Abstract
Plus-strand RNA viruses serve as templates for translation and then transcription by newly synthesized RdRp. A ribosome-binding tRNA-shaped structure (TSS) and upstream hairpin H4 in the 3′ UTR of Turnip crinkle virus (TCV) play key roles in translation and transcription. Second-site mutations generated to compensate for altering the critical asymmetric internal loop of H4 included a three-to-two base alteration in the terminal loop of a 3′ proximal hairpin (Pr) located downstream of the TSS. Unlike the non-deleterious three base alteration, single mutations in Pr loop were detrimental for RdRp transcription while enhancing translation and RdRp binding. One deleterious mutation in the Pr loop altered the structures of both the TSS and H4. These complex interactions in the 3′ UTR support a compact structural arrangement likely permitting RdRp access to a number of residues within a 195-base region including the 3′ end that are necessary for efficient transcription initiation.
Introduction
Replication and translation are key steps in the amplification of viral genomes. For plant RNA viruses, the rate of these processes and control of maximal virus levels are particularly important as innate defenses such as RNA silencing target double-stranded regions in unprotected viral genomes, and progeny migration through intercellular junctions requires maintenance of living cells (Culver and Padmanabhan, 2007). After discarding the viral capsid, plus-strand RNA viruses must be rapidly translated by cellular ribosomes to produce RNA-dependent RNA polymerase (RdRp) for transcription of complementary minus-strands followed by synthesis of progeny plus-strands. Translation and replication are incompatible activities on a single template due to the opposing directions of translating ribosomes and transcribing RdRp (Barton et al., 1999; Gamarnik and Andino, 1998). Mechanisms are therefore necessary to suppress transcription by newly synthesized RdRp until translation produces sufficient levels of the polymerase, and then repress translation when transcription of complementary strands initiates. Such precise control would require finely tuned switches that likely rely on information at least partially specified by the structure of the RNA genome. The increasing number of short and long range RNA:RNA interactions with known or presumptive roles in virus replication and/or translation (Alvarez et al., 2005; Diviney et al., 2008; Hu et al., 2007; Romero-López and Berzal-Herranz, 2009; Serrano et al., 2006; Wu et al., 2009) has led to the view of the RNA genome as a 3D molecule with a fluid conformation for promoting or inhibiting conflicting activities (Edgil and Harris, 2006; Miller and White, 2006; Simon and Gerhke, 2009; Wu et al., 2009).
Recent discoveries that RNA structures required for translation and replication overlap, either at the 5′ or 3′ end of the molecule (Isken et al., 2004; Stupina et al., 2008; Villordo and Gamarnik, 2008; Wu and White, 1999; Gamarnik and Andino, 1998; Liu et al., 2009), suggests mechanisms whereby conformational shifts induced by binding of translation or replication factors inhibit the alternate process until local levels of the binding factor are reduced. This evolving, complex view of basic viral processes necessitates the use of simple model systems to identify core elements and local and distal interacting regulatory sequences. TCV is a small, plus-strand RNA virus with limited coding capacity, making it a useful model for determining RNA structure-function relationships. The single 4054 nt genomic RNA contains only 5 ORFs, one of which (p88) requires readthrough of an amber termination codon (Fig. 1; Hacker et al., 1992). p28 and p88 are the virus-encoded subunits of the RdRp; movement proteins p8 and p9 are translated from the larger of two subgenomic RNAs, while the CP ORF is translated from the smaller subgenomic RNA (Qu and Morris, 2000). TCV is also associated with a small (356 nt), untranslated satellite RNA (satC), composed of a second TCV satRNA, satD, and a 3′ region derived mainly from the 3′ terminus of TCV genomic RNA (Simon and Howell, 1986). Studies that utilize satC have led to the identification of 3′ elements shared with TCV that function specifically in replication (Guo et al., 2009; Simon and Howell, 1986; Sun and Simon, 2006).
Fig. 1.
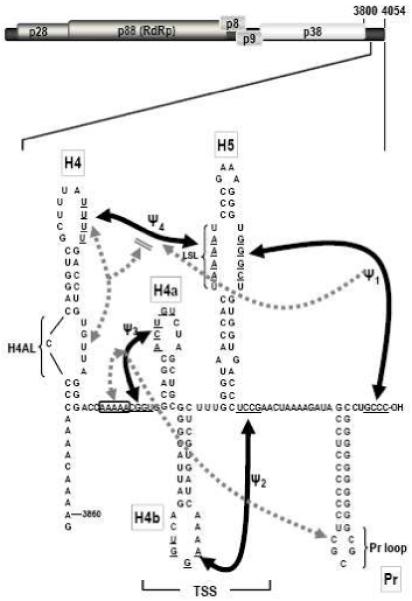
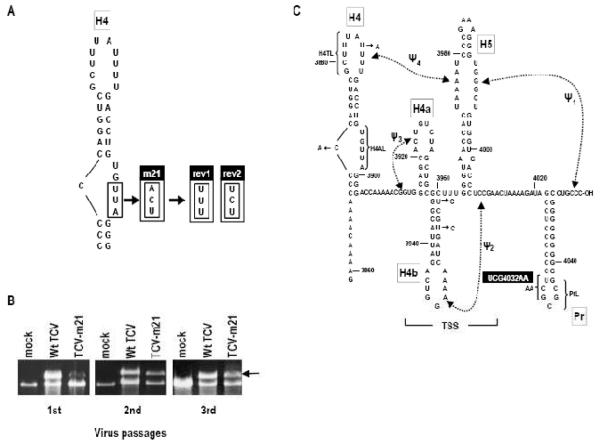
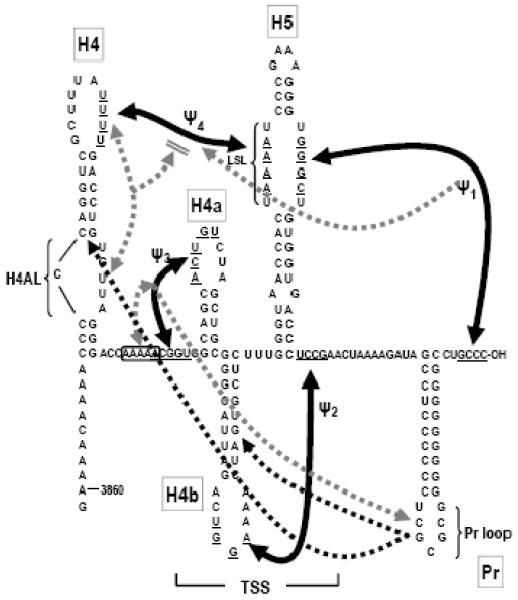
Interactions in the 3′ UTR of TCV. Top, genome organization of TCV genomic RNA. P28 and p88 are required for viral replication, p8 and p9 are proteins that assist in virus cell-to-cell movement and p38 is the single capsid protein. Lower, secondary and tertiary interactions in the 3′ UTR of TCV. The hairpins and canonical Watson-Crick interactions (black lines) were mapped genetically, and are structurally conserved in CCFV. Dotted lines indicate that disrupting an element affected the structure indicated by the arrowhead. Bi-directional arrows indicate that disruption of one element affected the structure of the other. Disruption of H4AL caused a strengthening of Ψ4 (Yuan et al., 2009). The TSS region folds into a tRNA-shaped structure (TSS) that binds to 60S ribosomal subunits and serves as a stable scaffold for interactions between its large symmetrical loop (LSL) and surrounding sequences (McCormack et al., 2008; Stupina et al., 2008; Yuan et al., 2009). Binding of RdRp to the fragment shown causes a conformational shift affecting H4 through at least Pr loop. Names of the hairpins are boxed.
The 3′ UTR of TCV contains a series of hairpins that also form in the most closely related carmovirus, Cardamine chlorotic fleck virus (CCFV) (Fig. 1). From 3′ to 5′, these hairpins are: Pr, which in satC functions as a core promoter (Song et al., 1995; Sun and Simon, 2006) but in TCV requires upstream sequences for function (Sun and Simon, 2006); H5, which is present in all carmoviruses and contains a conserved large symmetrical loop (LSL; Zhang et al., 2004); the juxtaposed hairpins H4a and H4b and upstream hairpin H4, which altogether are unique to TCV and CCFV (McCormack et al., 2008). TCV and CCFV H4 and Pr are highly conserved in both sequence and structure, unlike other hairpins that occupy similar positions in the two viruses but are only structurally conserved (McCormack et al., 2008). Four pseudoknots in this region have also been identified that are important for TCV accumulation and can putatively form in CCFV. Ψ1 connects the 3′ terminus with the 3′ side of the H5 LSL (Zhang et al., 2006b) and is conserved throughout the Tombusviridae (Na and White, 2008; Pogany et al., 2003); Ψ2, which connects the loop of H4b and sequence just downstream of H5, is important for accumulation of TCV and satC (Guo et al., 2009; McCormack et al., 2008; Zhang et al., 2006a); Ψ3, an H-type pseudoknot in the H4a region, is stabilized by adjacent upstream adenylates and is required for accumulation of TCV but not satC (Guo et al., 2009; Yuan et al., 2009); and Ψ4, which connects the loop of H4 and the 5′ side of the H5 LSL (Stupina et al., 2008; Yuan et al., 2009). The region encompassed by Ψ3 and Ψ2 containing H4a, H4b and H5, forms an internal tRNA-shaped structure (TSS) in TCV but not satC (McCormack et al., 2008; Stupina et al., 2008). The TSS was originally discovered using in silico methodology (McCormack et al., 2008) and confirmed experimentally using NMR and small angle X-ray scattering (Zuo et al., 2010). The TSS, which comprises a portion of the TCV 3′ proximal translational enhancer, binds to the P-site of 80S ribosomes through the 60S subunit (Stupina et al., 2008). The TSS is also a highly stable scaffold for interacting with surrounding sequences through the H5 LSL (Yuan et al., 2009).
Alterations in the hairpin H4 asymmetric internal loop (H4AL) negatively affected translation and transcription and reduced virus accumulation to near undetectable levels (Stupina et al., 2008; Yuan et al., 2009). The structure of H4AL was also affected by RdRp binding to the 3′ region, which potentiates a wide-spread conformational shift disrupting elements required for efficient ribosome binding. These results suggested that H4 plays a critical role in the virus life cycle, possibly by mediating the shift between translation and replication that is proposed to occur when RdRp binds to the 3′ end.
To further explore the importance of H4 for accumulation of TCV and search for interactions between the hairpin and local or distal regions, we examined progeny of TCV H4AL mutants inoculated onto plants for second site mutations. A number of mutations were located in the 3′ UTR as well as in discrete structures within the CP ORF. Examination of a three base to two base second site alteration within the loop of the Pr hairpin as well as additional Pr loop mutations revealed the critical importance of the TCV Pr loop, which participates in complex interactions in the 3′ UTR of TCV that likely support a compact 3D structural arrangement that permits RdRp access to a number of elements within a 195 base region including the 3′ end that is necessary for efficient transcription initiation.
Results
TCV with multiple alterations in H4AL generated progeny with primary and second site mutations in plants
One approach for elucidating sequences that interact directly or indirectly with a particular region is to identify compensatory second site mutations that arise spontaneously in mutant viruses due to the error-prone nature of viral RdRp. For the current study, a previously characterized H4AL alteration was used (m21) that reduces viral accumulation by 5-fold in protoplasts (see Fig. 3B; Stupina et al., 2008). m21 contains three altered bases in H4AL, converting the 3′ side of the asymmetric loop sequence from 5′ UGUUA to 5′ UGACU (Fig. 2A). As in Arabidopsis protoplasts, turnip plants inoculated with m21 accumulated reduced levels of virus, which increased to near wt levels following three passages (Fig. 2B). Enhanced accumulation of the mutant viral RNA population suggested either that primary site reversions had occurred during the prolonged propagation, or that second site mutations had arisen that compensated for the original alterations, or both.
Fig. 3.
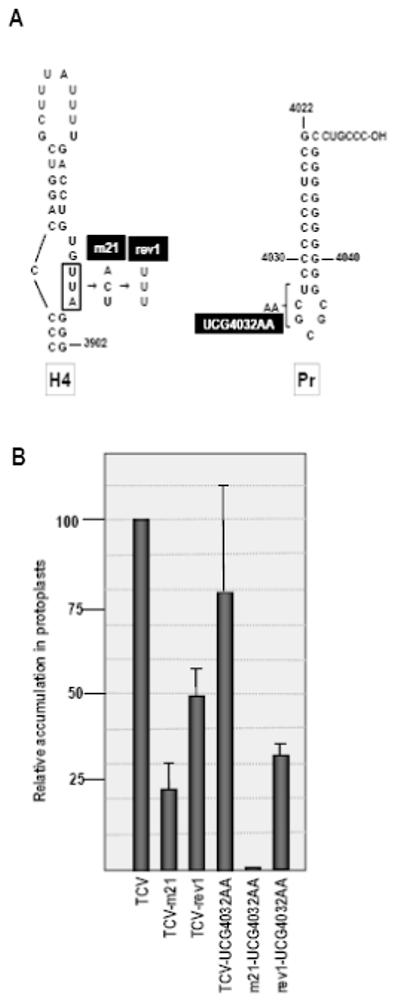
Effect of Pr loop second site mutations on accumulation of wt and mutant TCV. (A) UCG4032AA second site alterations were combined with either wt TCV or TCV containing m21 (TCV-m21) or rev1 (TCV-rev1) sequences. (B) RNA transcripts were assayed for accumulation in protoplasts along with the parental TCV constructs. Values are averages of three independent experiments. Standard deviation is shown.
Fig. 2.
Location of primary and second site alterations recovered in TCV-m21 accumulating after three passages through host plants. (A) Primary site alterations. Either the rev1 or rev2 sequence was found in all recovered clones. (B) Viral RNA levels in plants during different passages. Turnip plants were inoculated with in vitro synthesized transcripts of TCV-m21. Three-weeks postinoculation, RNA was extracted for ethidium-bromide stained gel analysis and used to inoculate fresh seedlings and the process repeated. Arrow denotes TCV genomic RNA and the faster migrating RNA is the 26S ribosomal RNA. (C) Second site alterations located in the 3′ UTR. Each alteration was found in a different clone. Black box denotes the alteration that was investigated further in this report.
The 5′ terminal 1564 nt and 3′ terminal 900 nt were separately cloned from progeny virus isolated three weeks into the third passage. No alterations were found in the 5′ region in the four clones sequenced. In contrast, all 13 3′ region clones had one of two unique sequences in the H4AL region requiring one or two base changes from the original m21 sequence (5′ UGACU[a3right]UGUUU [rev1] or UGUCU [rev2]). In addition, nine of the 13 clones had between one and three second site mutations located in the 3′ UTR and/or upstream CP ORF (Table 1). Of the clones with second site alterations, three contained single mutations, one contained two mutations, and four contained three mutations.
Table 1.
Second site mutations in progeny of TCV-m21
| Clone | Final H4AL sequencea |
Other mutations | Amino acid change? |
|
|---|---|---|---|---|
| Alteration | Location | |||
| 1 | UGUUU | C3872A | H4 | |
| U3885A | H4 | |||
| U3956C | H4b | |||
| 2 | UGUUU | U3330C | CP ORF | F→L |
| A3597G | CP ORF | N→S | ||
| U3959C | H4b | |||
| 3 | UGUUU | 4032UCG→AA | Pr | |
| 4 | UGUUU | A3475G | CP ORF | No change |
| A3709G | CP ORF | No change | ||
| U3741C | CP ORF | V→A | ||
| 5 | UGUUU | C3239U | CP ORF | L→F |
| C3630U | CP ORF | P→L | ||
| 6 | UGUUU | U3329C | CP ORF | F→L |
| 7 | UGUUU | G3561A | CPORF | G→E |
| 8 | UGUCU | A3420G | CP ORF | Q→R |
| U3508G | CP ORF | No change | ||
| U3510G | CP ORF | V→G | ||
| 9 | UGUCU | G3620A | CP ORF | E→K |
| 10,11 | UGUUU | none | ||
| 12,13 | UGUCU | none | ||
wt sequence is UGUUA; rn21 sequence is UGACU
The second site alterations in the CP ORF will be presented elsewhere (M. Young, X. Yuan, K. Shi, and A.E. Simon, unpublished). Second site mutations in the 3′ UTR were found opposite the original mutated sequence in the H4AL (C3872A), in the H4 terminal loop (U3885A), in two positions in the stem of H4b (U3956C, U3959C), or a replacement of three bases (UCG) in the Pr lower stem and loop with two adenylates (UCG4032AA) (Fig. 2C). For this report, we examined the latter alteration, since it suggested a previously unknown relationship between H4 and Pr. This was unexpected since disrupting Ψ3 caused conformational changes in Pr loop without any discernable structural changes to H4AL or any region between Ψ3 and Pr loop. At the time, this result was interpreted as suggesting that the Pr loop interacted specifically with the Ψ3 region (Yuan et al., 2009).
Mutations in Pr loop enhance translation and reduce transcription
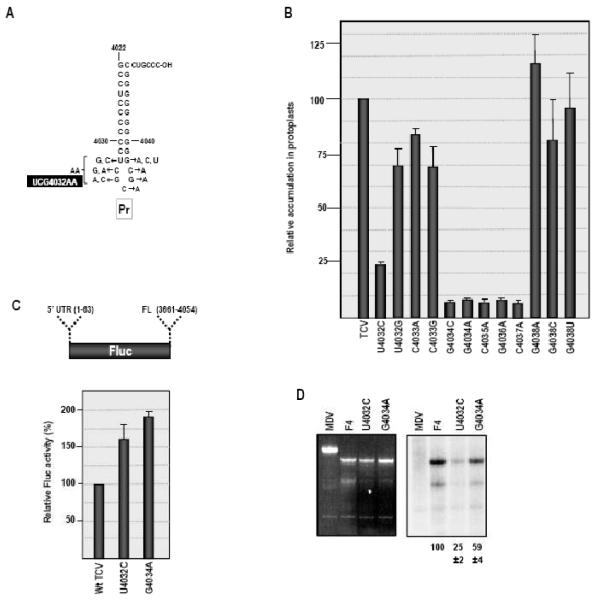
The Pr loop second site mutations were found in TCV containing the rev1 sequence in H4AL (TCV-rev1; Fig. 3A). Since UCG4032AA could have arisen before, during or after the 2 nt alteration from the m21 sequence to rev1, the following three constructs were generated: wt TCV+UCG4032AA, TCV-m21+UCG4032AA and TCV-rev1+UCG4032AA. Mutant and wt TCV were inoculated into Arabidopsis protoplasts and levels of accumulating virus determined by RNA gel blots at 40 hours postinoculation (hpi). TCV-m21 accumulated to 21% of wt levels, while TCV-rev1 levels were enhanced, accumulating to 50% of wt TCV (Fig. 3B). UCG4032AA reduced the average accumulation of wt TCV by 20% and reduced TCV-rev1 levels by 36%. In contrast, UCG4032AA reduced accumulation of TCV-m21 to below detectable levels. These results suggest that the three base changes in UCG3042AA arose in some combination during the transition of TCV-m21 to TCV-rev1. In addition, the incompatibility of these Pr mutations with the m21 alteration supports the existence of a relationship between H4 and Pr loop.
Since the current result suggested a possible direct or indirect interaction between Pr loop and H4AL, we wanted to explore further the sequence and structural requirements of Pr loop. Single base alterations were generated in Pr loop and flanking U4032:G4038 residues in wt TCV, and mutant virus accumulation was assayed for in protoplasts (Fig. 4A). Alteration of U4032 to C, which should create a strong C4032:G4038 base-pair, was not well tolerated, with mutant TCV accumulating to only 20% of wt TCV levels (Fig. 4B). In contrast, altering U4032 to G, producing a G-G mismatch, retained 70% of wt TCV activity. Similarly, altering G4038 to C, U or A had no significant negative effect on TCV accumulation (Fig. 4B). This suggests that only weak U:G or U:A pairings or no canonical base-pairing is well tolerated at this location.
Fig. 4.
Pr loop sequence is important for viral RNA replication. (A) Mutations generated in the Pr loop region of TCV. The second site change UCG4032AA mutation is also shown. (B) Accumulation of mutant TCV in protoplasts. Mutations shown in (A) were generated in full-length TCV cDNA and RNA transcripts assayed for accumulation in protoplasts. Values are averages of three independent experiments. Standard deviation is shown. (C) Effect of mutations on translation in vivo. Two of the Pr loop mutations were engineered into a construct containing a single luciferase reporter ORF flanked by the 3′ 393 nt and the precise 5′ UTR of TCV. Transcripts were transformed into protoplasts along with a control RLuc transcript and luciferase assayed at 18 hpi. Relative values shown are from three independent experiments and standard deviations are shown. (D) In vitro transcription of F4 RNA wt and mutant RNA fragments (positions 3859-4054 nt) using purified recombinant TCV RdRp and radioactive nucleotides. A representative gel and autoradiogram is shown. Below the lanes are the normalized values from three independent experiments. Standard deviations are shown.
To determine if reduced accumulation of TCV-U4032C in protoplasts was due to disruption of transcription or translation, U4032C was generated in a single luciferase construct containing the 5′ UTR and the 3′ 393 nt of TCV and translation was assayed for in protoplasts. RNA transcripts containing U4032C produced 60% more luciferase than wt transcripts (Fig. 4C), suggesting that the alteration did not reduce but rather enhanced translation. In contrast, when mutant TCV RNA fragments (F4+U4032C) were subjected to in vitro transcription using purified recombinant TCV RdRp, transcription was reduced by 75% compared with wt F4 (Fig. 4D). These results strongly suggest that reduced accumulation of TCV-U4032C is caused, at least in part, by a defect in minus-strand synthesis.
To determine the importance of the remaining residues in Pr loop, additional single base alterations were generated at each loop position and mutant virus was assayed for accumulation in protoplasts (Fig. 4A). Altering C4033 to A or G reduced TCV accumulation by less than 30% (Fig. 4B). However, altering single bases from G4034 through C4037 caused similar, substantial reductions in TCV levels, with viral genomic RNA accumulating to less than 10% of wt. One of these TCV Pr mutants, G4034A, was selected for further analysis since this alteration was present in the UCG4032AA second site changes that were not significantly detrimental to wt TCV accumulation. As with U4032C, G4034A enhanced translation of luciferase in vivo (Fig. 4C) and reduced transcription of minus strands in vitro (Fig. 4D). Since G4034A had less of an effect on transcription than U4032C, G4034A likely has additional negative properties beyond repression of minus-strand synthesis as measured in vitro.
Alterations in the Pr loop affect the structure of H4b as well as the RdRp-mediated conformational shift
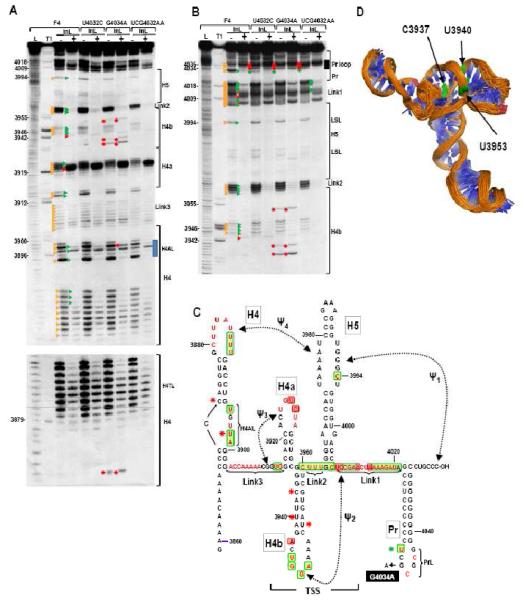
To explore possible structural interactions between H4AL and Pr loop, in-line probings were conducted on Pr loop mutants F4+UCG4032AA, F4+U4032C and F4+G4034A (Fig. 5). In-line probing examines cleavages of the phosphate backbone mediated by 2′ hydroxyl groups, which require in-line topology between the oxygen nucleophile, the phosphate acceptor and the oxyanion leaving group. This topology only exists if the nucleoside can rotate relative to the phosphate-sugar backbone, which does not occur if the nucleoside is overly constricted by canonical or non-canonical hydrogen bonding. The in-line cleavage pattern of wt F4 was very similar to that previously published (Yuan et al., 2009), but some differences were also found. These differences were mainly qualitative, with some previous weak cleavages being more prominent. The precise in-line cleavage pattern is sensitive to minor differences in temperature (X. Yuan and A. E. Simon, unpublished) and assay conditions and thus wt and mutant transcripts are always assayed simultaneously. Outside of the Pr loop region, F4+UCG4032AA and F4+U4032C had no significant discernable differences with wt F4 in the cleavage pattern of any residue, with the exception of position 4032, whose strong cleavage was eliminated in both mutants (Fig. 5B). The conformational change in this residue supports the prediction that U4032C generates a strong C:G pair at this location, significantly reducing the flexibility of C4032. In contrast, the single alteration in F4 containing the highly detrimental mutation G4034A (F4+G4034A) caused conformational changes in several upstream locations: 1) the strong cleavage at U4032 was eliminated; 2) a new cleavage was found at position C3873 at the 5′ base of the H4 upper stem; 3) the stem of TSS hairpin H4b contained new cleavages at positions C3937, U3940, and U3953.
Fig. 5.
In-line structure probing of wt and Pr loop mutants. (A) In-line cleavage of F4 containing U4032C, G4034A or second site alteration UCG4032AA, with (+) and without (−) RdRp. Representative gels are shown. Positions of the most prominent cleavages in wt F4 (F4) are denoted by orange triangles. Red triangles and green triangles denote residues that exhibited greater or lesser cleavage, respectively, in the presence of RdRp. Red and green asterisks denote residues that show enhanced and reduced cleavage, respectively, in mutants compared with wt F4. Lower gel is a longer exposure of the upper gel that reveals an additional difference in F4+G4034A. L, OH-generated ladder; T1, Rnase T1 digest (specific for guanylates); InL, cleavages generated after 14 h incubation of RNAs at 21°C. Incubation in the presence of RdRp was reduced to 1 h. Blue box denotes the H4AL region. (B) Longer run of the samples shown in (A) to enlarge the region from H4b through Pr loop. Black box denotes the Pr loop region. (C) Location of flexible bases in wt F4. Prominent cleaved residues are in red. Additional residues cleaved in F4+G4034A (with and/or without RdRp binding) are labeled with red asterisks and reduced cleavage is denoted by a green asterisk. Green and red boxes denote residues with reduced or enhanced cleavage, respectively, upon RdRp binding to wt F4. Position of the G4034A mutation is shown. (D) Location of the new cleavages in F4+G4034A H4b in the NMR/SAXS structure of the TSS. Residues with enhanced flexibility are in green. Figure courtesy of Yun-Xing Wang and Xiaobing Zuo (NCI).
In the presence of the TCV RdRp, a widespread conformational shift occurs in wt F4 that substantially alters the structure of the fragment including pseudoknot (Ψ3), which is necessary for robust ribosome binding (Yuan et al., 2009; Fig. 5). This RdRp-induced conformational shift results in significant changes in the flexibility of wt F4 RNA, with a distinctive in-line cleavage pattern evident after only 1 h of incubation with the RdRp (compared with 14 h required for a similar overall level of cleavage in the absence of RdRp). The cleavage patterns of F4+UCG4032AA and F4+U4032C outside of the Pr loop region were virtually identical to the wt F4 pattern, with the exception that positions 4018 to 4020 at the 3′ edge of Link 1 were less flexible in F4+UCG4032AA. In F4+G4034A, the new cleavages in H4b and H4 were maintained in the presence of RdRp, and the conformation of H4AL was altered with a stronger cleavage evident at position U3898. Altogether, the in-line probing results indicate that G4034A in the Pr loop affects, either directly or indirectly, the conformation of the upstream TSS and H4 regions. In addition, although G4034A is present in the UCG4032AA second site mutations, fragments containing the latter alterations did not exhibit any of the additional conformational changes associated with G4034A. Furthermore, while mutations that disrupt Ψ3 affected the conformation of Pr loop (Yuan et al., 2009), G4034A had no discernable effect on the Ψ3/H4a region.
RdRp binding is enhanced to Pr loop mutants
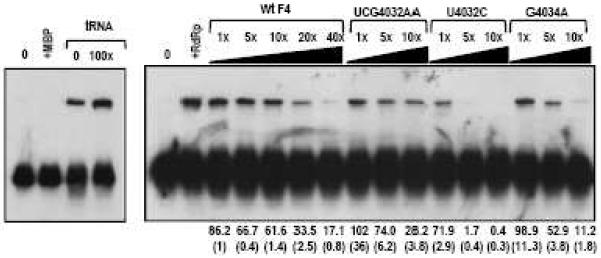
A previously designed electrophoretic mobility shift competition assay was used to determine if the Pr loop mutations affect RdRp binding to F4 (Sun and Simon, 2006). This assay utilizes unlabelled F4 and F4 mutant RNAs as competitors for RdRp binding to radiolabeled TCV satellite satD minus strands (satD−). As shown in Fig. 6, F4+UCG4032AA and wt F4 are similar competitors for binding to satD-. In contrast, F4+U4032C and F4+G4034A complete substantially better than wt F4 for binding to TCV RdRp, with U4032C being the strongest competitor. These results indicate that the entire sequence of Pr loop is not required for efficient RdRp binding. In addition, the ability of RdRp to bind these mutant F4 fragments is inversely correlated with template activity in RdRp-mediated in vitro transcription assays (Fig. 4D).
Fig. 6.
RdRp binding to Pr loop mutants. Electrophoretic mobility shift competition assays were conducted using radiolabeled satD minus strands (satD−) and increasing amounts of unlabeled competitor RNAs. Left panel, controls with no RdRp (0), maltose binding protein (+MBP) or TCV RdRp (brackets) with 0 or 100X yeast tRNA as competitor. Right panel, no RdRp (0); plus RdRp alone (+RdRp); or plus RdRp and increasing amounts of wt and mutant competitor RNAs (brackets). The assays were conducted four times. Average values for percent shifted RNA are given and standard deviation is in parentheses.
Discussion
The search for second site mutations that compensate for deleterious primary mutations is a well established approach for identifying putative interacting regions in RNA viruses, such as Ψ2 in TCV-associated satC (Zhang et al., 2006a). The critical importance of the asymmetric internal loop of H4 in both translation and replication (Stupina et al., 2008; Yuan et al., 2009) suggested that this element might have short or long range interactions with other elements (e.g., at the 5′ end).
All 3′ region clones derived from multiple passages of TCV-m21 in turnip contained one or two base changes in H4AL (rev1 and rev2), converting the original three base m21 alteration to sequences that more closely resembled the wt sequence. Most clones also contained second site changes in the 3′ UTR and/or upstream CP ORF (Table 1). The three base alteration in Pr loop (UCG4032AA) was surprising since previous results strongly suggested an interaction between the Pr and Ψ3 (Yuan et al., 2009), which had no known connection with H4. UCG4032AA had only a modest negative effect on wt TCV and a moderate negative effect on TCV-rev1 accumulation, while strongly suppressing accumulation of TCV-m21. Since the Pr loop alterations were not compensatory with TCV-m21, the base changes in Pr loop likely occurred during the primary site reversion to the rev1 sequence.
Since UCG4032AA did not significantly affect the accumulation of wt TCV, we wanted to explore whether any base specificity exists in the Pr loop region in TCV. U4032, which is located opposite G4038 at the base of the Pr stem, is highly flexible in the structure of the F4 fragment but loses its flexibility in the presence of the TCV RdRp (Yuan et al., 2009). All alterations at either positions 4032 or 4038 that eliminated canonical Watson-Crick base pairing or created a weak U:A pair were well tolerated. However, a strong C:G pair at this location reduced accumulation by over 75%. Uracil residues are frequently located at the ends of helixes and have been suggested to serve as helix breakers (Nissen et al., 2001). The ability of residues that do not generate a Watson-Crick pair in this location to support high levels of virus accumulation supports a non-specific, helix breaking role for U4032 and indicates that paired bases in this location are not required and can be inhibitory for virus accumulation. As shown in Fig. 7, U4032 and G4038 are now placed in the Pr loop and not at the base of the Pr stem.
Fig. 7.
Complex interactions in the 3′ UTR of TCV. See legend to Fig. 1. New interactions between Pr loop and H4 and the TSS elucidated in this study are shown by black dotted lines. The precise make-up of these interactions and whether the interactions are direct or indirect, remain unresolved. In addition, U4032 and G4038 are now placed in the Pr loop and not at the base of the Pr stem, since paired residues in this location are not required and can be inhibitory for viral accumulation.
Alterations at position 4033 in Pr loop had only a modest (25%) reduction in TCV levels (Fig. 4B). In contrast, changing any of the adjacent Pr loop residues (4034GCGC) reduced accumulation in protoplasts by over 90%. Interestingly, this region includes the G4034A alteration found in UCG4032AA, indicating that G4034A is only detrimental in the context of adjacent wt residues. The importance of maintaining specific Pr loop residues supports previous findings that TCV was fully functional when associated with the Pr of Cardamine chlorotic fleck virus (CCFV), which shares the identical Pr loop with TCV, but did not accumulate with the unrelated Pr of Japanese iris necrotic ring virus (JINRV; McCormack et al., 2008).
Our previous findings suggested an interaction between Ψ3/H4a and Pr loop (Yuan et al., 2009). Our current results indicate that at least one mutation in Pr loop disrupts the structure of the upstream TSS, and H4 just outside of H4AL. These interactions, depicted in Fig. 7, could be either direct or indirect. Untranslated satC, which does not contain H4 or a replication requirement for Ψ3 (Guo et al., 2009), can tolerate a wide variety of sequences in its Pr loop (Carpenter and Simon, 1998; Song et al., 1995; Stupina et al., 1997). SatC replicates poorly with the Pr of TCV (Zhang et al., 2006b), supporting the hypothesis that the TCV Pr is constituted to fulfill additional function(s) besides promoting replication that includes upstream interactions. This hypothesis is also supported by finding that G4034A causes distinctive structural changes in the stems of H4b and H4. In the solution structure of the TSS, H4b is centrally located in a coaxially stacked, quasi-continuous helix composed of H4a, Ψ3 and Ψ2 (Fig. 5D). Pr loop may function to stabilize the stacked helices, possibly assisting in TSS interaction with H4. It should be noted, however, that the vast majority of residues in the TSS and flanking regions maintain their structural identity in F4+G4034A and thus enhanced flexibility of H4b residues in this mutant do not lead to significant pleiotropic effects on the overall structure in the region. This supports previous findings that the TSS is a particularly stable scaffold for interactions with surrounding sequences (Yuan et al., 2009). Interestingly, second site mutations were also recovered in the H4b stem, which might enhance the flexibility of the TSS stacked helices to assist in the interaction between mutated H4 and the TSS.
The Pr loop of JINRV (5′ GUGGCA), as with all other Carmovirus Pr loop sequences, is unrelated to that of TCV and CCFV. While 14 carmoviruses (all except for Galinsoga mosaic virus, whose membership in the Carmovirus genus is in question; Ciuffreda et al., 1998) contain Pr, H5 and Ψ1, only 10 contain a hairpin equivalent to H4b (with 7 able to putatively form Ψ2), and only 5 also contain an H4a equivalent (Table 2). Only CCFV and TCV have H4 and the potential for forming Ψ3 and Ψ4, and the CCFV combination of hairpins and pseudoknots is fully functional when replacing the equivalent region in TCV (McCormack et al., 2008). These differences among Carmoviruses suggest that most members of this genus, as well as satC, differ from TCV and CCFV in how their 3′ terminal Pr hairpins participate in 3′ end structure and virus replication. The base changes and base deletions that produced the current satC Pr from the TCV Pr following the recombination events that generated the satRNA, evolved a promoter that is more capable of promoting complementary strand synthesis in the absence of upstream TCV-specific interactions compared with the progenitor TCV Pr (Sun et al., 2006). TCV does not detectably accumulate with the Pr hairpin of satC (Zhang et al., 2006b), supporting the hypothesis that the interaction of Pr with upstream elements is needed for the TCV genomic RNA’s unique additional requirements. Additional support for this hypothesis comes from our previous finding that the TCV Pr promotes abortive cycling by the RdRp only in the context of upstream TCV sequences and not when associated with satC (Nagy et al., 1997), which lacks the ability to form a TSS or bind to ribosomes (Guo et al., 2009). Impeding the initial progression of the RdRp, which leads to the synthesis of short abortive products, could therefore be a consequence of interaction between the Pr and the TCV TSS.
Table 2.
Conservation of elements in the 3′ UTR of Carmoviruses
| Name | Accession # | Nucleotide identity |
Pr | H5 | H4ba | H4ac | H4 | Ψ 1 | Ψ 2b | Ψ 3 | Ψ 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Turnip crinkle virus (TCV) | NC_003821 | 100 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| TCV satellite C (satC) 3′ related region | 90 | Yes | Yes | Yes | Yes | No | Yes | Yes | No | No | |
| Cardamine chlorotic fleck virus (CCFV) | NC_001600 | 66 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Japanese iris necrosis virus (JINRV) | NC_002187 | 55 | Yes | Yes | Yes | Yes | No | Yes | Yes | No | No |
| Soybean yellow mottle mosaic virus (SYMMV) | NC_011643 | 54 | Yes | Yes | Yes | Yes | No | Yes | Yes | No | No |
| Pelargonium line pattern virus (PLPV) | NC_007017 | 54 | Yes | Yes | Yes | Yes | No | Yes | No | No | No |
| Carnation mottle virus (CarMV) | NC_001265 | 55 | Yes | Yes | Yes | No | No | Yes | Yes | No | No |
| Nootka lupine vein clearing virus (NLVCV) | NC_009017 | 53 | Yes | Yes | Yes | No | No | Yes | Yes | No | No |
| Calibrachoa mottle virus (CbMV) | GQ244431 | 53 | Yes | Yes | Yes | No | No | Yes | Yes | No | No |
| Saguaro cactus virus (SCV) | NC_001780 | 54 | Yes | Yes | Yes | No | No | Yes | No | No | No |
| Pelargonium flower break virus (PFBV) | NC_005286 | 53 | Yes | Yes | Yes | No | No | Yes | No | No | No |
| Cowpea mottle virus (CPMoV) | NC_003535 | 55 | Yes | Yes | No | No | No | Yes | No | No | No |
| Angelonia flower break virus (AFBV) | NC_007733 | 54 | Yes | Yes | No | No | No | Yes | No | No | No |
| Melon necrotic spot virus (MNSV) | NC_001504 | 53 | Yes | Yes | No | No | No | Yes | No | No | No |
| Hibiscus chlorotic ringspot virus (HCRSV) | NC_003608 | 56 | Yes | Yes | No | No | No | Yes | No | No | No |
| Galinsoga mosaic virus (GaMV) | NC_001810 | 53 | No | Yesd | No | No | No | Yes | No | No | No |
hairpin within 7 bases 5′ of H5.
3-4 nt complement between H4b loop sequence and sequence within 2 nt of the 3′ of base of H5
hairpin immediately 5′ of H4b.
structurally related to the Tombusvirus equivalent of H5 (SL3; Pogany et al., 2003)
G4034A also caused enhanced translation and enhanced binding to the RdRp, suggesting that positioning of the Pr within the 3D structure of the region sterically hinders cis- or trans-acting contacts. Curiously, while F4 with Pr loop alterations bind more tightly to the polymerase (Fig. 6), this did not correlate with enhanced transcription of the template in vitro (Fig. 4D). One possibility is that the RdRp is unable to efficiently access the 3′ end, which participates in Ψ1 with the H5 LSL, when bound to F4 containing a spatially altered Pr. A second possibility is that the RdRp is bound too tightly to its promoter, affecting release and initiation of transcription of mutant templates. The compact arrangement of hairpins, pseudoknots and non-canonical interactions that characterize the TCV 3′ structure likely prevents early RdRp binding to the region before sufficient quantities of the polymerase have been synthesized; when bound, the compact topology of the region may permit simultaneous RdRp access to multiple elements including the Pr hairpin and the 3′ end, which may be required for the appropriate RdRp-mediated conformational switch that is proposed to restrict translation and promote transcription.
It is not known how the UCG4032AA alteration is able to maintain the function of the Pr loop in its interaction with upstream sequences in an otherwise wt TCV. Pairs of adenylates that are not involved in stems of secondary structure elements are frequently conserved and buried within the 3D structure of large RNAs while participating in significant, non-canonical tertiary contacts with consecutive and stacked Watson-Crick base pairs, known as A-minor motifs (Nissen et al., 2001; Lescoute and Westhof, 2006). Two of the four types of A-minor motif interactions are not specific for adenylates, but contacts with the receptor RNA minor groove helix are optimized when the base is an adenine. A-minor motifs stabilize the tertiary structure of RNA and serve as important signals for protein recognition (Lescoute and Westhof, 2006). UCG4032AA may therefore represent a second solution to the stable, non-canonical packing of the Pr loop with upstream elements in wt TCV.
Materials and Methods
Isolation of Second Site Mutations
TCV-m21, containing a 3 base alteration in the asymmetric loop of H4 (3897UUA to ACU) was used to search for second site mutations. Plants (Turnip cv Just Right) and the two true leaf stage were mechanically inoculated with in vitro transcribed TCV-m21 RNA (2 μg for each of two leaves), as described previously (Kong et al., 1997). Total RNA was extracted at 21 days post inoculation (dpi) and used to re-inoculate seedlings (5 μg for each of two leaves). The process was repeated a total of three times. Total RNA isolated from the third passage was used for RT-PCR amplification of fragments corresponding to the 5′ end 1564 b or 3′ end 900 b, which were cloned and subjected to sequencing.
Construction of TCV mutants
All mutants were constructed by oligonucleotide-mediated site-directed mutagenesis starting with pTCV66, which contains full-length wt TCV (TCV-ms isolate) sequence located downstream of a T7 RNA polymerase promoter. All mutants were subjected to regional sequencing to confirm the mutation sites. Plasmids containing wt and TCV mutants were linearized with SmaI and used as templates for in vitro transcription using T7 RNA polymerase.
Protoplast Preparation, Inoculation and RNA Gel Blots
Protoplasts were prepared from callus cultures of Arabidopsis thaliana ecotype Col-0 as previously described (Zhang et al., 2006a). To assay for accumulation of TCV, 20 μg of uncapped in vitro transcribed wt or mutant TCV RNA was inoculated onto protoplasts using polyethylene glycol (PEG) and total RNA extracted at 40 hpi was denatured and subjected to electrophoresis through 1% agarose gels, transferred to a nitrocellulose membrane and viral RNA detected using a random 32P-labeled TCV-specific probe. Levels of TCV gRNA were normalized to ribosomal RNA.
In Vivo Translation Assays
A previously described reporter construct, consisting of the 5′ UTR of TCV (positions 1-63) upstream of Fluc and the 3′ end region of TCV (positions 3661-4054) downstream of Fluc, was used to assay for translation in vivo (Stupina et al., 2008). Specific 3′ mutations were engineered into the construct by PCR amplification of mutant fragments using oligonucleotides containing the mutations, which were inserted into the NruI/SspI site in place of the wt fragment. All constructs were confirmed by sequencing. Plasmids were linearized with SspI and used as templates for in vitro transcription using T7 RNA polymerase. Uncapped, in vitro transcribed RNA (30 μg) was inoculated onto Arabidopsis protoplasts along with 10 μg of uncapped in vitro transcripts containing internal control Rluc. Protoplasts were harvested at 18 hpi and cell extracts prepared for luciferase measurements using the dual-luciferase reporter assay system (Promega) and a TD20/20 luminometer (Turner Designs) according to the instructions of the manufacturer.
In-Line Structure Probing
In-line structure probing was carried out as previously described (Yuan et al., 2009). Briefly, wt F4 and mutant RNA fragments were synthesized from plasmids in vitro using T7 RNA polymerase, purified following agarose gel electrophoresis, were 5′-end labeled using T4 polynucleotide kinase and [γ-32P]-ATP and then re-purified through 5% polyacrylamide gels. 5′ labeled RNA fragments were heated to 75°C and slow-cooled to room temperature. RNA (5 pmol) was then incubated at 25°C in 50 mM Tris-HCl (pH 8.5) and 20 mM MgCl2 for 14 h without RdRp or for 1 h in the presence of 7.5 pmol of purified RdRp. RdRp was expressed in E. coli and purified as a recombinant protein with maltose binding protein (MBP), as previously described (Rajendran et al. 2002). RNA cleavage ladders were generated by incubating 5′- end labeled RNA in 1 μg yeast tRNA, 50 mM NaHCO3/Na2CO3 (pH 9.2) and 1 mM EDTA for 5 min at 95°C. RNase T1 digests were generated by incubating denatured 5′ end labeled RNA in 1 μg yeast tRNA, 20 mM sodium citrate (pH 5.0), 1 mM EDTA, 7 M urea and 1 U RNase T1 (Ambion) for 15 min at room temperature. All reactions were ethanol precipitated, heated at 95°C for 5 min and then subjected to electrophoresis through denaturing 8 M urea-8% polyacrylamide gel followed by autoradiography.
In Vitro RdRp Assays
TCV66 was used as a template for PCR amplification to obtain wt or mutant fragments containing the T7 RNA polymerase promoter and genomic sequences corresponding to positions 3859 to 4054. Transcripts synthesized by T7 RNA polymerase were subjected to in vitro RdRp assays using purified recombinant TCV RdRp, as previously described (Yuan et al., 2009). Briefly, 1 μg of purified RNA was added to a 25 μl reaction mixture containing 50 mM Tris-HCl (pH 8.2), 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM each of ATP, CTP, GTP, 0.01 mM UTP, 10 μCi of [α-32P] UTP, and 2 μg of RdRp. After 90 min incubation at 20°C, 1 μg of tRNA was added, and the mixture was subjected to phenol-chloroform extraction and ammonium acetate-isopropanol precipitation. Radiolabeled products were subjected to electrophoresis through denaturing 8 M urea-5% polyacrylamide gel. The gel was then stained with ethidium bromide to detect the template levels and subjected to autoradiography.
Electrophoretic Mobility Shift Competition Assays
Each reaction contained approximately 10 ng of 5′ end-labeled satD minus strands, 1 μg of RdRp, and competitor RNAs in excess, as indicated. The RNAs and RdRp were incubated together for 30 minutes at room temperature in a buffer containing 50 mM Tris pH 8.2, 10 mM MgCl2, 10 mM dithiothreitol, and 10% glycerol. 1.5 μg of maltose binding protein was used as a protein control and yeast tRNA was used as a control competitor. Reactions were subjected to electrophoresis on a 1% agarose gel in an ice bath at 25 mA for 35 minutes. The gel was then dried and subjected to autoradiography.
Acknowledgements
This work was supported by grants from the U.S. Public Health Service (GM 061515-05A2/G120CD) and National Science Foundation (MCB-0615154) to A.E.S. M. Young was supported by NIH training grant T32 AI51967-01. We are grateful to Yun-Xing Wang and Xiaobing Zuo (NCI) for Fig. 5D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alvarez DE, Lodeiro M, Ludueña SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the Dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Morasco BJ, Flanegan JB. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CD, Simon AE. Analysis of sequences and putative structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 1998;26:2426–2432. doi: 10.1093/nar/26.10.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda P, Rubino L, Russo M. Molecular cloning and complete nucleotide sequence of galinsoga mosaic virus genomic RNA. Arch. Virol. 1998;143:173–180. doi: 10.1007/s007050050277. [DOI] [PubMed] [Google Scholar]
- Culver JN, Padmanabhan MS. Virus-induced disease: altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 2007;45:221–243. doi: 10.1146/annurev.phyto.45.062806.094422. [DOI] [PubMed] [Google Scholar]
- Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008;82:9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Lin W, Zhang J, Simon AE, Kushner DB. Structural plasticity and rapid evolution in a viral RNA revealed by in vivo genetic selection. J. Virol. 2009;83:927–939. doi: 10.1128/JVI.02060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker DL, Petty IT, Wei N, Morris TJ. Turnip crinkle virus genes required for RNA replication and virus movement. Virology. 1992;186:1–8. doi: 10.1016/0042-6822(92)90055-t. [DOI] [PubMed] [Google Scholar]
- Hu B, Pillai-Nair, Hemenway C. Long-distance RNA-RNA interactions between terminal elements and the same subset of internal elements on the potato virus X genome mediate minus- and plus-strand RNA synthesis. RNA. 2007;13:267–280. doi: 10.1261/rna.243607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Yu H, Behrens S-E. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Wang J, Simon AE. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell. 1997;9:2051–2063. doi: 10.1105/tpc.9.11.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. The A-minor motifs in the decoding recognition process. Biochimie. 2006;88:993–999. doi: 10.1016/j.biochi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Bioch. Biophys. Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JC, Yuan X, Yingling YG, Zamora RE, Shapiro BA, Simon AE. Structural domains within the 3′ UTR of Turnip crinkle virus. J. Virol. 2008;82:8706–8720. doi: 10.1128/JVI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, White KA. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H, White KA. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology. 2006;345:305–316. doi: 10.1016/j.virol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Carpenter CD, Simon AE. A novel 3′ end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Fabian MR, White KA, Nagy PD. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22:5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Morris TJ. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran KS, Pogany J, Nagy PD. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 2002;76:1707–1717. doi: 10.1128/JVI.76.4.1707-1717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P, Pulido MR, Sáiz M, Martínez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- Simon AE, Gerhke L. RNA conformational changes in the life cycles of RNA viruses, viroids, and virus-associated RNAs. BBA. 2009;1789:571–583. doi: 10.1016/j.bbagrm.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, Howell SH. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′ end of the helper virus genome. EMBO J. 1986;5:3423–3428. doi: 10.1002/j.1460-2075.1986.tb04664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Simon AE. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Kasprzak W, Shapiro BA, Dinman JD, Simon AE. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Simon AE. A cis-replication element functions in both orientations to enhance replication of Turnip crinkle virus. Virology. 2006;352:39–51. doi: 10.1016/j.virol.2006.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Pogany J, Na H, Nicholson BL, Nagy PD, White KA. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathogens. 2009;5:e1000323. doi: 10.1371/journal.ppat.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, White KA. A primary determinant of cap-independent translation is located in the 3′-proximal region of the Tomato bushy stunt virus genome. J. Virol. 1999;73:8982–8988. doi: 10.1128/jvi.73.11.8982-8988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Shi K, Meskauskas A, Simon AE. The 3′ End of Turnip crinkle virus contains a highly interactive structure with a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15:1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhang J, Simon AE. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 2004;78:7619–7633. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang G, Guo R, Shapiro BA, Simon AE. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J. Virol. 2006a;80:9181–9191. doi: 10.1128/JVI.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang G, McCormack JC, Simon AE. Evolution of virus-derived sequences for high-level replication of a subviral RNA. Virology. 2006b;351:476–488. doi: 10.1016/j.virol.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Wang J, Yu P, Eyler D, Xu H, Starich MR, Tiede DM, Simon AE, Kasprzak W, Schwieters CD, Shapiro BA, Wang Y-W. The cap-independent translational enhancer and ribosome binding structure element in 3′ UTR of Turnip crinkle virus RNA folds into a tRNA-like shape in solution. Proc. Natl. Acad. Sci. USA. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]