Abstract
The observation made twenty years ago that type IB topoisomerases bound DNA helix-helix juxtapositions was unexpected, given the controlled helical rotation mechanism of the enzyme. In this issue, Patel et al. (2010) provide an elegant structural explanation for this interaction.
Topoisomerases are essential enzymes that regulate DNA supercoiling and remove knots and tangles from the genome (Leppard and Champoux, 2005, Liu et al., 2009, Deweese and Osheroff, 2009). Type I topoisomerases act by creating transient single-stranded breaks in the double helix, followed by passage of the opposite intact strand through the break (type IA) or by controlled rotation of the helix around the break (type IB). Type II topoisomerases act by creating transient double-stranded breaks and interconvert topological structures by passing an intact double helix through the DNA gate.
Because topoisomerases modulate the topological structure of DNA, it is not surprising that they can distinguish the topological state of their substrate (Leppard and Champoux, 2005, Liu et al., 2009, Deweese and Osheroff, 2009). It has long been known that the type IB and type II topoisomerases act on both negatively supercoiled (underwound) and positively supercoiled (overwound) DNA molecules, and prefer either substrate to relaxed molecules that lack torsional/axial stress. In contrast, type IA topoisomerases preferentially act on negatively supercoiled DNA and display little activity towards overwound molecules. This preference for underwound substrates reflects the fact that the type IA enzymes utilize a single-stranded DNA passage mechanism and consequently require single-stranded DNA (which is unlikely to form in overwound molecules) for activity.
In an effort to characterize the mechanism by which topoisomerases distinguish the topological state of DNA, Zechiedrich and Osheroff (1990) utilized electron microscopy to visualize enzyme-DNA complexes. They observed that eukaryotic topoisomerase II preferentially bound DNA at regions of helix-helix juxtaposition. This binding preference seemed a logical mechanism for sensing topology, because DNA crossovers are more prevalent in supercoiled molecules, irrespective of the directionality of torsional stress. Furthermore, it is consistent with the catalytic mechanism of type II topoisomerases, which requires that these enzymes simultaneously interact with two helices during the double-stranded DNA passage reaction.
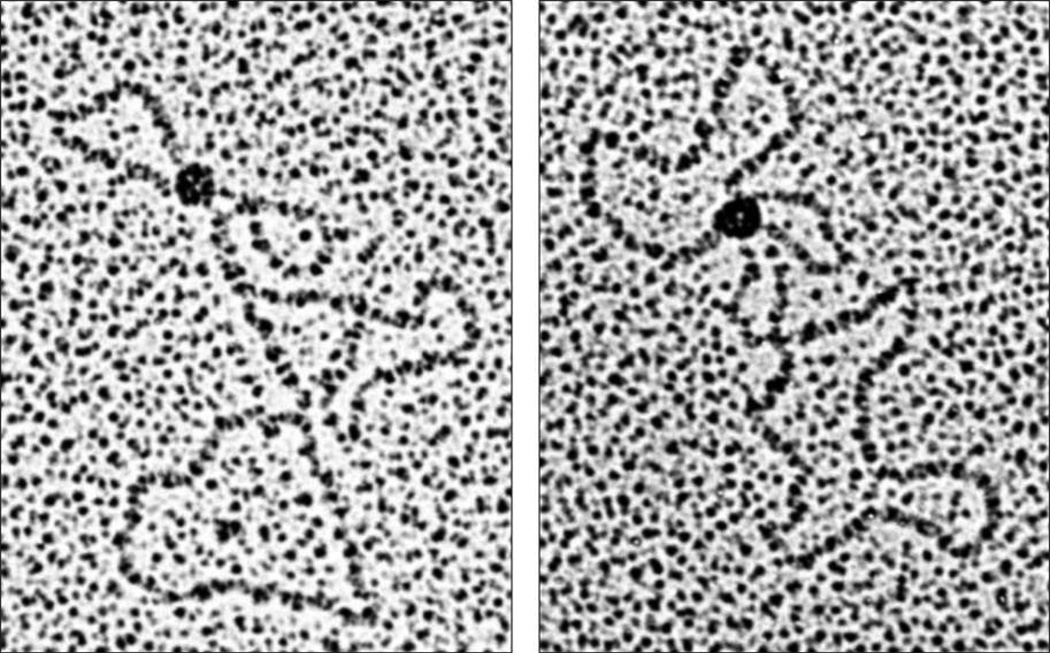
Zechiedrich and Osheroff (1990) also found that eukaryotic topoisomerase IB preferentially bound DNA at regions of helical crossovers in plasmids (Figure 1) and could generate intramolecular synapses in linear molecules. This observation was unexpected, as the type IB enzymes have no catalytic mandate to interact with more than a single DNA helix while removing superhelical twists. Although the mechanistic repercussions of the finding were unclear, three later studies came to similar conclusions regarding topoisomerase IB-DNA interactions. Madden et al. (1995) reported that an active site mutant of human topoisomerase IB preferentially bound underwound or overwound plasmids compared to relaxed molecules, and thus concluded that the enzyme most likely interacted at regions of helix-helix juxtaposition. Shuman et al. (1997) visualized poxvirus topoisomerase IB-DNA complexes by electron microscopy and found that the enzyme formed intramolecular loop structures in which noncontiguous DNA segments were synapsed. Moreno-Herrero et al., (2005), utilizing atomic force microscopy, also observed that poxvirus topoisomerase IB could introduce intramolecular synapses in DNA molecules.
Figure 1.
Binding of eukaryotic topoisomerase IB to helix-helix juxtapositions in circular plasmids (pBR322, 4.6 kb). Conditions are described in Zechiedrich and Osheroff (1990).
While the above studies provided novel insights into the interactions of topoisomerase IB with DNA, they were unable to address two critical issues regarding the recognition of helix-helix crossovers by the enzyme. First, they could not distinguish whether the DNA synapses formed by topoisomerase IB were generated by a single enzyme molecule with two intrinsic nucleic acid binding sites or by two separate DNA-bound enzymes bridged by protein-protein interactions. Second, they provided no direct insight into the role that DNA synapse formation played in enzyme function. Both of these questions are addressed in the groundbreaking crystallographic study of Deinoccocus radiodurans topoisomerase IB by Patel et al. (2010) that is reported in the current issue. This bacterial type IB enzyme is related to eukaryotic nuclear topoisomerase IB, but is similar in size and structure to the poxvirus family of type IB topoisomerases discussed above (Krogh and Shuman, 2002).
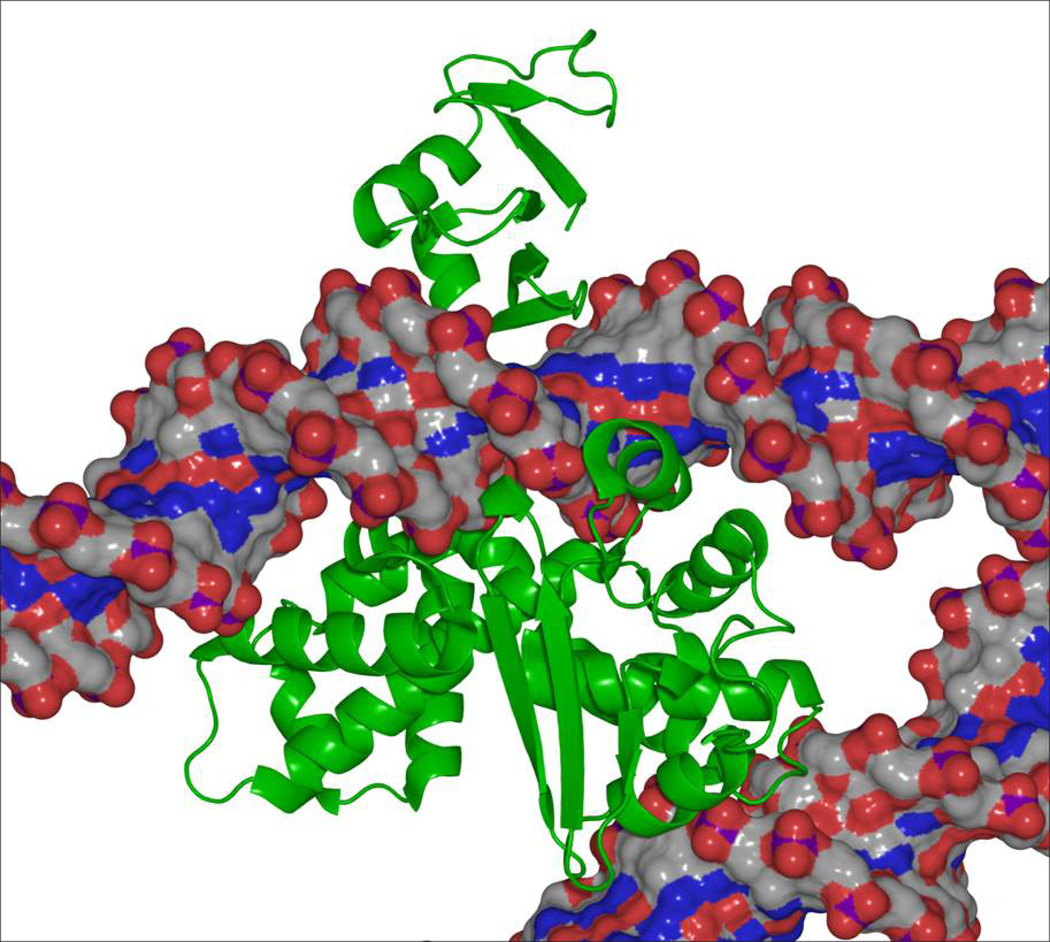
Patel et al. (2010) describe the structure of a complex between D. radiodurans topoisomerase IB and a 12-bp duplex that is not a substrate for cleavage by the enzyme. In contrast to previous structures of bacterial/poxvirus type IB enzymes (Perry, et al., 2006, Perry, et al., 2010), the DNA was bound at a secondary site that is ~30 Å from the active site amino acid residues and the catalytic DNA binding site (Figure 2). The secondary DNA binding site is located within lobe 1 of the C-terminal domain of D. radiodurans topoisomerase IB (aa 106–129), and the DNA interface is comprised of a helix-loop-helix structure. Protein-DNA interactions are stabilized by a network of direct and water-mediated hydrogen bonds. The amino acid residues that make up the interaction domain appear to be conserved across the bacterial, poxvirus, and nuclear topoisomerase IB enzymes, implying that the secondary DNA site contributes to enzyme function. Furthermore, a model of the poxvirus enzyme with both the catalytic and secondary DNA sites filled suggests a mechanism by which a single molecule of topoisomerase IB can recognize, stabilize, or induce helix-helix synapses (Figure 2).
Figure 2.
A model of poxvirus topoisomerase IB (green) in which both the catalytic and secondary DNA sites are filled. The enzyme consists of two domains that form a C-shaped protein clamp around the catalytic DNA helix. The N-terminal domain of poxvirus topoisomerase IB is at the top, and the C-terminal domain is between the two DNA molecules (gray/red/blue). The DNA molecule at the top represents the catalytic helix that is cleaved in the active site of the enzyme. The molecule at the bottom fills the newly discovered secondary DNA binding site described by Patel et al. (2010). Note that the two DNA molecules are held at an angle, such that the paths of the two helices intersect one another. Figure is courtesy of Dr. A. Mondragón, Northwestern University.
To address the functional role of the secondary DNA binding site, Patel et al. (2010) mutagenized amino acid residues in poxvirus topoisomerase IB that mediate DNA interactions. In all cases, mutant enzymes relaxed negatively supercoiled plasmid molecules at rates that were ~33–100% that of wild-type topoisomerase IB. In addition, rates of single-turnover cleavage of a duplex “suicide substrate” were indistinguishable from those of the unmodified enzyme. Thus, it appears that the bacterial/poxvirus type IB enzymes do not require recognition of helix-helix juxtapositions by the secondary DNA binding site for normal catalytic activity. It is not known whether this conclusion extends to the nuclear type IB enzymes. However, biochemical studies with human topoisomerase IB suggest that if synapse binding plays a role in sensing DNA geometry during catalysis, other factors also must be involved. Indeed, even though the enzyme binds negatively and positively supercoiled molecules with similar affinities (Maden et al., 1995, Frohlich, et al., 2007), it relaxes and cleaves positively supercoiled substrates much more efficiently (McClendon and Osheroff, 2006, Frohlich, et al., 2007).
Despite the lack of a catalytic effect, Patel et al. (2010) found that mutagenesis of the secondary DNA binding site blocked the ability of poxvirus topoisomerase IB to stabilize synapse formation and promote plectonemic braiding of the DNA. These data suggest that the secondary DNA binding site on topoisomerase IB may play a role in stabilizing higher order nucleic acid structures in the cell. It will be fascinating to observe the physiological effects of mutant type IB topoisomerases that lack the ability to bind DNA at the secondary site.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Deweese JE, Osheroff N. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich RF, Veigaard C, Andersen FF, McClendon AK, Gentry AC, Andersen AH, Osheroff N, Stevnsner T, Knudsen BR. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Shuman S. Proc. Natl. Acad. Sci. USA. 2002;99:1853–1858. doi: 10.1073/pnas.032613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppard JB, Champoux JJ. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- Liu Z, Deibler RW, Chan HS, Zechiedrich L. Nucleic Acids Res. 37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden KR, Stewart L, Champoux JJ. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon AK, Osheroff N. Biochemistry. 2006;45:3040–3050. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, Holtzer L, Koster DA, Shuman S, Dekker C, Dekker NH. Nucleic Acids Res. 2005;33:5945–5953. doi: 10.1093/nar/gki906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Yakovleva L, Shuman S, Mondragón A. Structure. 2010 doi: 10.1016/j.str.2010.03.007. this issue, xxx-xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD, Van Duyne GD. Mol. Cell. 2006;23:343–354. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD, Van Duyne GD. Structure. 2010;18:127–137. doi: 10.1016/j.str.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Bear DG, Sekiguchi J. EMBO J. 1997;16:6584–6589. doi: 10.1093/emboj/16.21.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Osheroff N. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]