Abstract
The estrogen response element (ERE) consensus sequence is AGGTCAnnnTGACCT, where nnn is known as the tri-nucleotide spacer sequence. Studying 1017 high-confidence ERα-bound loci, we found that genomic EREs are enriched for spacers composed of C(A/T)G, suggesting that the spacer may influence receptor binding and transcriptional responses. We designed consensus EREs containing variable spacer sequences and compared ERα binding in gel shift assays and enhancer function in reporter assays. We found that ERα-ERE binding affinity is modulated by the tri-nucleotide spacer sequence and is favored by spacer sequences of CTG > GCC > TTT. Similarly, luciferase reporter assays indicated that the estrogen-stimulated transcriptional response is modulated by the spacer and parallels the gel shift data: CTG > GCC > TTT. Reporter assays demonstrated that the spacer sequence also modulates the sensitivity of EREs to repression engendered by the receptor antagonist hydroxytamoxifen. These experiments indicate that the sequence of the tri-nucleotide spacer is non-random at receptor-bound genomic loci, influences ERα-DNA binding affinity, and modulates transactivation potential of the receptor-ligand-DNA complex. This work has implications for understanding which genomic EREs are targeted by ERα, should improve computational prediction of functional EREs within genomic sequences, and describes novel sequence determinants of the estrogen response.
Keywords: estrogen, estrogen receptor, estrogen response element, tri-nucleotide spacer
1. Introduction
Estrogens are steroid hormones with widespread physiologic effects including important functions in human bone, reproductive organs, and the cardiovascular system. Estrogens also promote the development of some breast and uterine cancers [1]. The estradiol (E2)-mediated proliferative response in MCF-7 breast cancer cells is predominantly mediated by the estrogen receptor-α (ERα/NR3A1), a ligand-activated transcription factor [2]. ERα regulates the transcription of target genes through direct binding to its cognate recognition sites, known as estrogen receptor response elements (EREs), or by modulating the activity of other DNA-bound transcription factors at alternative DNA sequences [3–10]. Receptor-mediated functions have recently been demonstrated to occur across great genomic distances (tens to hundreds of kilobases) and even across chromosomes [11–21].
Estrogen receptor dimers were initially described to bind to the canonical 13 bp ERE, GGTCAnnnTGACC, a palindromic inverted repeat (IR) separated by any three nucleotides (nnn) originally identified from conserved sequence alignments of the estrogen-sensitive Xenopus laevis vitellogenin and the chicken apo-VLDL genes [22, 23]. The consensus ERE sequence was subsequently extended to a 15 bp inverted repeat (AGGTCAnnnTGACCT) when the flanking sequences were noted to contribute to dimer binding-affinity [24]. Once full human genomic sequence data became available, several groups of investigators combined bioinformatic approaches (principally position weight matrices, or PWMs) with large-scale gene expression studies in order to identify E2-responsive and possibly ERα-regulated genes of interest [25–28]. The PWMs were designed using fewer than 20 promoter-proximal EREs detected in humans [24]. It was soon recognized that functional EREs generally do not conform to the consensus sequence in vivo [29, 30] and promoter analyses identified functional EREs containing single, double, and triple nucleotide substitutions from the consensus ERE motif [24, 30].
Experimental data have indicated decreased estrogen receptor binding to variant ERE sequences in vitro [25, 31] although binding affinity does not relate linearly with transactivation potential [32, 33]. The altered binding affinity of ERα with variant EREs has been attributed to nucleotide substitutions in the ERE half-site(s) which deviate from the consensus sequence or to variable spacing and/or orientation between the half-sites [24, 30]. In addition to EREs, additional determinants of estrogen receptor function may include local non-ERE DNA sequences, DNA methylation status, regional chromatin composition and post-translational modifications, cofactor interactions, and the nature of the receptor ligand that is engaged [11, 12, 34–38].
Studies in MCF-7 cells have indicated that only ~1,000–10,000 loci are bound by ERα in response to E2 treatment [11, 13–15, 38] and demonstrate that many more putative EREs exist in the human genome than are bound by ERα in any given cell type [39]. The published human genome reveals 2310 perfect EREs (13 bp core ERE sequences), 49,803 ERE sequences with only one bp deviation from the consensus sequence, and 265,482 loci that deviate by only two mismatches. Importantly, there is substantial cell type-specific determination of ERα binding sites which correlates with cell type-specific post-translational histone modifications at receptor-bound sites [39].
There is increasing evidence that multiple ERα-bound loci with varying DNA-binding affinities can cooperate to form a productive cis-regulatory module [20, 25, 38–41]. In order to comprehensively identify ERα-bound targets in MCF-7 cells, and to address the question of ERE sequence specificity, we recently employed chromatin immunoprecipitation (ChIP) experiments with whole genome DNA arrays (i.e. ChIP-on-chip) [11]. We combined these data with data from a similar study conducted by the Brown laboratory [13] in order to develop a list of high-confidence ERα-bound loci [42]. These immunoprecipitated chromatin fragments are likely to contain true estrogen responsive elements because 1) they were crosslinked to ERα in living cells (directly or via protein intermediaries), and 2) they were detected by two independent laboratories.
Our analysis of 1017 high-confidence ERα-bound ChIP sites indicated that approximately half of all ERα-bound loci do not have a discernable ERE and likely represent sites of ERα tethering via other transcription factors or sites that contain atypical estrogen response elements (i.e. tandem half-ERE sites) [42]. Further, we found that most ERE sequences at ERα-bound cis-regulatory elements are not consensus EREs [42]. Here, we demonstrate that the three bp spacer between the inverted ERE half-sites, rather than being random nucleotides, is enriched for selected sequences at in vivo receptor targets. We demonstrate that the tri-nucleotide spacer sequence modulates estrogen receptor binding affinity in vitro and modulates E2- and receptor antagonist-mediated responses in luciferase reporter assays. This work has implications for understanding which genomic EREs are targeted by the ERα in vivo and should improve computational prediction of functional EREs within genomic sequences. Further, these data suggest that diverse ERE sequences will demonstrate variable E2-mediated transcriptional responses in part because of variations in their respective tri-nucleotide spacer sequences.
2. Materials and methods
2.1. Cell Culture
MCF-7 cells (ATCC) were grown as described [43]. Cells were changed to E2-depleted, phenol red-free media consisting of MEM alpha (Gibco) with 10% charcoal/dextran-stripped calf serum, insulin, penicillin G, streptomycin, and L-glutamine (all Gibco), for 72 hours prior to treatments. Where indicated, treatments included vehicle control (100% EtOH) and estradiol (10 or 100 nM, Sigma). Telomerase-immortalized Human Endometrial Stromal Cells (HESC cells), a generous gift from Dr. Graciela Krikun, were grown in the same media used for the MCF-7 cells. HESC cells have normal chromosome numbers and structures [44].
2.2. Preparation of Nuclear Extracts and EMSA
HESC nuclear extracts (NE) were purified using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the manufacturer’s instructions. HESC cells have no demonstrable ERα activity using sensitive luciferase reporter assays and no ERα protein detected by Western blot analysis (data not shown). However, HESC cell nuclei have cofactors that promote the binding of recombinant ERα (rERα, Affinity Bioreagents) to target DNA in EMSA and these factors enhance binding when compared to recombinant ERα alone. EMSA experiments were therefore conducted using HESC nuclear extracts combined with rERα. Protein determinations were performed using the Micro BCA assay (Pierce) and 5 μg of nuclear extract (with protease inhibitors, Roche) plus rERα (400 fmol) were run in each lane of a 5% acrylamide gel in TBE/glycerol buffer. Oligonucleotide probes were labeled using the Biotin 3’ End DNA Labeling Kit (Pierce). Each Biotin-labeled probe was used at 20 fmol/lane and binding reactions were performed per LightShift Chemiluminescent EMSA Kit instructions (Pierce). For super-shift assays, relevant antibody was used as indicated (400 ng/reaction): anti-ERα Ab-10 (LabVision) and anti Sp1 H-225 sc-14027 (Santa Cruz). A complete list of oligonucleotide sequences used as probes for EMSA is presented in the supplementary materials (Table S5).
2.3. Luciferase Reporter Assays
Luciferase reporter assays were performed using the Luciferase Assay System (Promega) according to the manufacturer’s instructions. Single copy ERE-containing regulatory elements were cloned into pGL2-Promoter (Promega) and all constructs were sequence verified prior to use in reporter assays. Reporter constructs were transfected into MCF-7 cells using the TransIT-LT1 Transfection Reagent (Mirus). Cotransfection with a β-galactosidase-expressing plasmid (Promega) enabled normalization of transfection efficiency across samples using a β-galactosidase assay kit (Promega) according to the manufacturer’s instructions.
2.4. Computational Detection of ERE Sequences
Genome-wide location analysis for ERα, and E2-dependent gene expression profiling, were performed by two independent groups as previously described [11, 13]. 1017 ERα-bound genomic loci common to both datasets (shared loci defined as falling within 1 kb of the center of each locus) were interrogated for ERE-like sequences. Starting at the center of each high-confidence ERα-bound locus, we extracted genomic sequences 1 kb in each direction (human genome 18, build 36.1). Because the chromatin shear size in ChIP experiments was optimized to average ~500 bp, we estimated that interrogating sequences of average size 2 kb would have a reasonable likelihood of capturing most sequences directly bound by ERα in the ChIP assays. Transcription Element Search Software (TESS) was used to identify ERE sequences [45]. Default settings were used with variation in Maximum Allowable String Mismatch of 10% and 20%. The TESS software will identify binding sites using consensus strings from the TRANSFAC, JASPAR, IMD, and CBIL-GibbsMat databases. Repetitive DNA elements were determined using RepeatMasker V3.1 at the default settings (http://www.repeatmasker.org).
2.5. Statistical analysis
Comparisons between two groups were made using a two-tailed t-test with P values indicated. Statistical analysis of the base pair distributions in the ERE spacer sequences was performed using the Pearson’s chi-square to test for goodness of fit. In all cases, the probability value to identify statistical significance was P<0.05.
3. Results
3.1. Computational detection of ERE sequences from ERα-bound loci in MCF-7 cells
Recently, we performed location analysis for ERα in MCF-7 cells using ChIP-on-chip with whole genome tiling arrays and combined these data with gene expression profiling in response to E2 exposure [11]. The location analysis identified 1615 genomic targets of ERα and revealed that the majority (~80%) of ERα-bound loci reside > 10 kb from any annotated transcription start site. Of 1615 loci that were bound by ERα in our analysis, 1017 (~60%) were also detected by the Brown group [13]. A list of the genomic coordinates for these highest-confidence ERα-bound loci appears in the supplementary materials (Table S1). ChIP-on-chip data were validated by ChIP-PCR for over 20 sites and revealed E2-dependent recruitment (>2 fold) of ERα at all of the loci that were tested [42].
Using Transcription Element Search Software (TESS) [45], we performed an analysis of the 1017 ERα-bound loci (average length 2 kb) for the presence of ERE sequences using two stringencies of ERE detection: ≤ 10% nucleotide deviation (≤ 2 mismatched residues within the core 15 bp ERE), and 10–20% nucleotide divergence (3–4 mismatched residues) from the 15 bp consensus ERE sequence (AGGTCAnnnTGACCT). We identified a total of 646 ERE sequences (Table S2) from 509 ERα-bound loci, indicating that ~50% of receptor-bound sites did not have a discernable ERE sequence. 391 (~77%) of the ERE-containing ChIP sites contained a single ERE sequence, 101 (~20%) contained two distinct ERE sequences, and 17 loci (~3%) contained three or more distinct ERE sequences within 2 kb of the center of their respective ChIP sites. In addition, we found that a considerable proportion of EREs lay within repetitive DNA elements (Table 1). Our recent data indicate that repetitive element EREs, and Alu elements in particular, may contribute considerably to ERα-mediated transcriptional responses [42].
Table 1.
Base Frequency Matrix as a Function of Nucleotide Position.
The tri-nucleotide spacer sequence is conserved at ERα-bound ERE sequences. Base frequency matrix as a function of nucleotide position for 646 ERE sequences. Transcription Element Search Software (TESS) was used to analyze 1017 ERα-bound loci at two different stringencies of detection. High stringency (0–10% deviation from the consensus ERE sequence) and low stringency (10–20% deviation from the consensus ERE) results are shown in Table 1A and 1B, respectively. These data are pooled (Table 1C) to reveal a total of 646 ERE sequences residing within 509 receptor-bound loci. The distribution of ERE sequences within repetitive elements is indicated for each stringency. The tri-nucleotide spacer sequences are non-randomly distributed at all stringencies of detection (see main text) even when repetitive element EREs are excluded from the analysis (see Table S4).
| A. Stringency 0–10%, 162 total EREs, 26 (17%) of EREs in repetitive elements. | |||||||||||||||
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| %A | 77.4% | 3.0% | 0.6% | 2.4% | 0.6% | 94.5% | 14.0% | 28.7% | 14.6% | 1.8% | 1.8% | 87.8% | 3.0% | 4.9% | 6.1% |
| %C | 3.0% | 0.0% | 1.8% | 2.4% | 94.5% | 1.2% | 43.3% | 17.1% | 24.4% | 0.0% | 1.2% | 7.3% | 93.9% | 91.5% | 13.4% |
| %G | 15.2% | 90.9% | 97.0% | 7.3% | 1.8% | 1.8% | 25.0% | 22.6% | 49.4% | 0.6% | 94.5% | 2.4% | 0.0% | 0.0% | 3.7% |
| %T | 4.3% | 6.1% | 0.6% | 87.8% | 3.0% | 2.4% | 17.7% | 31.7% | 11.6% | 97.6% | 2.4% | 2.4% | 3.0% | 3.7% | 76.8% |
| B. Stringency 10–20%, 484 total EREs, 169 (36%) of EREs in repetitive elements. | |||||||||||||||
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| %A | 76.6% | 8.7% | 5.4% | 5.8% | 2.1% | 75.9% | 12.9% | 36.7% | 11.8% | 5.0% | 12.9% | 82.6% | 3.9% | 6.6% | 6.4% |
| %C | 3.1% | 1.2% | 11.4% | 6.2% | 74.9% | 3.9% | 57.1% | 13.7% | 17.8% | 9.1% | 4.8% | 9.8% | 75.7% | 86.1% | 20.1% |
| %G | 16.2% | 81.5% | 78.8% | 7.1% | 3.9% | 15.6% | 17.6% | 18.0% | 58.3% | 4.1% | 80.1% | 3.1% | 17.2% | 1.2% | 3.5% |
| %T | 4.1% | 8.5% | 4.4% | 80.9% | 19.1% | 4.6% | 12.4% | 31.5% | 12.0% | 81.7% | 2.3% | 4.6% | 3.1% | 6.0% | 69.9% |
| C. Stringency 0–20%, 646 total EREs, 195 (30%) of EREs in repetitive elements. | |||||||||||||||
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| %A | 76.8% | 7.3% | 4.2% | 5.0% | 1.7% | 80.7% | 13.2% | 34.7% | 12.5% | 4.2% | 10.1% | 83.9% | 3.7% | 6.2% | 6.3% |
| %C | 3.1% | 0.9% | 9.0% | 5.3% | 79.9% | 3.3% | 53.6% | 14.6% | 19.5% | 6.8% | 3.9% | 9.1% | 80.3% | 87.5% | 18.4% |
| %G | 15.9% | 83.9% | 83.4% | 7.1% | 3.4% | 12.1% | 19.5% | 19.2% | 56.0% | 3.3% | 83.7% | 2.9% | 12.8% | 0.9% | 3.6% |
| %T | 4.2% | 7.9% | 3.4% | 82.7% | 15.0% | 4.0% | 13.8% | 31.6% | 11.9% | 85.8% | 2.3% | 4.0% | 3.1% | 5.4% | 71.7% |
3.2. The majority of ERα-bound loci contain non-consensus ERE sequences
The sequence requirements for ERα binding to chromatin in vivo are surprisingly flexible. Table 1 demonstrates the base frequency at each position in the 15 bp ERE sequence for each stringency assayed (Table 1A and 1B). The base frequencies are pooled for all EREs that were detected (0–20% nucleotide divergence from consensus) in Table 1C. At all stringencies assayed, the 13 bp core bases were more highly conserved than the flanking sequences located at positions 1 and 15 of the EREs. It is noteworthy that, even at high stringency of detection, almost all possible single base substitutions were detected in our dataset. These data are consistent with in vitro data (electrophoretic mobility shift assays, EMSAs) indicating that all single bp deviations from consensus are capable of binding to the estrogen receptor, although with variable affinity [25]. When ERE detection criteria were relaxed to permit 10–20% base divergence from the consensus sequence, all forms of nucleotide substitutions were permissive for receptor binding though some substitutions were less common than others (Table 1B and 1C). For example, from all EREs that were detected, position 2 is rarely (<1%) cytosine, whereas position 13 is guanine in 12.8% of EREs (Table 1C).
Ignoring the tri-nucleotide spacer sequence, 348 different ERE sequences were detected from the group of 646 total EREs (Table S3). There was equal representation of an imperfect ERE (16 examples of GGGTCAnnnTGACCT) and a perfect consensus ERE (16 examples of AGGTCAnnnTGACCT) (Table S3). Excluding analysis of the less conserved positions 1 and 15 in the 646 ERE sequences that we identified, we detected 51 (~8%) perfect core consensus EREs (GGTCAnnnTGACC). Thus, of the 2310 perfect consensus ERE sequences detected in the published sequence of the human genome, our highest-confidence location analysis revealed receptor occupancy at only 51 (2.2%) of these sites in MCF-7 cells. These data demonstrate that ERα binds to widely variant EREs in MCF-7 cells and that many “perfect” EREs are not receptor-bound in these cells under these culture conditions.
3.3. The tri-nucleotide spacer sequence is conserved at ERα-bound ERE sequences
The tri-nucleotide spacer sequence between the two ERE half-sites does not make important base contacts with the estrogen receptor’s DNA-binding domain (DBD) [46–48] and has historically been described as nnn, meaning that any three bp sequence will suffice. Our data indicated that, at all stringencies of ERE detection, the tri-nucleotide spacer was conserved at receptor-bound EREs. Specifically, positions 7–9 were preferentially C(A/T)G at ERα-bound loci; this spacer sequence was found at more than 41% of EREs (Table 2 and Table S3). When compared to the expected equal distribution of bases at each position, a statistically significant non-random distribution of sequences at positions 7–9 was indicated by the chi-square test with P values of 1.9E–14, 2.04E–96, and 7.97E–106 for stringencies 0–10% (A), 10–20% (B), and 0–20% (C), respectively (Table 1). The observed conservation of the central triad sequence remained even when all repetitive element EREs were excluded from the analysis (Table S4). While the molecular justification for this triad sequence preference is unclear, these data suggested that the 3 bp spacer has functional significance, possibly modulating ERα-ERE binding and subsequent transcriptional responses.
Table 2.
The spacer sequence C(A/T)G is detected at over 41% of ERα-bound ERE sequences. Shown are the ten most common ERE half-site and spacer sequences from 646 EREs detected within high-confidence ERα-bound loci. Over 41% of the spacer sequences were CAG or CTG whereas the expected prevalence of C(A/T)G spacers would be 3.1% if these residues were random nucleotides. Similar findings were noted when repetitive element EREs were excluded from the analysis (Table S4).
| 5′ ERE half-site | Spacer Sequence | 3′ ERE half-site | ||||||
|---|---|---|---|---|---|---|---|---|
| Seq. | # | % | Seq. | # | % | Seq. | # | % |
| AGGTCA | 150 | 23.22 | CAG | 150 | 23.22 | TGACCT | 163 | 25.23 |
| AGGTTG | 65 | 10.06 | CTG | 116 | 17.96 | TGAGCC | 42 | 6.50 |
| GGGTCA | 56 | 8.67 | GGG | 17 | 2.63 | TGCCCT | 36 | 5.57 |
| AGCTCA | 35 | 5.42 | GAG | 15 | 2.32 | TGAGCT | 36 | 5.57 |
| ATGTCA | 31 | 4.80 | GCC | 15 | 2.32 | TGACCC | 35 | 5.42 |
| AGGGCA | 30 | 4.64 | AGG | 14 | 2.17 | CAACCT | 34 | 5.26 |
| AAGTCA | 19 | 2.94 | GGC | 14 | 2.17 | TGACCA | 23 | 3.56 |
| AGGTTA | 15 | 2.32 | CAC | 12 | 1.86 | TGACAT | 22 | 3.41 |
| AGGCCA | 12 | 1.86 | GGT | 12 | 1.86 | TGACTT | 19 | 2.94 |
| TGGTCA | 12 | 1.86 | AGC | 11 | 1.70 | TAACCT | 17 | 2.63 |
3.4. The tri-nucleotide spacer sequence modulates ERα-ERE binding affinity
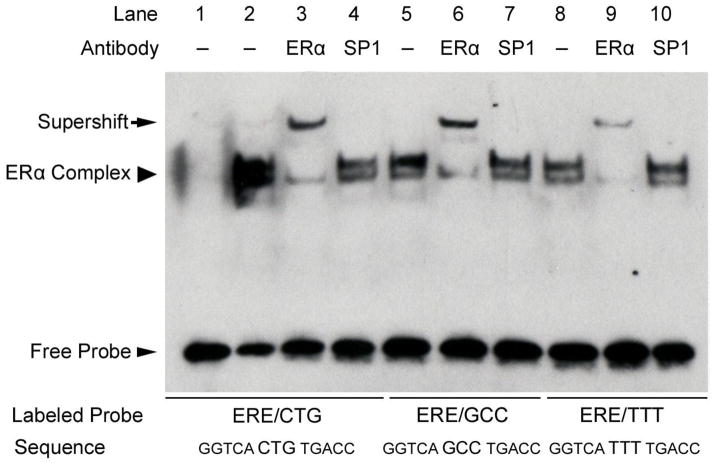
The observed preference of ERα for C(A/T)G-containing ERE sequences in vivo suggested the possibility that this sequence is more avidly bound by the receptor than EREs with alternative spacer sequences. In order to address this question, we performed EMSAs of three ERE sequences, each with identical consensus ERE half-sites but each with variant ERE tri-nucleotide spacer sequences (specifically, GGTCACTGTGACC, GGTCAGCCTGACC, and GGTCATTTTGACC). As can been seen from Fig. 1, binding affinity of ERα was greatest for CTG-containing and least for TTT-containing consensus ERE sequences (Fig. 1, lanes 2, 5, and 8). Specificity of the ERα-containing complexes is shown by supershift of the labeled probes using a monoclonal anti-ERα antibody (Fig. 1, lanes 3, 6, and 9) but was not seen when using an irrelevant antibody against Sp1 (Fig. 1, lanes 4, 7, and 10).
Fig. 1.
The tri-nucleotide spacer sequence modulates ERα-ERE binding affinity. EMSA of ERα binding to consensus ERE sequences with variable tri-nucleotide spacer sequences. An ERα-containing complex bound to all three ERE sequences (arrowhead, lanes 2, 5, and 8) and was confirmed by supershift (arrow) using a monoclonal antibody that recognizes ERα (lanes 3, 6, and 9). Receptor binding affinity for the sequences favored tri-nucleotide spacer sequences of CTG > GCC > TTT. The non-specific antibody recognizing Sp1 had no effect on the ERα-containing complexes bound to these probes (lanes 4, 7, and 10). Shown is a representative experiment performed at least three times.
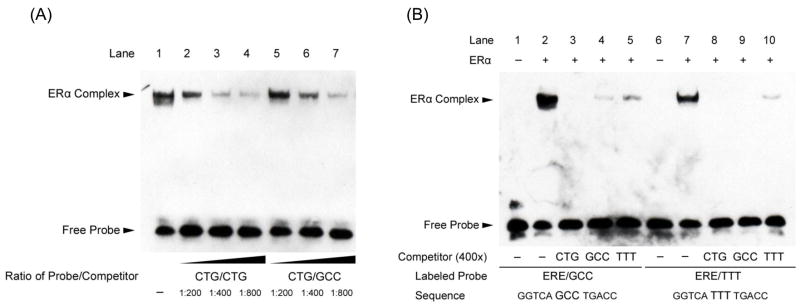
We employed titration experiments using unlabeled competitor probes in EMSA analyses in order to better-assess receptor-DNA affinity for each of the variant EREs described in Fig. 1. Fig. 2A shows a representative EMSA experiment in which the labeled probe was GGTCACTGTGACC. Titration of increasing concentrations of unlabeled competitor probes indicated that unlabeled ERE competitor with spacer sequence CTG was most effective at competing for labeled CTG-containing ERE probe (Fig. 2A, lanes 2, 3, and 4), unlabeled GCC-containing ERE competed with intermediate affinity (Fig. 2A, lanes 5, 6, and 7), and the unlabeled TTT-containing ERE competed with the least affinity for the CTG-containing probe (not shown).
Fig. 2.
Binding affinity for the ERE with a CTG spacer sequence is greater than for EREs with GCC or TTT spacer sequences. (A) ERα binding to the consensus ERE sequence (GGTCACTGTGACC) is shown in lane 1. Competition using serial dilutions of the same unlabeled DNA sequence (lanes 2, 3, and 4) or unlabeled sequences with the variant spacer sequence GCC (lanes 5, 6, and 7) is shown. The unlabeled competitor with ERE spacer CTG demonstrated higher affinity binding to ERα than did the GCC-spaced ERE sequence. (B) Complementary EMSA experiments confirm that ERα preferentially binds EREs with a CTG tri-nucleotide spacer sequence. ERα binding to two consensus EREs with variant tri-nucleotide spacers is shown: GGTCAGCCTGACC (lanes 2–5) and GGTCATTTTGACC (lanes 7–10). Competition using 400 fold excess of the indicated unlabeled ERE sequences is also shown and reveals that relative efficiency of competition follows the order CTG (lanes 3 and 8) > GCC (lanes 4 and 9) > TTT (lanes 5 and 10). Shown are representative experiments performed at least three times.
Complementary EMSA experiments were performed to demonstrate the relative estrogen receptor binding affinities for each of the variant ERE spacer sequences shown above. When the labeled ERE probe contained the intermediate affinity GCC spacer sequence (GGTCAGCCTGACC, Fig. 2B, lane 2), competition using unlabeled ERE probe with spacer sequence CTG was the strongest, spacer GCC was intermediate, and spacer TTT the weakest competitor (Fig. 2B, lanes 3, 4, 5, respectively). Similarly, when the labeled ERE probe contained the lowest affinity TTT spacer sequence (GGTCATTTTGACC, Fig. 2B, lane 7), competition using unlabeled ERE probe with spacer sequences CTG and GCC were superior competitors compared to the TTT-containing ERE (Fig. 2B, lanes 8, 9, 10, respectively).
3.5. The tri-nucleotide spacer sequence modulates the transcriptional response mediated by ERα
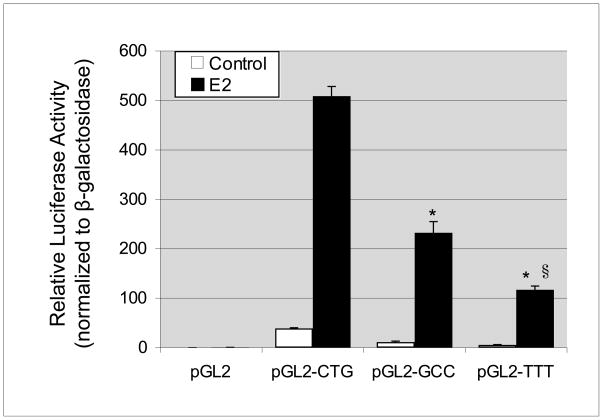
In order to assess the potential of variant ERE spacer sequences to influence receptor-mediated transactivation, we cloned consensus EREs with variant spacers into a luciferase reporter construct (pGL2-promoter). ERα-expressing MCF-7 cells were transfected with each variant ERE-reporter construct and data were normalized for transfection efficiency using β-galactosidase expressing plasmid (Promega). We observed that basal and E2-stimulated ERE-driven reporter activity was highest for the sequence GGTCACTGTGACC, intermediate for GGTCAGCCTGACC, and lowest for GGTCATTTTGACC (Fig. 3). Combined, our data indicate that the ERE spacer sequence affects both ERα-DNA binding affinity and overall transcriptional responses mediated by the estrogen receptor.
Fig. 3.
The tri-nucleotide spacer sequence modulates transcriptional response to ERα. Luciferase reporter assays of single copy consensus EREs with variable tri-nucleotide spacer sequences were performed in MCF-7 cells. Basal and E2-stimulated luciferase values are shown normalized to co-transfected β-galactosidase expressing plasmid. Basal and E2-stimulated luciferase activities were negligible for empty vector (pGL2) and highest for the ERE with spacer sequence CTG, followed by spacer GCC and then spacer TTT. Values are the average of three experiments, performed in triplicate, with SEM indicated. *P<0.01 when compared to E2-treated CTG reporter. §P<0.01 when compared to E2-treated GCC reporter.
3.6. The tri-nucleotide spacer sequence modulates ERE sensitivity to repression by an estrogen receptor antagonist
The observation that the tri-nucleotide spacer sequence can modulate the overall transcriptional response to estrogen (Fig. 3) prompted us to test whether this sequence can influence the transcriptional response to receptor antagonists as well. The selective estrogen receptor modulator tamoxifen is metabolized by the liver to the active metabolite hydroxytamoxifen (OHT), a competitive antagonist of the estrogen receptor which blocks many E2-mediated transcriptional responses in receptor-positive breast cancer cells [49]. Tamoxifen is standard anti-estrogen therapy for estrogen-receptor positive breast cancers [50].
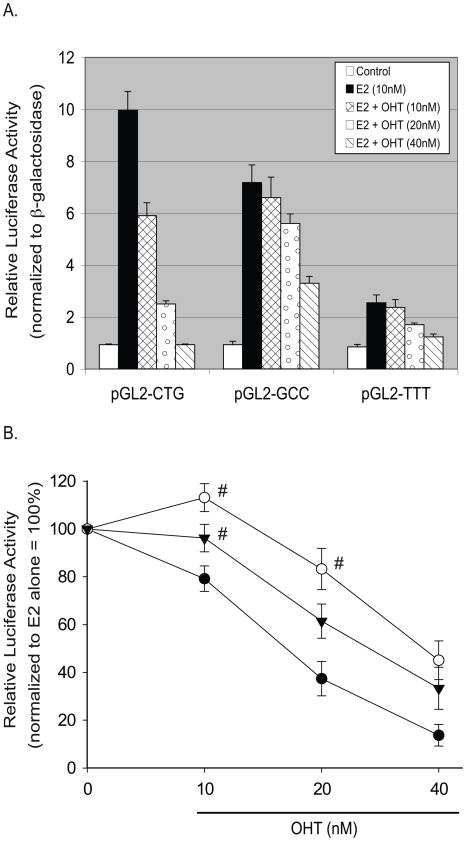
We examined the dose-dependent effects of OHT on E2-stimulated luciferase reporter constructs using perfect EREs with variant tri-nucleotide spacer sequences. Tissue concentrations of OHT depend upon drug dosage and metabolism and range from 11–72 nM [51]. At low-therapeutic doses of OHT (10 nM) we observed that E2-stimulated reporter activity was antagonized for the sequence GGTCACTGTGACC but not for the sequences GGTCATTTTGACC or GGTCAGCCTGACC (Fig. 4A). At higher doses (40 nM) OHT was effective in antagonizing the E2 response of all reporter constructs regardless of the spacer sequence (Figs. 4A and 4B).
Fig. 4.
The tri-nucleotide spacer sequence modulates ERE sensitivity to repression by an estrogen receptor antagonist. Luciferase reporter assays of single copy consensus EREs with variable tri-nucleotide spacer sequences were performed in MCF-7 cells. Basal and E2-stimulated luciferase values are shown normalized to co-transfected β-galactosidase expressing plasmid. (A) Basal and E2-stimulated luciferase activities were negligible for empty vector (pGL2) and highest for the ERE with spacer sequence CTG, followed by spacer GCC and then spacer TTT. Dose response co-treatments with the estrogen receptor antagonist OHT revealed highest OHT sensitivity of ERE sequences spaced by CTG, followed by TTT- and GCC-spaced response elements. Shown is a representative experiment performed in triplicate, with SEM indicated. (B) Comparative sensitivity of each ERE sequence to therapeutic doses of OHT relative to E2-stimulated cells. At low-therapeutic doses of OHT (10 nM), CTG-spaced EREs were significantly repressed whereas GCC- and TTT-spaced EREs were not. At higher doses of OHT (40 nM), all EREs were significantly repressed by the estrogen receptor antagonist. Shown are data from 3–5 biological repeats each performed in triplicate, with SEM indicated. P<0.05 compared to E2 alone for all treatments except where labeled with (#). #P>0.05 compared to E2 alone.
These data indicate that the tri-nucleotide spacer sequence modulates the response to E2 as well as to OHT, and that these responses are not always congruous: the TTT spaced ERE is least responsive to E2 (Figs. 3 and 4A), but intermediate in response to the antiestrogenic effects of low- and medium-therapeutic doses (20–40 nM) of OHT (Fig. 4B). These data raise the possibility that the estrogen response of endogenous genes may be differentially subject to regulation by tamoxifen depending not only on the ERE half-site base composition [52] but also upon the tri-nucleotide spacer sequences of their respective estrogen response elements, a finding that may be of clinical significance for patients receiving this therapy. These results may offer mechanistic explanation for cohorts of tamoxifen-resistant genes in individuals that, due to patient variation in the metabolism of tamoxifen, may demonstrate lower tissue levels of OHT [53–55].
4. Discussion
We observed in vivo enrichment of ERα binding at ERE sequences containing C(A/T)G tri-nucleotide spacer sequences and enhanced affinity for this spacer sequence in vitro, by EMSA analysis. Together, these experiments support the conclusion that enrichment of ERα binding at genomic C(A/T)G spacer-containing ERE sequences in MCF-7 cells was a result of enhanced affinity for the receptor at these sites. Consistent with these observations, testing in luciferase reporter assays indicated that the enhancer elements containing C(A/T)G spacers demonstrated stronger basal and E2-stimulated transactivation potential than identical half sites spaced with alternative tri-nucleotide spacer sequences of lower receptor-binding affinity.
Our data represent the largest collection of human ERE sequences ever reported from genomic loci each of which demonstrated consistent evidence of ERα binding in vivo [11, 13]. We observed the presence of full ERE sequences at approximately half of all receptor-bound loci. The absence of ERE sequences at many ERα-bound genomic loci may reflect widespread tethering of ERα to DNA targets via alternative transcription factors (i.e. AP-1, Sp1 [10]) or the presence of widely divergent ERα-binding motifs not detected using our motif searching software. Our findings indicate that most ERα-bound cis-regulatory elements are not canonical EREs and that considerable deviation from the consensus sequence can be permissive for receptor binding in vivo. Low affinity interactions between transcription factors and imperfect DNA binding sites have historically been difficult to detect in vivo. Recent data in yeast and rodents suggest that such interactions may be more common and provide more biological impact than had previously been suspected [56, 57]. Together with our data, these observations suggest numerous low-affinity or transient transcription factor-DNA interactions occurring with diverse DNA sequences to modulate transcriptional responses.
Our finding that the tri-nucleotide spacer sequences between ERE half-sites is non-random at ERα-bound loci was surprising. This observation held true even when repetitive element (i.e. repeat-masked) EREs were excluded. It has been suggested that DNA sequence serves as an additional “ligand” for transcription factor-containing protein complexes and can contribute to receptor dimerization at imperfect EREs, alter receptor conformation, and influence the net transcriptional response of an ERE [33, 58–61]. Our data suggest that, in addition to the ERE half-site sequences, ERα-mediated transcriptional responses are influenced by the tri-nucleotide spacer sequence. Further, these data indicate that the tri-nucleotide spacer sequence may add predictive value when scoring genomic sequences for functional ERE motifs. The data also suggest that variant spacer sequences and variant ERE half-sites may collectively modulate the overall magnitude of the estrogen response at divergent genomic loci and that both features of the intact ERE could play a role in determining target gene sensitivity to estrogen exposure. Our data were derived from ChIP experiments using ERα-specific antibodies. While there is substantial homology between the DNA-binding domains of ERα and ERβ and considerable overlap in the genomic loci that are targeted by each receptor, it remains to be seen whether the tri-nucleotide spacer sequence influences the transcriptional response to ERβ.
We observed that divergent tri-nucleotide spacer sequences confer variable sensitivity to repression mediated by the estrogen receptor antagonist OHT. While ligand and ERE half-site sequence(s) are each known to contribute to the ultimate transcriptional response [52], this report is the first to identify the tri-nucleotide spacer sequence as a modulator of estrogen/anti-estrogen responses. Variations in the metabolism of tamoxifen can lead to inconsistent tissue levels of OHT in diverse individuals [51] and there may exist circumstances when the effects of OHT are lost for cohorts of E2-responsive genes in some patients. For example, a subject who rapidly metabolizes OHT could conceivably have tissue levels of the drug that will not suppress TTT-spaced ERE enhancers but will suppress CTG-spaced EREs. These results raise the possibility that clinical loss of responsiveness to OHT therapy may, in some circumstances, result from active gene enhancers driven by ERE sequences with OHT-resistant tri-nucleotide spacers in subjects with lower therapeutic levels of OHT.
The molecular mechanisms by which ERE sequences, either in the half-sites or the intervening tri-nucleotide spacer sequences, modulate the transcriptional potential of receptor-bound elements remain incompletely understood. Changes in receptor-DNA binding affinity and/or the tertiary structure of DNA-bound receptors may independently influence the magnitude of the overall transcriptional response. While we have shown that the tri-nucleotide spacer sequence can affect DNA binding affinity, effects of the spacer sequence on estrogen receptor dimer tertiary structure remain to be shown. Mapping any such changes to altered coregulator composition or coregulator binding affinity are important next steps.
It remains plausible that the tri-nucleotide spacer sequence, which resides at the interface of the receptor dimers, may influence overall dimer conformation. It has been demonstrated that receptor dimers bend the DNA-double helix towards the major groove and that DNA bending can influence overall transcriptional response; however, the spacer sequence has never been implicated in this effect [62]. Notably, crystallographic data of receptor-bound EREs have indicated that the base pairs between the half-sites produce propeller twist of the adjacent half sites, orienting the edges of the half sites for optimal interaction with receptor residues [46]. It remains possible that sequence variations in the tri-nucleotide spacer could confer variable propeller twist and produce steric changes in the DNA-protein conformation that might energetically favor, or disfavor, receptor-ERE complex formation, stability, or interactions with coregulatory proteins. Factors that modulate cis-regulatory element (DNA) bending have been shown to influence the transcriptional responses to the estrogen receptor and to the AP-1 (Fos-Jun) family of transcription factors [33, 63, 64]. Similarly, there is a precedent for subtle conformational changes influencing transcriptional functions of the estrogen receptor; conformational changes induced by estrogen receptor “antagonists” such as tamoxifen account for altered coregulator recruitment leading to repression of gene transcription [65].
In addition to DNA sequence, there exist chromatin modifications that are necessary for ERα-mediated transcriptional responses which remain incompletely described. We recently reported that the gene for the variant histone H2A.Z is E2-responsive in MCF-7 cells and we found that H2A.Z protein expression is an independent predictor of breast cancer survival [11]. We also showed that H2A.Z is necessary for the E2-stimulated proliferative response in MCF-7 cells. These findings were supported by the demonstration that H2A.Z is cyclically incorporated into the enhancer and promoter regions of ERα gene targets and is important for gene induction by the liganded receptor [37]. Combined, these data argue for a feed-forward loop in breast cancer cells in which E2 stimulates H2A.Z production, which in turn maximizes ERα-mediated transcriptional responses. Although the precise DNA sequence determinants of histone placement along genomic DNA to form nucleosomes remain uncertain, recent evidence suggests that H2A.Z-containing nucleosomes are globally enriched at GC-rich sequences in euchromatin [66]. While specific interactions of histones with DNA sequences in the vicinity of EREs have not been investigated on a large scale, these observations raise the possibility that diverse EREs and/or divergent tri-nucleotide spacer sequences could influence transcriptional efficiency through variable effects on the recruitment of H2A.Z to nucleosomes.
5. Conclusion
The present work indicates that the sequence of the tri-nucleotide spacer is non-random at receptor-bound genomic loci, influences ERα-DNA binding affinity, and modulates transactivation potential of the receptor-ligand-DNA complex. This work has implications for understanding which genomic EREs are targeted by ERα, should improve computational prediction of functional EREs within genomic sequences, and describes novel sequence determinants of the estrogen response. Given that a minority of predicted ERE sequences is operative in any given cell type [39] and that diverse ERE sequences are capable of recruiting ERα in vivo, understanding which ERE sequences are functional in a given cell type and milieu remains challenging. The cell type-specific determinants of ERE utilization remain to be fully understood as are the mechanisms by which these determinants are maintained or modulated by the cellular milieu.
Supplementary Material
Listing and genomic coordinates for 1017 high-confidence ERα-bound ChIP sites detected by two independent groups. The ChIP ID numbers (ER_#s) correspond to loci published by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188 (hg18, NCBI Build 36.1). These ChIP loci were reassigned unique “Hit #s” after ERE motif analysis was performed (see main text and Table S2). All ChIP sites were independently detected by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97).
Listing of 646 ERE motifs detected within 509 high-confidence ERα-bound ChIP sites using the TESS software (see main text). Hit #s correspond to genomic coordinates listed in Table S1. Some genomic loci contain more than one ERE motif. ERE motifs that reside within repetitive DNA sequences (determined using RepeatMasker V3.1, http://www.repeatmasker.org/) are indicated. All ChIP sites were independently detected by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188, and by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97.
Prevalence of ERE half-site and middle triad sequences determined from 646 EREs detected within 509 high-confidence ERα-bound ChIP sites. Listed are prevalences of non-unique half sites (A and C), and non-unique triad spacers (B) found in 646 EREs detected from receptor-bound loci. Prevalences of whole EREs, ignoring the triad spacer sequences, are indicated in (D). ChIP sites were detected by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188 and independently by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97).
Base frequency matrix as a function of nucleotide position after excluding EREs residing within repetitive elements. Transcription Element Search Software (TESS) was used to analyze 1017 ERα-bound loci at two different stringencies of detection. Repeat masking was performed and all EREs residing within repetitive elements were excluded from the analysis. High stringency (0–10% deviation from the consensus ERE) and low stringency (10–20% deviation from the consensus ERE) results are shown in Table S4A and S4B, respectively. These data are pooled (Table S4C) to reveal 451 EREs residing outside of repeat-masked sequences. When compared to the expected equal distribution of bases at each position, a statistically significant non-random distribution of sequences at positions 7–9 was indicated by the chi-square test with P values of 7.7E–10, 4.2E–22, and 8.9E–31 for stringencies 0–10% (A), 10–20% (B), and 0–20% (C), respectively.
Oligonucleotide primers used for EMSA and cloning.
Acknowledgments
The authors are grateful to Drs. Graciela Krikun and Charles Lockwood for providing the immortalized human endometrial stromal cell line, and to Drs. Joshua R. Friedman and Robert N. Taylor for scientific discussions.
Abbreviations
- E2
17-β-estradiol/estrogen
- ERα
estrogen receptor-α
- ERE
estrogen response element
- OHT
4-OH-tamoxifen/hydroxytamoxifen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 2.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 3.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 4.Cerillo G, Rees A, Manchanda N, Reilly C, Brogan I, White A, Needham M. The oestrogen receptor regulates NFkappaB and AP-1 activity in a cell-specific manner. J Steroid Biochem Mol Biol. 1998;67(2):79–88. doi: 10.1016/s0960-0760(98)00078-8. [DOI] [PubMed] [Google Scholar]
- 5.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277(5331):1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 6.Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, Kushner PJ. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13(10):1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- 7.Khan S, Abdelrahim M, Samudio I, Safe S. Estrogen receptor/Sp1 complexes are required for induction of cad gene expression by 17beta-estradiol in breast cancer cells. Endocrinology. 2003;144(6):2325–2335. doi: 10.1210/en.2002-0149. [DOI] [PubMed] [Google Scholar]
- 8.Kim K, Barhoumi R, Burghardt R, Safe S. Analysis of estrogen receptor alpha-Sp1 interactions in breast cancer cells by fluorescence resonance energy transfer. Mol Endocrinol. 2005;19(4):843–854. doi: 10.1210/me.2004-0326. [DOI] [PubMed] [Google Scholar]
- 9.Krishnan V, Wang X, Safe S. Estrogen receptor-Sp1 complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J Biol Chem. 1994;269(22):15912–15917. [PubMed] [Google Scholar]
- 10.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol. 2008;41(5):263–275. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, Krausz TN, Olopade OI, Rimm DL, White KP. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102(33):11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 14.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3(6):e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104(12):4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupien M, Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer. 2009;16(2):381–389. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- 17.Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29(12):3413–3423. doi: 10.1128/MCB.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27(14):5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnett DH, Sheng S, Charn TH, Waheed A, Sly WS, Lin CY, Liu ET, Katzenellenbogen BS. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 2008;68(9):3505–3515. doi: 10.1158/0008-5472.CAN-07-6151. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Nawaz Z, Slingerland JM. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol Endocrinol. 2007;21(11):2651–2662. doi: 10.1210/me.2007-0082. [DOI] [PubMed] [Google Scholar]
- 21.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105(49):19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein-Hitpass L, Ryffel GU, Heitlinger E, Cato AC. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988;16(2):647–663. doi: 10.1093/nar/16.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker P, Germond JE, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12(22):8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29(14):2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18(6):1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 26.Tang S, Tan SL, Ramadoss SK, Kumar AP, Tang MH, Bajic VB. Computational method for discovery of estrogen responsive genes. Nucleic Acids Res. 2004;32(21):6212–6217. doi: 10.1093/nar/gkh943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourdeau V, Deschenes J, Laperriere D, Aid M, White JH, Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamalakaran S, Radhakrishnan SK, Beck WT. Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J Biol Chem. 2005;280(22):21491–21497. doi: 10.1074/jbc.M409176200. [DOI] [PubMed] [Google Scholar]
- 29.Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. Anatomy of the estrogen response element. Trends Endocrinol Metab. 2004;15(2):73–78. doi: 10.1016/j.tem.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 30.O’Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18(8):1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 31.Driscoll MD, Sathya G, Muyan M, Klinge CM, Hilf R, Bambara RA. Sequence requirements for estrogen receptor binding to estrogen response elements. J Biol Chem. 1998;273(45):29321–29330. doi: 10.1074/jbc.273.45.29321. [DOI] [PubMed] [Google Scholar]
- 32.Kulakosky PC, McCarty MA, Jernigan SC, Risinger KE, Klinge CM. Response element sequence modulates estrogen receptor alpha and beta affinity and activity. J Mol Endocrinol. 2002;29(1):137–152. doi: 10.1677/jme.0.0290137. [DOI] [PubMed] [Google Scholar]
- 33.Nardulli AM, Romine LE, Carpo C, Greene GL, Rainish B. Estrogen receptor affinity and location of consensus and imperfect estrogen response elements influence transcription activation of simplified promoters. Mol Endocrinol. 1996;10(6):694–704. doi: 10.1210/mend.10.6.8776729. [DOI] [PubMed] [Google Scholar]
- 34.Wu F, Khan S, Wu Q, Barhoumi R, Burghardt R, Safe S. Ligand structure-dependent activation of estrogen receptor alpha/Sp by estrogens and xenoestrogens. J Steroid Biochem Mol Biol. 2008;110(1–2):104–115. doi: 10.1016/j.jsbmb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 36.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23(13):1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009 doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. Unique ERalpha cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22(11):2393–2406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deschenes J, Bourdeau V, White JH, Mader S. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem. 2007;282(24):17335–17339. doi: 10.1074/jbc.C700030200. [DOI] [PubMed] [Google Scholar]
- 41.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 42.Mason CE, Feng-Jue S, Wang C, Session RM, Kallen RG, Sidell N, Yu T, Liu MH, Cheung E, Kallen CB. Location analysis for the Estrogen Receptor-α reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp1188. Epub ahead of print, Jan4, 2010, PMID: 20047966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Yu J, Kallen CB. Two estrogen response element sequences near the PCNA gene are not responsible for its estrogen-enhanced expression in MCF7 cells. PLoS ONE. 2008;3(10):e3523. doi: 10.1371/journal.pone.0003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krikun G, Mor G, Alvero A, Guller S, Schatz F, Sapi E, Rahman M, Caze R, Qumsiyeh M, Lockwood CJ. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology. 2004;145(5):2291–2296. doi: 10.1210/en.2003-1606. [DOI] [PubMed] [Google Scholar]
- 45.Schug J. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence. J.Wiley and Sons; 2008. [DOI] [PubMed] [Google Scholar]
- 46.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75(3):567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 47.Schwabe JW, Chapman L, Finch JT, Rhodes D, Neuhaus D. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure. 1993;1(3):187–204. doi: 10.1016/0969-2126(93)90020-h. [DOI] [PubMed] [Google Scholar]
- 48.Schwabe JW, Fairall L, Chapman L, Finch JT, Dutnall RN, Rhodes D. The cocrystal structures of two zinc-stabilized DNA-binding domains illustrate different ways of achieving sequence-specific DNA recognition. Cold Spring Harb Symp Quant Biol. 1993;58:141–147. doi: 10.1101/sqb.1993.058.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66(14):7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- 50.E.B.C.T.C. Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351(9114):1451–1467. [PubMed] [Google Scholar]
- 51.Kisanga ER, Gjerde J, Guerrieri-Gonzaga A, Pigatto F, Pesci-Feltri A, Robertson C, Serrano D, Pelosi G, Decensi A, Lien EA. Tamoxifen and metabolite concentrations in serum and breast cancer tissue during three dose regimens in a randomized preoperative trial. Clin Cancer Res. 2004;10(7):2336–2343. doi: 10.1158/1078-0432.ccr-03-0538. [DOI] [PubMed] [Google Scholar]
- 52.Krieg AJ, Krieg SA, Ahn BS, Shapiro DJ. Interplay between estrogen response element sequence and ligands controls in vivo binding of estrogen receptor to regulated genes. J Biol Chem. 2004;279(6):5025–5034. doi: 10.1074/jbc.M307076200. [DOI] [PubMed] [Google Scholar]
- 53.Osborne CK, Wiebe VJ, McGuire WL, Ciocca DR, DeGregorio MW. Tamoxifen and the isomers of 4-hydroxytamoxifen in tamoxifen-resistant tumors from breast cancer patients. J Clin Oncol. 1992;10(2):304–310. doi: 10.1200/JCO.1992.10.2.304. [DOI] [PubMed] [Google Scholar]
- 54.Osborne CK, Coronado E, Allred DC, Wiebe V, DeGregorio M. Acquired tamoxifen resistance: correlation with reduced breast tumor levels of tamoxifen and isomerization of trans-4-hydroxytamoxifen. J Natl Cancer Inst. 1991;83(20):1477–1482. doi: 10.1093/jnci/83.20.1477. [DOI] [PubMed] [Google Scholar]
- 55.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85(2):151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 56.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16(8):962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324(5935):1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loven MA, Likhite VS, Choi I, Nardulli AM. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor beta conformation. J Biol Chem. 2001;276(48):45282–45288. doi: 10.1074/jbc.M106211200. [DOI] [PubMed] [Google Scholar]
- 59.Loven MA, Wood JR, Nardulli AM. Interaction of estrogen receptors alpha and beta with estrogen response elements. Mol Cell Endocrinol. 2001;181(1–2):151–163. doi: 10.1016/s0303-7207(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 60.Wood JR, Greene GL, Nardulli AM. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol Cell Biol. 1998;18(4):1927–1934. doi: 10.1128/mcb.18.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15(7):1114–1126. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- 62.Nardulli AM, Shapiro DJ. Binding of the estrogen receptor DNA-binding domain to the estrogen response element induces DNA bending. Mol Cell Biol. 1992;12(5):2037–2042. doi: 10.1128/mcb.12.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kerppola TK, Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 64.Ramirez-Carrozzi VR, Kerppola TK. Dynamics of Fos-Jun-NFAT1 complexes. Proc Natl Acad Sci U S A. 2001;98(9):4893–4898. doi: 10.1073/pnas.091095998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 66.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19(6):967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Listing and genomic coordinates for 1017 high-confidence ERα-bound ChIP sites detected by two independent groups. The ChIP ID numbers (ER_#s) correspond to loci published by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188 (hg18, NCBI Build 36.1). These ChIP loci were reassigned unique “Hit #s” after ERE motif analysis was performed (see main text and Table S2). All ChIP sites were independently detected by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97).
Listing of 646 ERE motifs detected within 509 high-confidence ERα-bound ChIP sites using the TESS software (see main text). Hit #s correspond to genomic coordinates listed in Table S1. Some genomic loci contain more than one ERE motif. ERE motifs that reside within repetitive DNA sequences (determined using RepeatMasker V3.1, http://www.repeatmasker.org/) are indicated. All ChIP sites were independently detected by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188, and by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97.
Prevalence of ERE half-site and middle triad sequences determined from 646 EREs detected within 509 high-confidence ERα-bound ChIP sites. Listed are prevalences of non-unique half sites (A and C), and non-unique triad spacers (B) found in 646 EREs detected from receptor-bound loci. Prevalences of whole EREs, ignoring the triad spacer sequences, are indicated in (D). ChIP sites were detected by our group, Hua, S. et al, Mol Syst Biol. 2008;4:188 and independently by Carroll, J.S. et al, Nat Genet. 2006 Nov;38(11):1289–97).
Base frequency matrix as a function of nucleotide position after excluding EREs residing within repetitive elements. Transcription Element Search Software (TESS) was used to analyze 1017 ERα-bound loci at two different stringencies of detection. Repeat masking was performed and all EREs residing within repetitive elements were excluded from the analysis. High stringency (0–10% deviation from the consensus ERE) and low stringency (10–20% deviation from the consensus ERE) results are shown in Table S4A and S4B, respectively. These data are pooled (Table S4C) to reveal 451 EREs residing outside of repeat-masked sequences. When compared to the expected equal distribution of bases at each position, a statistically significant non-random distribution of sequences at positions 7–9 was indicated by the chi-square test with P values of 7.7E–10, 4.2E–22, and 8.9E–31 for stringencies 0–10% (A), 10–20% (B), and 0–20% (C), respectively.
Oligonucleotide primers used for EMSA and cloning.