Abstract
Thioredoxin reductase (TrxR) is a selenocysteine-containing flavoprotein that catalyzes the NADPH-dependent reduction of oxidized thioredoxin and plays a key role in regulating cellular redox homeostasis. In the present studies we examined the effects of 2-chloroethyl ethyl sulfide (CEES), a model sulfur mustard vesicant, on TrxR in lung epithelial cells. We speculated that vesicant-induced alterations in TrxR contribute to oxidative stress and toxicity. Treatment of human lung A549 epithelial cells with CEES resulted in a time- and concentration-dependent inhibition of TrxR. Using purified rat liver TrxR, we demonstrated that only the reduced enzyme was inhibited and that this inhibition was irreversible. The reaction of TrxR with iodoacetamide, which selectively modifies free thiol or selenol on proteins, was also markedly reduced by CEES, suggesting that CEES induces covalent modification of the reduced selenocysteine-containing active site in the enzyme. This was supported by our findings that recombinant mutant TrxR in which selenocysteine was replaced by cysteine, was marked less sensitive to inhibition by CEES, and that the vesicant preferentially alkylated selenocysteine in the C-terminal redox motif of TrxR. TrxR also catalyzes quinone redox cycling, a process that generates reactive oxygen species. In contrast to its inhibitory effects on thioredoxin reductase activity, CEES was found to stimulate redox cycling. Taken together, these data suggest that sulfur mustard vesicants target TrxR and that this may be an important mechanism mediating oxidative stress and tissue injury.
Keywords: oxidative stress, 2-chloroethyl ethyl sulfide, sulfur mustard, lung
Introduction
Sulfur mustard (2,2’-dichlorodiethyl sulfide) is a potent vesicant that has been used as a chemical warfare agent. Inhalation of sulfur mustard causes lung damage in humans and laboratory animals (1, 2). Pathological responses in humans include bronchial mucosal injury, inflammation, fibrosis and pneumonia (1). In rodents, lipid peroxidation, pulmonary edema and alveolar hemorrhage have also been observed, as well as increases in lung lavage fluid, protein, neutrophils and proinflammatory cytokines (3–6). Similar results are observed following intratracheal instillation of the half mustard, 2-chloroethyl ethyl sulfide (CEES) (7–8). The molecular mechanisms mediating sulfur mustard-induced lung toxicity are not well understood. The observation that CEES-induced lung injury can be attenuated by reducing agents or anti-oxidants, suggests that oxidative stress is key to the pathogenic process (7–9). This is supported by findings that antioxidants such as N-acetyl cysteine and glutathione-ethyl ester protect against CEES-induced cytotoxicity and apoptosis in Jurkat cells and human lymphocytes (10). CEES has also been reported to stimulate the production of reactive oxygen species (ROS) in Jurkat cells (10), and to modulate antioxidant enzyme activities (10–13). In human bronchial and small airway epithelial cells, CEES induces mitochondrial dysfunction and stimulates mitochondrial production of ROS; mitochondrial oxidative stress can be prevented with a catalytic metalloporphyrin antioxidant (14). These findings indicate that vesicants may function by inducing oxidative stress and altering cellular redox balance.
Mammalian thioredoxin reductase (TrxR) is a homodimeric flavoprotein that catalyzes the reduction of oxidized thioredoxin, as well as other redox-active proteins including glutaredoxin 2 and protein disulfide isomerase, and small molecules like 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and hydrogen peroxide (15). It is an essential antioxidant enzyme which plays a key role in maintaining cellular redox homeostasis (16, 17). Thioredoxin also functions as a disulfide reductase for a variety of enzymes, many of which are important in the control of DNA synthesis, antioxidant defense, signal transduction, and protein folding (for a review see (16)). TrxR is a selenoprotein containing a C-terminus cysteine-selenocysteine redox pair (18). Due to the low pKa value of selenol (pKa = 5.3), at physiological pH selenocysteine is ionized to a cysteinyl-selenol (Cys-Se−) which reacts with many electrophiles resulting in enzyme inhibition (19–22). Inhibition of TrxR prevents thioredoxin reduction and compromises cellular redox homeostasis leading to oxidative stress (19, 23, 24). The present studies demonstrate that CEES is a potent irreversible inhibitor of TrxR and that this is due to covalent modification of the selenocysteine residue in the C-terminal redox center of the enzyme. The findings that vesicants can target TrxR suggest a mechanism by which these compounds induce oxidative stress and toxicity in lung cells.
Materials and Methods
Chemicals and enzymes
Purified rat TrxR, recombinant E. coli TrxR, E. coli Trx, insulin, NADPH, menadione, DTNB, and protease inhibitor cocktail (P2714) were purchased from Sigma (St. Louis, MO). CEES was from Aldrich (Milwaukee, WI). N-(biotinoyl)-N′-(iodoacetyl) ethylenediamine (BIAM) and Amplex Red reagent were from Molecular Probes (Eugene, OR). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, penicillin/streptomycin were from Gibco (Rockville, MD) and horseradish peroxidase (HRP) - conjugated streptavidin from GE Healthcare (Piscataway, NJ). Chemiluminescence (ECL) immunoblot detection reagents were from Perkin Elmer (Waltham, MA).
Cell culture and treatments
Human lung carcinoma A549 cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were grown in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 µg/ml streptomycin and maintained in a humidified atmosphere of 5% CO2 at 37 °C. Stock solutions of CEES were prepared fresh in 95% ethanol and diluted immediately before use in serum-free DMEM.
Cell viability was determined using alamarBlue (BioSource International; Camarillo, CA) as previously described (25). In brief, A549 cells were plated into 96-well dishes (1.2 × 104 cells/well) and allowed to attach for 24 h. Cells were then rinsed and incubated in serum-free DMEM medium containing vehicle control (1% ethanol, v/v) or increasing concentrations of CEES (0.25–10 mM). After 24 h, 5% alamarBlue diluted in serum free-DMEM was added to the cultures. After an additional 4 h, alamarBlue reduction was assayed by fluorescence (excitation wavelength of 555 nm and emission wavelength of 590 nm) using a SpectraMax M5 spectrofluorometer (Molecular Devices; Sunnyvale, CA). Viability was expressed as the percentage of dye reduction in the presence of CEES relative to vehicle control. The concentration of CEES inhibiting cell viability by 50% was 5–7 mM after 24 h treatment.
For preparation of lysates, A549 cells (1.2 × 106 cells) were seeded into 10-cm culture dishes and treated with CEES (0.1–2 mM) or control. After 24 h, cells were washed twice with PBS, removed from the plates with a cell scraper, centrifuged (1600 g, 5 min), and pellets lysed by sonication in 0.6 mL PBS, pH 7.4, containing 0.1% Triton X-100 and protease inhibitor cocktail. The homogenates were centrifuged at 9,000 g for 10 min at 4°C to remove insoluble material. Protein concentrations in supernatants were determined using a DC protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
TrxR enzyme assays
TrxR activity was assayed using DTNB as the substrate (26). Reactions were run in a final volume of 100 µL in 50 mM potassium phosphate buffer, pH 7.0, containing 25 µg cell lysate or 100 nM purified TrxR, 1 mM EDTA, 50 mM KCl, 0.2 mg/ml bovine serum albumin, 0.25 mM NADPH, and 2.5 mM DTNB. Enzyme activity was monitored by increases in absorbance at 412 nm and calculated using a molar extinction coefficient for thionitrobenzoic acid of 13.6 mM−1 min−1 (27). Background TrxR-independent reduction of DTNB in cell lysates, determined in the presence of aurothiomalate (200 µM), was subtracted from each value.
DTNB is not a substrate for E. coli TrxR; the activity of E. coli TrxR was analyzed using an insulin reduction assay as described previously with some modifications (26). The reaction mixture (25 nM recombinant E. coli TrxR, 50 mM Tris-HCl, pH 7.6, 20 mM EDTA, 0.25 mM NADPH, 0.3 mM insulin, and 20 µM E. coli Trx) in a final volume of 200 µL was incubated at 37 °C for 30 min. The reaction was terminated by the addition of 200 µL of 8 M guanidine-HCl, 5 mM DTNB, and 200 µM Tris-HCl, pH 8.0. Enzyme activity was determined by changes in absorbance at 412 nm.
Expression of recombinant human mutant TrxR (hTrxRSec498Cys)
A bacterial expression construct of the human mutant TrxR, pET-28a–hTR1 (containing a full-length human TrxR1 gene in which TGA (Sec) codon was replaced by TGC codon and a His-tagged element in the N-terminus), was kindly provided by Dr. Anton Turanov (University of Nebraska) (28). The construct was expressed in E. coli and purified by Ni-NTA affinity chromatography (28). Expression of purified TrxR was confirmed by Western blotting using TrxR B2 antibody (Santa Cruz, Santa Cruz, CA) and by enzyme activity.
CEES treatment of purified TrxR; characterization of TrxR inhibition
TrxR (25–100 nM) was reduced with NADPH (0.25 mM) at room temperature in 50 mM potassium phosphate buffer, pH 7.0, 1 mM EDTA, and 50 mM KCl. After 5 min, CEES (1 – 2000 µM) or control was added, and the reaction mixture incubated for additional 30 min. The ability of CEES to inhibit TrxR activity was determined using the DTNB assay (rat TrxR and human mutant TrxR) or insulin reduction assay (E. coli TrxR) as described above.
For studies on the reversibility of TrxR inhibition, reaction mixtures containing CEES-modified TrxR were purified using Chroma Spin TE-10 columns (Clontech, Mountain View, CA) to remove unreacted CEES. The modified-TrxR was then analyzed for enzyme activity using the DTNB assay. For kinetic analysis, TrxR inhibition was characterized using increasing concentrations of DTNB (3 – 2500 µM) and CEES (0 – 25 µM) in the presence of 0.25 mM NADPH.
BIAM labeling of TrxR and Western blotting
CEES modified TrxR, purified as described above, was incubated in the dark with 50 µM BIAM (dissolved in 50 mM Tris-HCl buffer at pH 6.5 or pH 8.5, respectively) at 37 °C for 30 min. BIAM-labeled protein was then analyzed by gel electrophoresis on 10% polyacrylamide gels (Criterion XT Bis-Tris gels, Bio-Rad, Hercules, CA) and electroblotted onto nitrocellulose membranes. The extent of BIAM labeling of TrxR was analyzed using HRP-conjugated streptavidin as a probe, followed by ECL detection. After BIAM analysis, the blots were stripped and the membrane reprobed with antibody against TrxR (Santa Cruz) for analysis of total TrxR protein loading in each well. Densitometric analysis of bands on the membranes was performed using a FluorChem Image System (Alpha Innotech, San Leandro, CA).
Analysis of TrxR by LC-MS/MS
NADPH-reduced rat TrxR (1 µM) was incubated with or without CEES (250 µM) at room temperature in a final volume of 100 µL in 50 mM potassium phosphate, pH 7.0, 1 mM EDTA, and 50 mM KCl. After 1 h, the incubation mixture was desalted with Chroma Spin TE-10 columns to remove unreacted CEES. Five µL of the filtered solution was analyzed for TrxR activity using the DTNB assay. Aliquots of filtered solution (18.75 µL) were then subjected to SDS-PAGE on 7.5 % gels. After staining with Comassie Blue, bands containing TrxR were cut from the gels, disulfide bonds reduced with 20 mM DTT for one hour at 60 °C, and unmodified thiol/selenol groups alkylated with 40 mM iodoacetamide for 30 min in the dark at room temperature. TrxR in gels was then digested with Lys-C (Roche, Indianapolis, IN) as previously described (29). Subsequently, digested peptides were extracted from the gels with 100 µL formic acid/water/acetonitrile (5:35:60, v/v/v) and dried in a speed vacuum. Peptides were reconstituted in 0.1% TFA and analyzed by LC-MS/MS on a Dionex U3000 (Dionex, Sunnyvale CA) operated in nanoLC mode on line with a Thermo LTQ (Thermo Fisher, San Jose, CA) fitted with a C18 AQ (120 mm × 75 µm) emitter column packed with 3 µm, 200Å Magic C18 AQ (Michrom Bioresources Inc., Auburn, CA). The samples were first equilibrated in solvent A (0.1% formic acid in water) and then eluted with a linear gradient of 2 to 45% of solvent B (0.1% formic acid in acetonitrile) over 30 min at a flow rate of 200 nl/min and analyzed by electrospray ionization MS/MS. Mass spectrometry data were acquired using a data-dependent acquisition procedure with a cyclic series of a full scan followed by zoom scans and MS/MS scans of the five most intense ions with a repeat count of two and the dynamic exclusion duration of 30 sec.
Assays for NADPH oxidation and redox cycling activity by TrxR
Oxidation of NADPH in reaction mixes was determined by measuring decreases in absorbance at 340 nm. Reactions consisted of 100 nM TrxR, 0.25 mM NADPH, 1 mM EDTA, 50 mM KCl, and 50 mM potassium phosphate buffer (pH 7.0), in the presence and absence of CEES (1–250 µM), in a total volume of 100 µL. The same solution without CEES was used as a control. Redox cycling activity of TrxR was determined by measuring the formation of H2O2 in enzyme reactions with the Amplex Red assay as previously described (30). Reactions were run in a total volume of 100 µL in 50 mM potassium phosphate buffer, pH 7.8, 0.25 mM NADPH, 1 unit/mL HRP, CEES (1–250 µM) or control, and 50 µM Amplex Red reagent. The reaction was initiated by the addition of TrxR and product formation was determined fluorometrically on the SpectraMax M5 spectrofluorometer (Molecular Devices; Sunnyvale, CA) using an excitation wavelength of 530 nm and emission wavelength of 587 nm. To analyze the effect of CEES-TrxR adducts on ROS production, reactions were also run in the presence of 100 µM menadione, a quinone known to actively redox cycle with TrxR (31).
Data analysis
TrxR activity, NADPH oxidation, and H2O2 formation were monitored over 30 min, and the initial velocities analyzed using SoftMax Pro software (Molecular Devices). IC50, Km, Vmax, Ki, and t1/2 values were determined by the non-linear regression method of curve fitting using Prism 5 software (GraphPad Inc, San Diego, CA). Statistical differences were determined using the Student’s t-test. A value of P<0.05 was considered significant.
Results
Effects of CEES on TrxR activity in lung epithelial cells
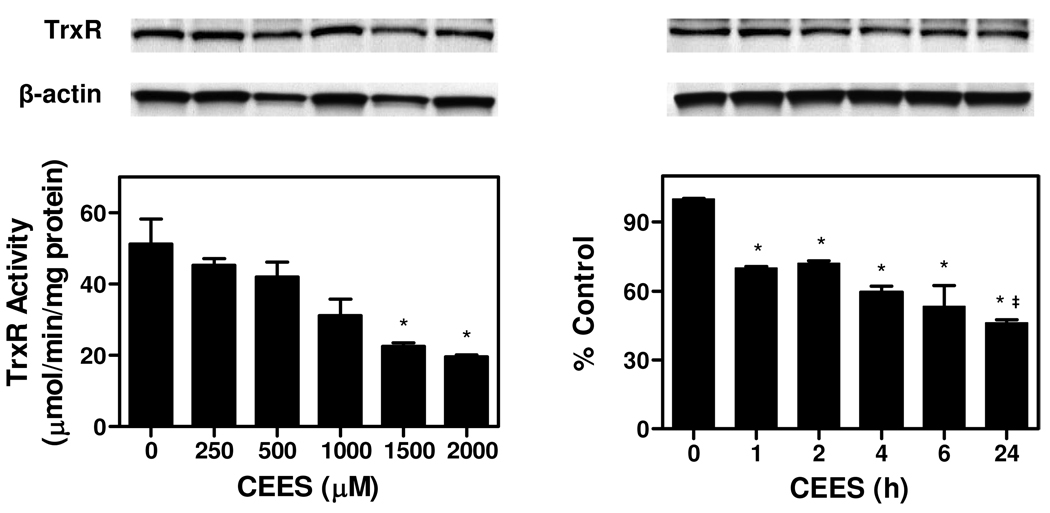
In initial experiments we examined the effects of CEES on TrxR activity in A549 lung epithelial cells. CEES was found to cause a concentration- and time-dependent inhibition of enzyme activity (Figure 1). Maximal inhibition was observed at 1.5–2 mM CEES and after 24 h. At 1.5 mM, CEES reduced the activity of TrxR by approximately 55%. Significantly more inhibition was observed at 24 h than 1 h with 1.5 mM CEES. Treatment of A549 cells with concentrations of CEES up to 2 mM did not significantly induce cytotoxicity, as determined by alamarBlue assay. Immunoblot analysis showed that there are no significant changes in TrxR protein expression following CEES treatment (Figure 1), suggesting that CEES inhibits TrxR by directly altering enzyme function.
Figure 1. Effects of CEES on TrxR in A549 cells.
Left panels: Effects of increasing concentration of CEES on TrxR protein expression and TrxR activity. Cells were incubated in serum-free medium with increasing concentrations of CEES or vehicle control. After 24 h, cells were lysed and analyzed for TrxR protein expression by Western blotting (upper panel) and TrxR activity using the DTNB assay (lower panel). After stripping, the blots were reprobed with antibody to β-actin as a protein loading control. TrxR activity was expressed as micromoles product/min/mg protein. Right panels: Time course of CEES inhibition. Cells were treated with 1.5 mM CEES for increasing periods of time and then refed with fresh serum-free medium. After 24 h, cells were lysed and assayed for TrxR protein expression and TrxR activity as indicated above. Data are mean ± SE (n = 3). *Significantly different (P < 0.05) from vehicle-treated control. ‡ Significantly different (P < 0.05) from 1 h CEES treatment.
Effects of CEES on purified TrxR
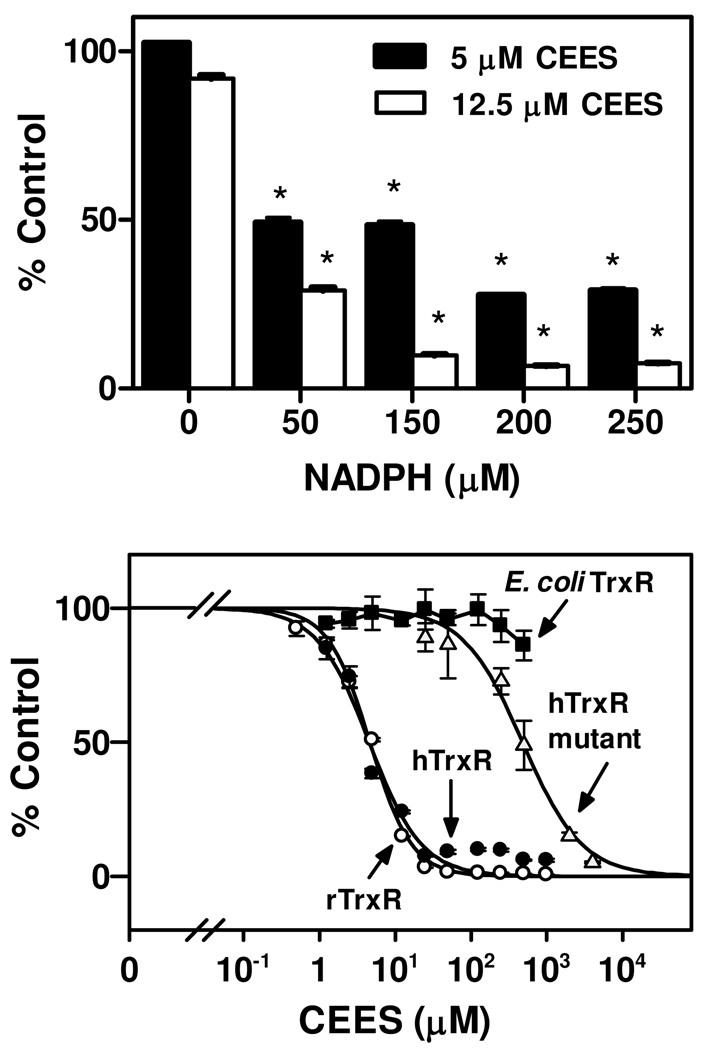
We next examined mechanisms mediating TrxR inhibition by CEES using purified rat liver enzyme. The effects of TrxR redox status on enzyme inhibition by CEES were investigated by pre-incubating the enzyme with NADPH. As shown in Figure 2, in the absence of NADPH pretreatment, CEES had no effect on TrxR activity. In contrast, a marked inhibition of enzyme activity was observed when TrxR was reduced with NADPH (50 – 250 µM). These data indicate that inhibition of TrxR by CEES is dependent on the redox state of TrxR, and that reduction of TrxR is required for CEES-induced inactivation. Inhibition of reduced-TrxR activity by CEES was found to be concentration-dependent (Figure 2). The IC50 value of CEES inhibition was 4.6 ± 0.2 µM (mean ± SE, n = 3) for purified rat TrxR. We also analyzed the effects of CEES on human TrxR, a recombinant mutant form of human TrxR and E. coli TrxR. In human A549 cell lysates, which contain wild type TrxR, CEES was found to be an effective enzyme inhibitor with an IC50 value (4.5 ± 0.6 µM) similar to rat TrxR. The mutant enzyme, in which selenocysteine (Sec498) was replaced with cysteine, was significantly less sensitive to CEES (IC50 = 477 ± 59 µM) suggesting that the selenocysteine containing C-terminal is key for enzyme inhibition. By comparison, E. coli TrxR, which lacks a C-terminal redox center, was not inhibited by CEES (IC50 > 1000 µM). These results indicate that the selenium-containing C-terminal redox motif of TrxR is a target for CEES alkylation.
Figure 2. Inhibition of TrxR by CEES.
Upper panel: Effects of the redox state of TrxR on enzyme inhibition by CEES. Purified rat liver TrxR (100 nM) was pre-incubated without and with increasing concentrations of NADPH at room temperature. After 5 min, CEES was added to the enzyme reaction mixture. After an additional 30 min, NADPH was added to a final concentration of 250 µM and TrxR activity determined by the DTNB assay. Data are expressed as mean ± SE (n = 3). *Significantly different (P < 0.05) from respective CEES treatment in the absence of NADPH. Lower panel: Effects of increasing concentrations of CEES on TrxR activity. Rat liver TrxR (rTrxR), human TrxR from A549 cells (hTrxR), hTrxR mutant or E. coli TrxR were treated with 250 µM NADPH for 5 min followed by CEES. After 30 min, enzyme activity was assayed using either the DTNB assay (rTrxR (○), hTrxR (●), hTrxR mutant (Δ)) or insulin reduction assay (E. coli TrxR (■)). Each value is the mean ± SE (n = 3).
Irreversible inhibition of TrxR by CEES
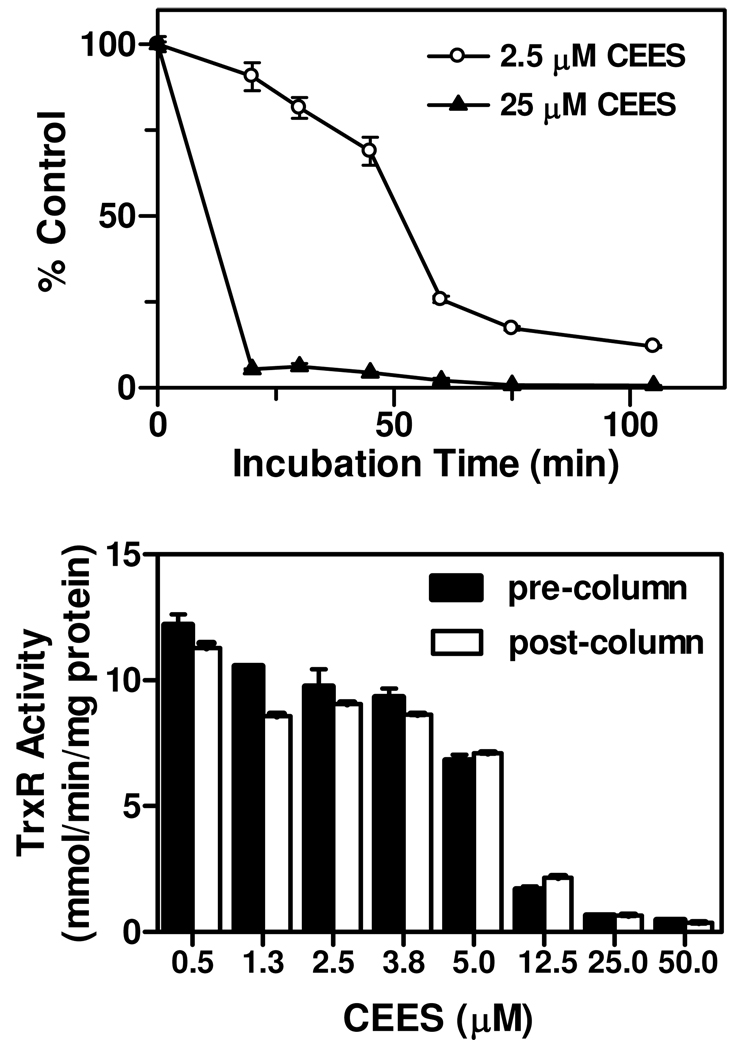
In further experiments, reduced-TrxR was incubated with CEES for increasing periods of time prior to initiating the enzyme reaction. Using 2.5 µM and 25 µM CEES, inhibition of 50% of TrxR activity was found to require approximately 45 min and 3 min, respectively (Figure 3). Time-dependent inactivation of TrxR implies that CEES causes an irreversible inhibition of the enzyme. CEES is thought to have a relatively short half-life (<5 min); our findings that 2.5 µM CEES required extended periods of time to inhibit the enzyme suggests that the kinetics of inhibition were complex since concentrations of CEES in reaction mixes over time were very low. Irreversible inhibition of TrxR by CEES was further assessed by examining the reversibility of TrxR in enzyme assays. In these studies, the recovery of TrxR enzyme activity was analyzed after separating free CEES from reaction mixtures using Chroma Spin TE-10 columns. Figure 3 shows that CEES-inhibition of TrxR activity was not reversible even after removal of unbound CEES.
Figure 3. Characterization of the effects of CEES on TrxR activity.
Upper panel: Time course of TrxR inhibition by CEES. Purified rat liver TrxR (100 nM), pre-reduced with NADPH (250 µM), was then incubated with 5 or 50 µM CEES for increasing periods of time and then assayed for enzyme activity. Lower panel: Irreversible inhibition of TrxR by CEES. Reduced-TrxR was incubated with CEES for 30 min at room temperature. TrxR in the reaction mixture was then purified using Chroma Spin TE-10 columns to remove unreacted CEES. The remaining TrxR activity was then determined by the DTNB assay. Data are mean ± SE (n = 3).
We also analyzed the effects of CEES on the kinetic parameters of TrxR using DTNB as the substrate. CEES was found to alter the Vmax of the enzyme without major changes in its Km (Table 1), indicating a noncompetitive type inhibition. The Ki value for CEES was 3.7 ± 0.3 µM (mean ± SE, n = 3) with respect to DTNB.
Table 1.
Changes in TrxR kinetic constants following treatment with CEES.
| CEES (µM)* |
||||
|---|---|---|---|---|
| 0 | 5 | 10 | 25 | |
| Vmax (mmol/min/mg protein) | 44.8 ± 0.5# | 21.5 ± 0.3 | 17.8 ± 0.2 | 3.3 ± 0.1 |
| Km (µM) | 224.4 ± 8.9 | 171.4 ± 7.6 | 182.7 ± 8.7 | 197.7 ± 8.9 |
Reduced-TrxR (100 nM) was treated with increasing concentrations of CEES. After 30 min, TrxR activity was assayed using increasing concentrations of the substrate DTNB.
Data are mean ± SE (n = 3).
Alkylation of the selenol/thiol redox motif in TrxR by CEES
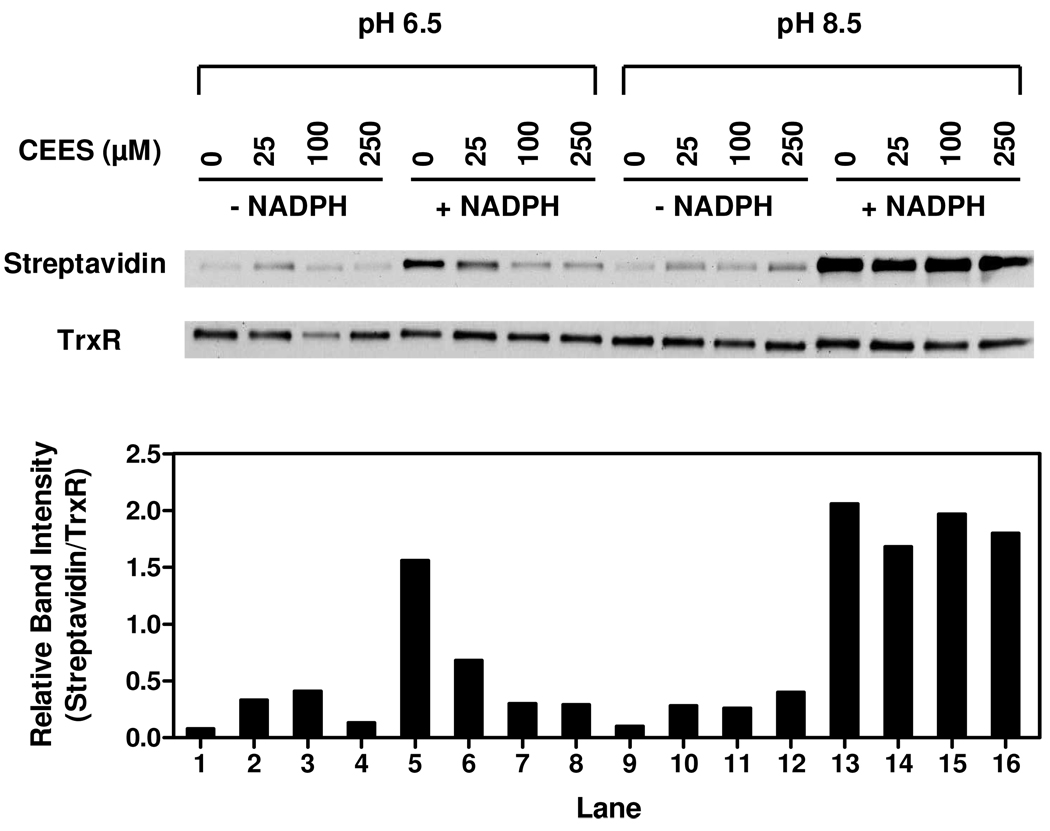
Our findings that CEES inhibited reduced, but not oxidized TrxR, suggested that it may act by modifying selenocysteine and/or cysteine residues in the enzyme. To investigate this we used a BIAM labeling technique. Previous studies have shown that BIAM selectively reacts with cysteine and/or selenocysteine in TrxR in a pH-dependent manner (32). Whereas at pH 6.5, only –SeH is alkylated by BIAM due to the low pKa (5.3) of selenol on selenocysteine, at pH 8.5, both -SH and –SeH are labeled by BIAM. The labeling efficiency of TrxR by BIAM at pH 6.5 and pH 8.5 is shown in Figure 4. No significant differences between vehicle control and CEES-treated enzyme were noted in labeling of oxidized TrxR at either pH (Figure 4). These data suggest that cysteine or selenocysteine in oxidized TrxR are not targets for CEES modification. This is consistent with our findings that only NADPH-reduced TrxR is inhibited by CEES. In contrast, in control samples of reduced-TrxR, increased BIAM labeling was evident at both pHs. At pH 6.5, CEES treatment of TrxR significantly decreased BIAM labeling while only a small decrease in BIAM labeling was evident at pH 8.5 (Figure 4). These results indicate that NADPH reduces the selenenylsulfide which facilitates alkylation of CEES.
Figure 4. Effects of CEES on labeling of TrxR by BIAM.
Reduced and non-reduced rat liver TrxR was treated with CEES or vehicle control. After 30 min, CEES-alkylated TrxR was purified using Chroma Spin TE-10 columns to remove unbound vesicant and then labeled with BIAM at either pH 6.5 or pH 8.5. After an additional 30 min, samples were analyzed by Western blotting. Upper panel: Western blots showing TrxR probed with strepavidin-conjugated HRP or antibody to TrxR. Lower panel: Densitometric analysis of labeling efficiency of TrxR by BIAM.
Identification of alkylated residues in TrxR
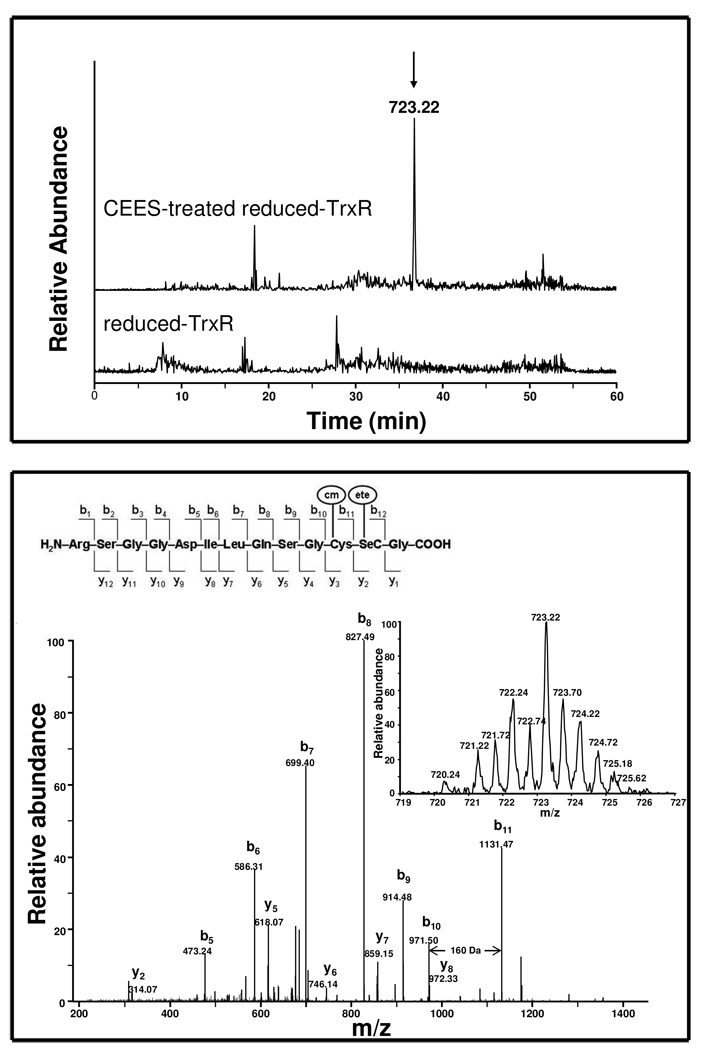
We next used LC-MS/MS to identify residues in TrxR alkylated by CEES following SDS-PAGE purification and in gel Lys-C digestion. During the digestion process, the samples were also treated with iodoacetamide to protect unreacted thiol/selenol groups. Iodoacetamide treatment can result in the addition of carbamoylmethyl groups (H2NCOCH2―, mass increase of 57 Da) to the enzyme on cysteine or selenocysteine residues; the appearance of these modified residues would indicate that they were not modified by CEES. Alternatively, CEES would lead to the addition of an ethylthioethyl group (C2H5SCH2CH2―; mass increase of 88 Da) on targeted residues. Based on distinctive isotope patterns of selenium containing peptides (from zoom scan of parent ions) and by searching for critical b and y ions in MS/MS, we identified a series of distinct modifications on the peptide, RSGGDILQSGCUG. The unique forms present only in CEES treated samples are listed in Table 2. Interestingly, one of the alkylated selenium-containing peptides, a doubly charged ion at m/z 723.22, corresponds to a mass addition of 145 Da to the RSGGDILQSGCUG peptide (1300.43, [M + H]+). The 145 Da increase correlated with unmodified peptide plus one carbamoylmethyl group and one ethylthioethyl group. MS/MS sequence analysis showed that the difference between b10 and b11 is 160 Da, indicating a mass increase of 57 Da on a cysteine residue (Figure 5, lower panel). This suggests that a cysteine residue is alkylated by a single carbamoylmethyl group and that the cysteine in the C-terminal redox motif of TrxR is not a target for CEES modification. A mass increase of 88 Da on y2 ion (314 Da) was observed in this modified peptide relative to the theoretical mass of the respective ion (226 Da) on unmodified peptide, indicating that a selenocysteine was modified with an ethylthioethyl group. In addition, fragment y5 through y8 ions increased by a mass of 145 Da, providing further evidence for the addition of one carbamoylmethyl and one ethylthioethyl on these ions. No peaks with mass increases corresponding to two ethylthioethyl-modifications (plus 176 Da) in the C-terminal selenocysteine-containing peptide were detected. These findings suggest that inhibition of TrxR by CEES resulted from specific ethylthioethyl alkylation of the selenocysteine, but not the cysteine residue, in the C-terminal redox center of TrxR.
Table 2.
Modified forms of selenium-containing C-terminal peptide identified in CEES-treated reduced-TrxR following Lys-C digestion.
| A A sequence | m/z | Charge | [M + H]+ | Δ Mass* | Peak area | Modifications# |
|---|---|---|---|---|---|---|
| RSGGDILQSGCUG | 714.72 | 2 | 1428.44 | + 128.01 | 1922850 | C + 57; U + 71 |
| RSGGDILQSGCUG | 723.22 | 2 | 1445.44 | + 145.01 | 2048004 | C + 57; U + 88 |
| RSGGDILQSGCUG | 731.18 | 2 | 1461.36 | + 160.93 | 13000752 | C + 57; U + 104 |
| RSGGDILQSGCUG | 743.78 | 2 | 1486.56 | + 186.13 | 11284893 | C + 57; no Se; + 131 at C-terminus |
| RSGGDILQSGCUG | 738.20 | 2 | 1475.40 | + 174.97 | trace | C + 71; U + 104 |
Compared to theoretical mass of unmodified RSGGDILQSGCUG, [M + H]+: 1300.43
Based on MS/MS results.
Figure 5. LC-MS/MS analysis of TrxR-CEES adducts.
Reduced-rat liver TrxR (1 µM) was incubated with or without CEES (250 µM) at room temperature. After 1 h, the reaction mixture was desalted and the CEES-modified protein was further purified by SDS-PAGE. The purified protein was reduced with DDT, reacted with iodoacetamide, and digested with Lys-C in the gel. The resulting peptides were analyzed using LC-MS/MS. Carbamoylmethyl (cm); ethylthioethyl (ete). Upper panel: Extracted ion chromatograph showing a specific peak as shown by the arrow (m/z = 723.22, r.t. 36.8 min) in CEES-treated but not in control sample. Lower panel: MS/MS spectrum of the specific peak in CEES-treated sample which was found to be a doubly charged RSGGDILQSGC(cm)U(ete)G peptide. Insets show the zoom scan spectrum and theoretical fragments of the doubly charged ion. Matched b and y fragments are marked. The mass difference between b10 and b11 is 160 Da indicating a carbamoylmethylated cysteine. No other evidence indicated that other cysteines in thioredoxin reductase were modified by CEES by searching cysteine-CEES adducts using the GPM and BioWorks software.
Effects of CEES on redox-cycling of TrxR
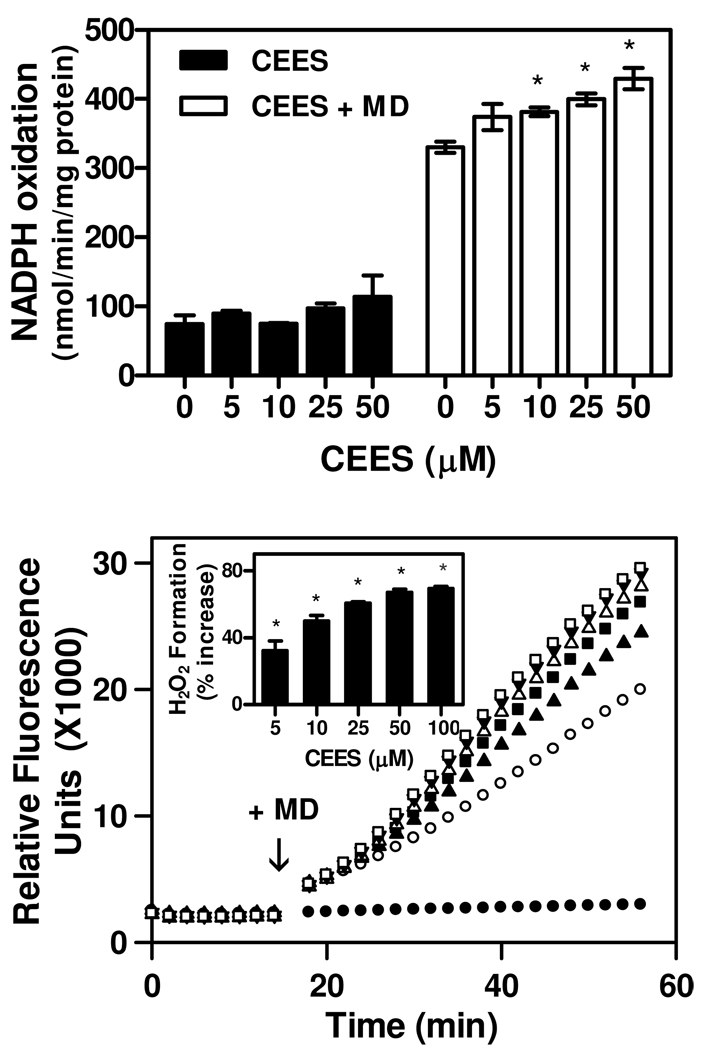
In addition to reducing thioredoxin, TrxR is also known to mediate chemical redox cycling (33). Thus, in an NADPH-dependent reaction, TrxR catalyzes the one electron reduction of redox active chemicals including bipyridylium herbicides and quinones; the subsequent reaction of the chemical radical with molecular oxygen yields the parent compound and superoxide anion (30, 34). Superoxide anion rapidly dismutates to hydrogen peroxide and, in the presence of redox active metals, forms highly toxic hydroxyl radicals (33). We found that the quinone menadione (2-methyl-1,4-napthoquinone), but not CEES, readily generated hydrogen peroxide by redox cycling with TrxR (Figure 6, lower panel). This was associated with a concomitant increase in NADPH utilization (Figure 6, upper panel). The formation of H2O2 by TrxR was inhibited by catalase, but not superoxide dismutase (data not shown). Under conditions where CEES inhibited TrxR activity, menadione continued to redox cycle with TrxR. In fact, CEES treatment significantly stimulated menadione redox cyling by TrxR. These data indicate that thioredoxin reduction and quinone redox cycling by TrxR occur by distinct mechanisms.
Figure 6. Effects of CEES on NADPH oxidation and redox cycling by TrxR.
Upper panel: Effects of CEES on NADPH oxidation. NADPH utilization in reaction mixes was followed by decreases in absorbance at 340 nm. The reaction mixes consisted of purified rat liver TrxR (200 nM), NADPH (0.25 mM), CEES or vehicle control, in the presence or absence of menadione (100 µM). Data are mean ± SE (n = 3). *Significantly different (P < 0.05) from vehicle-treated control. Lower panel: Effects of CEES treatment on menadione generated H2O2 by purified rat liver TrxR. Reduced-TrxR was incubated with increasing concentrations of CEES or vehicle (EtOH) control. The production of H2O2 was monitored using the Amplex Red assay. After 14 min, 100 µM menadione (MD) or DMSO control (●) was added to the reaction mixture (shown by the arrow) and H2O2 formation monitored continuously for an additional 40 min. (○) 0 µM CEES; (▲) 5 µM CEES; (■) 10 µM CEES; (Δ) 25 µM CEES; (▼) 50 µM CEES; (□) 100 µM CEES. The vehicle control sample without menadione was also treated with 100 µM CEES. Inset shows the effects of CEES on menadione generated H2O2 relative to vehicle treated TrxR. Activity was determined as an increase in the rate of formation of H2O2. Results are expressed as mean ± SE (n = 3). *Significantly different (P < 0.05) from vehicle treated samples in the presence of 100 µM menadione.
Discussion
The present studies demonstrate that TrxR is a target for CEES resulting in inhibition of enzyme activity. This was observed in human type II lung epithelial cells, and in highly purified enzyme preparations from rat liver. Using the purified enzyme, the inhibitory effects of CEES were found to be NADPH-dependent and irreversible. This latter conclusion is based on the time-dependence of enzyme inhibition and its non-competitive nature with respect to the TrxR substrate DTNB, as well as the observations that TrxR activity remained suppressed even after free CEES was removed from enzyme assays, that pretreatment of TrxR with CEES prevented binding of iodoacetamide, and the identification of CEES alkylation products in the critical redox active motif of the enzyme. Since TrxR is a key enzyme for maintaining redox homeostasis, our findings suggest that altered redox balance may be an important mechanism of CEES-induced vesicant toxicity in the lung.
A number of electrophilic compounds have been identified as inhibitors of TrxR including dinitrohalobenzenes, quinones, various chemotherapeutic agents and dietary components, and endogenous lipid peroxidation products such as 4-hydroxynonenal (19–21, 23, 31, 35, 36). Alkylating agents such as the nitrogen mustards and nitrosoureas also inhibit TrxR (37). Like CEES, most of these agents are irreversible inhibitors of TrxR and their effects are NADPH-dependent, indicating a requirement for the reduced enzyme (19–21, 35). TrxR is a homodimeric flavoprotein containing a conserved N-terminal redox-active disulfide and a C-terminal redox-active selenenylsulfide, which forms a dithiol and selenolthiol, respectively, in the reduced enzyme. According to the proposed catalytic mechanism model, redox-active motifs within each subunit of the enzyme communicate (16). Thus, reducing equivalents derived from NADPH are transferred to FAD and then to the N-terminal redox center in each subunit which subsequently transfers electrons to the C-terminal selenosulfide of the next subunit before delivery to TrxR substrates (38). The selenol group on selenocysteine is highly nucleophilic with a low pKa (5.3). At physiological pH, it is a reactive selenolate (-Se−) and its location in the open C-terminus of TrxR makes it a target for irreversible alkylation by electrophilic compounds which are inhibitors of TrxR enzyme activity (16). The present studies demonstrate that CEES also targets selenocysteine in TrxR. This is based on the findings that 1) CEES is an efficient inhibitor of human and rat TrxR which contain selenocysteine, but not human mutant enzyme (Sec498Cys) or E. coli TrxR which do not contain the selenocysteine redox motif; 2) CEES is more effective in blocking BIAM binding at pH 6.5 when compared to pH 8.5, due to the low pKa for selenocysteine; and that 3) CEES-derivatized selenocysteine, but not cysteine, was detected following LC-MS/MS analysis of peptide fragments from reduced, but not oxidized, TrxR treated with CEES. We speculate that by modifying selenocysteine, CEES interferes with the transfer of reducing equivalents to TrxR substrates and/or substrate binding to the enzyme, and that this leads to enzyme inhibition. Preferential alkylation of TrxR on selenocysteine has been demonstrated for several TrxR inhibitors including 1-chloro-2,4-dinitrobenzene, 4-hydroxynonenal, curcumin and arsenic trioxide (21, 23, 35, 36, 39). In contrast to our findings with CEES, these inhibitors alkylated cysteine, as well as selenocysteine, in the C-terminal redox motif, suggesting that they function by mechanisms distinct from CEES. It is possible that these other inhibitors interfere with the formation of critical dithiol intermediates in the TrxR reaction (21, 23, 35, 36). It appears that 5-iodoacetamidofluorescein, another TrxR inhibitor, does not alkylate the C-terminal redox active cysteine suggesting that its actions may be similar to CEES (39). At the present time we cannot rule out the possibility that CEES interacts with other amino acid residues in TrxR important in regulating enzyme activity. We also found that CEES can target mutant TrxR lacking selenocysteine, although this was at least two orders of magnitude less efficient. TrxR has been reported to catalyze enzyme substrate reduction via N- or C-terminal redox active cysteines independent of selenocysteine (40). The reaction of CEES with these amino acids and/or others may also be important in mediating its TrxR inhibitory activity. This is supported by findings that vesicants can react with many cysteine residues in proteins, and to a lesser extent with histidine, glutamic acid and aspartic acid (41–43). E. coli TrxR functions by a mechanism distinct from mammalian TrxR’s where active site cysteine residues are located in the central NADPH domain; these may be protected from CEES and this may account for the inability of this vesicant to inhibit E. coli enzyme activity. Further studies are required to identify additional CEES-TrxR adducts in the rat liver enzyme and to elucidate their roles in regulating TrxR activity.
Consistent with other TrxR inhibitors, the effects of CEES were dependent on reduced TrxR (19–21, 35). NADPH is thought to act on TrxR by reducing selenenylsulfide to highly reactive selenols and thiols in the C-terminal redox center, a process triggering a conformational change in the catalytic center of the enzyme, facilitating substrate binding and reactivity with electrophilic compounds (44–46). This is supported by recent analysis of the crystal structures of selenocysteine-containing rat TrxR1, which revealed that the oxidized enzyme possesses a C-terminal selenenylsulfide motif in a trans-configuration, a site distant from the N-terminal disulfide/dithiol motif required for optimal substrate binding and electron transfer during catalysis (47). Reduction by NADPH triggers a conformational change which brings the selenocysteine residue to the enzyme surface, a process that facilitates substrate binding and catalytic activity (47). It also presents a reactive target for various electrophiles and provides a mechanism for the selective inactivation of reduced TrxR by CEES (48).
Inhibition of TrxR by 1-chloro-2,4-dinitrobenzene has been reported to be associated with large increases in NADPH oxidase activity (23, 49). This was subsequently identified as redox cycling by TrxR, a process whereby the enzyme mediates the NADPH-dependent one electron reduction of the nitroaromatic compound into a nitro anion radical (49). The reaction of this radical with molecular oxygen leads to the formation of ROS, regenerating the parent compound in the process (23). These earlier studies demonstrated that redox cycling can occur despite modification of the selenocysteine in TrxR and inhibition of enzyme activity. Subsequently, it was shown that TrxR mediates the NADPH-dependent redox cycling of many redox active agents including nitroaromatic compounds, quinones, curcumin, flavonoids and paraquat (20, 30, 31, 35, 50). As observed with 1-chloro-2,4-dinitrobenzene, many of these chemicals inactive TrxR. Our data showing the CEES-inactivated TrxR can continue to mediate redox cycling of menadione are consistent with these studies. A question arises as to the mechanism mediating redox cycling of the inhibited enzyme. Modification of selenocysteine by CEES, as well as the neighboring cysteine by other inhibitors in the C-terminal redox motif of TrxR, leading to inhibition of enzyme activity, indicates that substrate reduction and redox cycling occur via distinct mechanisms. It has been proposed that in the inhibited enzyme, FAD can still be reduced by NADPH, possibly forming a flavin-thiolate charge transfer complex in the N-terminal disulfide (31, 51). Presumably, the proximity of redox active chemicals to one or more of these targets permits their one electron reduction, a process that initiates redox cycling (31, 51). Of particular interest was our finding that treatment of TrxR with higher concentrations of CEES further stimulated menadione redox cycling. Similar results have been reported for 1-chloro-2,4-dinitrobenzene as measured by increases in NADPH oxidase activity (23). It is possible that this increases the number of modified amino acids in TrxR; resulting conformational changes may allow greater access of redox active chemicals to the active site on the enzyme. However, other mechanisms can not be excluded including changes that allow increased electron transfer to the substrate.
We hypothesize that both inhibition of TrxR and redox cycling contribute to CEES-induced oxidative stress. Inhibition of TrxR has been shown to deplete cells of reduced thioredoxin (21, 52). As a key player in redox regulation, reduced thioredoxin is key for maintaining the integrity of many redox sensitive signaling molecules and proteins. Moreover, both thioredoxin and TrxR function as antioxidants and can also scavenge ROS (53, 54). Interfering with these processes contributes to oxidative stress. Increasing ROS production by TrxR redox cycling can also lead to oxidative stress. As indicated above, 1-chloro-2,4-dinitrobenzene and various quinones inhibit TrxR; these compounds also redox cycle with the enzyme (23, 31). Although CEES is effective in inhibiting TrxR, its does not inhibit redox cycling by the enzyme. Thus, TrxR can generate ROS following exposure to redox cycling chemicals such as menadione. The fact that CEES modifies selenocysteine to inhibit TrxR activity suggests that this amino acid does not participate in the redox cycling reaction. It should be noted that redox cycling is not limited to exogenous chemicals. For example, catechol estrogens have been reported to effectively redox cycle and generate ROS (55, 56) and it is possible that TrxR redox cycles these estrogen metabolites thus contributing to oxidative stress.
In summary, our studies provide a mechanistic basis for vesicant-induced oxidative stress. We show that CEES is an effective inhibitor of TrxR in intact cells, cell lysates, and purified enzyme. Inhibition requires a reduced enzyme and appears to be due to modification of selenocysteine in the C-terminal redox motif of the enzyme. Inhibition of TrxR prevents substrate reduction, a process that disrupts cellular redox homeostasis; however, it does not prevent chemical redox cycling and subsequent generation of ROS. Each process alone or in combination can cause oxidative stress and contribute to tissue injury. There are few effective countermeasures against vesicant-induced toxicity; the fact that they react with TrxR suggests that this enzyme may be a target for therapeutic development.
Acknowledgments
This work was supported in part by National Institutes of Health grants CA100994 (JDL), CA093798 (DEH), ES005022 (JDL, DLL), ES004738 (DLL, JDL), CA132624 (DLL, JDL), AR055073 (JDL, DLL, DEH), F32ES017389 (YJ) and GM034310 (DLL, JDL). This work was also funded in part by the National Institutes of Health CounterACT Program through the National Institute of Arthritis and Musculoskeletal and Skin Diseases (award #U54AR055073 to JDL). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the federal government.
Abbreviations
- TrxR
thioredoxin reductase
- CEES
2-chloroethyl ethyl sulfide
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- ROS
reactive oxygen species
- BIAM
N-(biotinoyl)-N′-(iodoacetyl) ethylenediamine
- DMEM
Dulbecco’s modified Eagle’s medium
- HRP
horseradish peroxidase
- ECL
chemiluminescence
References
- 1.Beheshti J, Mark EJ, Akbaei HM, Aslani J, Ghanei M. Mustard lung secrets: long term clinicopathological study following mustard gas exposure. Pathol. Res. Pract. 2006;202:739–744. doi: 10.1016/j.prp.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Ghanei M, Harandi AA. Long term consequences from exposure to sulfur mustard: a review. Inhal. Toxicol. 2007;19:451–456. doi: 10.1080/08958370601174990. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DR, Yourick JJ, Moeller RB, Petrali JP, Young GD, Byers SL. Pathologic Changes in Rat Lungs Following Acute Sulfur Mustard Inhalation. Inhal. Toxicol. 1996;8:285–297. [Google Scholar]
- 4.Anderson DR, Byers SL, Clark CR, Schlehr JA. Biochemical alterations in rat lung lavage fluid following acute sulfur mustard inhalation. Inhal. Toxicol. 1997;9:43–51. [Google Scholar]
- 5.Anderson DR, Taylor SL, Fetterer DP, Holmes WW. Evaluation of protease inhibitors and an antioxidant for treatment of sulfur mustard-induced toxic lung injury. Toxicology. 2009;263:41–46. doi: 10.1016/j.tox.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 6.Weber WM, Kracko DA, Lehman MR, Irvin CM, Blair LF, White RK, Benson JM, Grotendorst GR, Cheng Y, McDonald JD. Inhalation exposure systems for the development of rodent models of sulfur mustard-induced pulmonary injury. Toxicol. Mech. Methods. 2010;20:14–24. doi: 10.3109/15376510903483730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das SK, Mukherjee S, Smith MG, Chatterjee D. Prophylactic protection by N-acetylcysteine against the pulmonary injury induced by 2-chloroethyl ethyl sulfide, a mustard analogue. J. Biochem. Mol. Toxicol. 2003;17:177–184. doi: 10.1002/jbt.10076. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee D, Mukherjee S, Smith MG, Das SK. Evidence of hair loss after subacute exposure to 2-chloroethyl ethyl sulfide, a mustard analog, and beneficial effects of N-acetyl cysteine. J. Biochem. Mol. Toxicol. 2004;18:150–153. doi: 10.1002/jbt.20020. [DOI] [PubMed] [Google Scholar]
- 9.Powell KL, Boulware S, Thames H, Vasquez KM, MacLeod MC. 2,6-Dithiopurine blocks toxicity and mutagenesis in human skin cells exposed to sulfur mustard analogues, 2-chloroethyl ethyl sulfide and 2-chloroethyl methyl sulfide. Chem. Res. Toxicol. 2010;23:497–503. doi: 10.1021/tx9001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han S, Espinoza LA, Liao H, Boulares AH, Smulson ME. Protection by antioxidants against toxicity and apoptosis induced by the sulphur mustard analog 2-chloroethylethyl sulphide (CEES) in Jurkat T cells and normal human lymphocytes. Br. J. Pharmacol. 2004;141:795–802. doi: 10.1038/sj.bjp.0705591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Rajaratnam V, Mukherjee S, Smith M, Das SK. Modulation of the expression of superoxide dismutase gene in lung injury by 2-chloroethyl ethyl sulfide, a mustard analog. J. Biochem. Mol. Toxicol. 2006;20:142–149. doi: 10.1002/jbt.20128. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee D, Mukherjee S, Smith MG, Das SK. Signal transduction events in lung injury induced by 2-chloroethyl ethyl sulfide, a mustard analog. J. Biochem. Mol. Toxicol. 2003;17:114–121. doi: 10.1002/jbt.10068. [DOI] [PubMed] [Google Scholar]
- 13.Qui M, Paromov VM, Yang H, Smith M, Stone WL. Inhibition of inducible Nitric Oxide Synthase by a mustard gas analog in murine macrophages. BMC Cell Biol. 2006;7 doi: 10.1186/1471-2121-7-39. 1471-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould NS, White CW, Day BJ. A role for mitochondrial oxidative stress in sulfur mustard analog 2-chloroethyl ethyl sulfide-induced lung cell injury and antioxidant protection. J. Pharmacol. Exp. Ther. 2009;328:732–739. doi: 10.1124/jpet.108.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 16.Gromer S, Urig S, Becker K. The thioredoxin system--from science to clinic. Med. Res. Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 17.Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc. Natl. Acad. Sci. U.S.A. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang J, Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J. Am. Chem. Soc. 2006;128:1879–1885. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Papp LV, Fang J, Rodriguez-Nieto S, Zhivotovsky B, Holmgren A. Inhibition of Mammalian thioredoxin reductase by some flavonoids: implications for myricetin and quercetin anticancer activity. Cancer Res. 2006;66:4410–4418. doi: 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Chew EH, Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12288–12293. doi: 10.1073/pnas.0701549104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoesel LM, Flierl MA, Niederbichler AD, Rittirsch D, McClintock SD, Reuben JS, Pianko MJ, Stone W, Yang H, Smith M, Sarma JV, Ward PA. Ability of antioxidant liposomes to prevent acute and progressive pulmonary injury. Antioxid. Redox. Signal. 2008;10:973–981. doi: 10.1089/ars.2007.1878. [DOI] [PubMed] [Google Scholar]
- 23.Nordberg J, Zhong L, Holmgren A, Arner ES. Mammalian thioredoxin reductase is irreversibly inhibited by dinitrohalobenzenes by alkylation of both the redox active selenocysteine and its neighboring cysteine residue. J. Biol. Chem. 1998;273:10835–10842. doi: 10.1074/jbc.273.18.10835. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Zhang J, Xu T. Cyclophosphamide as a potent inhibitor of tumor thioredoxin reductase in vivo. Toxicol. Appl. Pharmacol. 2007;218:88–95. doi: 10.1016/j.taap.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 25.Seyfried J, Wullner U. Inhibition of thioredoxin reductase induces apoptosis in neuronal cell lines: role of glutathione and the MKK4/JNK pathway. Biochem. Biophys. Res. Commun. 2007;359:759–764. doi: 10.1016/j.bbrc.2007.05.176. [DOI] [PubMed] [Google Scholar]
- 26.Luthman M, Holmgren A. Rat liver thioredoxin and thioredoxin reductase: purification and characterization. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
- 27.Holmgren A. Bovine thioredoxin system: Purification of thioredoxin reductase from calf liver and thymus and studies of its function in disulfide reduction. J. Biol. Chem. 1977;252:4600–4606. [PubMed] [Google Scholar]
- 28.Turanov AA, Su D, Gladyshev VN. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J. Biol. Chem. 2006;281:22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- 29.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- 30.Gray JP, Heck DE, Mishin V, Smith PJ, Hong JY, Thiruchelvam M, Cory-Slechta DA, Laskin DL, Laskin JD. Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H:paraquat oxidoreductase: identification of the enzyme as thioredoxin reductase. J. Biol. Chem. 2007;282:7939–7949. doi: 10.1074/jbc.M611817200. [DOI] [PubMed] [Google Scholar]
- 31.Cenas N, Nivinskas H, Anusevicius Z, Sarlauskas J, Lederer F, Arner ES. Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian selenoprotein. J. Biol. Chem. 2004;279:2583–2592. doi: 10.1074/jbc.M310292200. [DOI] [PubMed] [Google Scholar]
- 32.Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 33.Nordberg J, Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 34.Criddle DN, Gillies S, Baumgartner-Wilson HK, Jaffar M, Chinje EC, Passmore S, Chvanov M, Barrow S, Gerasimenko OV, Tepikin AV, Sutton R, Petersen OH. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J. Biol. Chem. 2006;281:40485–40492. doi: 10.1074/jbc.M607704200. [DOI] [PubMed] [Google Scholar]
- 35.Fang J, Lu J, Holmgren A. Thioredoxin reductase is irreversibly modified by curcumin: a novel molecular mechanism for its anticancer activity. J. Biol. Chem. 2005;280:25284–25290. doi: 10.1074/jbc.M414645200. [DOI] [PubMed] [Google Scholar]
- 36.Cassidy PB, Edes K, Nelson CC, Parsawar K, Fitzpatrick FA, Moos PJ. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006;27:2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- 37.Witte AB, Anestal K, Jerremalm E, Ehrsson H, Arner ES. Inhibition of thioredoxin reductase but not of glutathione reductase by the major classes of alkylating and platinum-containing anticancer compounds. Free Radic. Biol. Med. 2005;39:696–703. doi: 10.1016/j.freeradbiomed.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun QA, Wu Y, Zappacosta F, Jeang KT, Lee BJ, Hatfield DL, Gladyshev VN. Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. J. Biol. Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 40.Lothrop AP, Ruggles EL, Hondal RJ. No selenium required: reactions catalyzed by mammalian thioredoxin reductase that are independent of a selenocysteine residue. Biochemistry. 2009;48:6213–6223. doi: 10.1021/bi802146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noort D, Hulst AG, de Jong LP, Benschop HP. Alkylation of human serum albumin by sulfur mustard in vitro and in vivo: mass spectrometric analysis of a cysteine adduct as a sensitive biomarker of exposure. Chem. Res. Toxicol. 1999;12:715–721. doi: 10.1021/tx9900369. [DOI] [PubMed] [Google Scholar]
- 42.Noort D, Hulst AG, Jansen R. Covalent binding of nitrogen mustards to the cysteine-34 residue in human serum albumin. Arch. Toxicol. 2002;76:83–88. doi: 10.1007/s00204-001-0318-2. [DOI] [PubMed] [Google Scholar]
- 43.Yeo TH, Ho ML, Loke WK. Development of a liquid chromatography-multiple reaction monitoring procedure for concurrent verification of exposure to different forms of mustard agents. J. Anal. Toxicol. 2008;32:51–56. doi: 10.1093/jat/32.1.51. [DOI] [PubMed] [Google Scholar]
- 44.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biterova EI, Turanov AA, Gladyshev VN, Barycki JJ. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15018–15023. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritz-Wolf K, Urig S, Becker K. The structure of human thioredoxin reductase 1 provides insights into C-terminal rearrangements during catalysis. J. Mol. Biol. 2007;370:116–127. doi: 10.1016/j.jmb.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Q, Sandalova T, Lindqvist Y, Arner ES. Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1. J. Biol. Chem. 2009;284:3998–4008. doi: 10.1074/jbc.M807068200. [DOI] [PubMed] [Google Scholar]
- 48.Tonissen KF, Di Trapani G. Thioredoxin system inhibitors as mediators of apoptosis for cancer therapy. Mol. Nutr. Food Res. 2009;53:87–103. doi: 10.1002/mnfr.200700492. [DOI] [PubMed] [Google Scholar]
- 49.Arner ES, Bjornstedt M, Holmgren A. 1-Chloro-2,4-dinitrobenzene is an irreversible inhibitor of human thioredoxin reductase: loss of thioredoxin disulfide reductase activity is accompanied by a large increase in NADPH oxidase activity. J. Biol. Chem. 1995;270:3479–3482. doi: 10.1074/jbc.270.8.3479. [DOI] [PubMed] [Google Scholar]
- 50.Cenas N, Prast S, Nivinskas H, Sarlauskas J, Arner ES. Interactions of nitroaromatic compounds with the mammalian selenoprotein thioredoxin reductase and the relation to induction of apoptosis in human cancer cells. J. Biol. Chem. 2006;281:5593–5603. doi: 10.1074/jbc.M511972200. [DOI] [PubMed] [Google Scholar]
- 51.Arner ES. Superoxide production by dinitrophenyl-derivatized thioredoxin reductase--a model for the mechanism and correlation to immunostimulation by dinitrohalobenzenes. Biofactors. 1999;10:219–226. doi: 10.1002/biof.5520100220. [DOI] [PubMed] [Google Scholar]
- 52.Carvalho CM, Chew EH, Hashemy SI, Lu J, Holmgren A. Inhibition of the human thioredoxin system: a molecular mechanism of mercury toxicity. J. Biol. Chem. 2008;283:11913–11923. doi: 10.1074/jbc.M710133200. [DOI] [PubMed] [Google Scholar]
- 53.Zhong L, Holmgren A. Mammalian thioredoxin reductases as hydroperoxide reductases. Methods Enzymol. 2002;347:236–243. doi: 10.1016/s0076-6879(02)47023-1. [DOI] [PubMed] [Google Scholar]
- 54.Das KC, Das CK. Thioredoxin, a singlet oxygen quencher and hydroxyl radical scavenger: redox independent functions. Biochem. Biophys. Res. Commun. 2000;277:443–447. doi: 10.1006/bbrc.2000.3689. [DOI] [PubMed] [Google Scholar]
- 55.Roy D, Cai Q, Felty Q, Narayan S. Estrogen-induced generation of reactive oxygen and nitrogen species, gene damage, and estrogen-dependent cancers. J. Toxicol. Environ. Health B Crit. Rev. 2007;10:235–257. doi: 10.1080/15287390600974924. [DOI] [PubMed] [Google Scholar]
- 56.Rivera-Portalatin NM, Vera-Serrano JL, Prokai-Tatrai K, Prokai L. Comparison of estrogen-derived ortho-quinone and para-quinol concerning induction of oxidative stress. J. Steroid Biochem. Mol. Biol. 2007;105:71–75. doi: 10.1016/j.jsbmb.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]