1021/bi060287y. [DOI] [PubMed] [Google Scholar]
- 20.Schramm VL. Enzymatic transition states: thermodynamics, dynamics and analogue design. Arch. Biochem. Biophys. 2005;433:13–26. doi: 10.1016/j.abb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Yu DG, Sawitzke JA, Ellis H, Court DL. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7207–7212. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mount DW. Bioinformatics: Sequence and Genome Analysis. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2004. [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 25.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1992. [Google Scholar]
- 27.Penefsky HS. A centrifuged-column procedure for the measurement of ligand binding by beef heart F1. Methods Enzymol. 1979;56:527–530. doi: 10.1016/0076-6879(79)56050-9. [DOI] [PubMed] [Google Scholar]
- 28.Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–108. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 29.Scatchard G. The attractions of proteins for small molecules and ions. Ann. N.Y. Acad. Sci. 1949;51:660–672. [Google Scholar]
- 30.Johnson KA. The Enzymes. New York: Academic Press, Inc.; 1992. Transient-state kinetic analysis of enzyme reaction pathways; pp. 1–61. [Google Scholar]
- 31.Bordwell FG. Equilibrium acidities in dimethyl sulfoxide solution. Acc. Chem. Res. 1988;21:456–463. [Google Scholar]
- 32.Bruice TC, Benkovic SJ. Chemical basis for enzyme catalysis. Biochemistry. 2000;39:6267–6274. doi: 10.1021/bi0003689. [DOI] [PubMed] [Google Scholar]
- 33.Freist W, Sternbach H, Pardowitz I, Cramer F. Accuracy of protein biosynthesis: quasi-species nature of proteins and possibility of error catastrophes. J. Theor. Biol. 1998;193:19–38. doi: 10.1006/jtbi.1998.0672. [DOI] [PubMed] [Google Scholar]
- 34.Schimmel P. Hazards of deducing enzyme structure-activity relationships on the basis of chemical applications of molecular biology. Acc. Chem. Res. 1989;22:232–233. [Google Scholar]
- 35.Freist W, Sternbach H, Pardowitz I, Cramer F. Accuracy of protein biosynthesis: quasi-species nature of proteins and possibility of error catastrophes. J. Theor. Biol. 1998;193:19–38. doi: 10.1006/jtbi.1998.0672. [DOI] [PubMed] [Google Scholar]
- 36.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W. H. Freeman and Co.; 1999. [Google Scholar]
- 37.Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J. Biol. Chem. 2005;280:18326–18335. doi: 10.1074/jbc.M501622200. [DOI] [PubMed] [Google Scholar]
- 38.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.