Abstract
We have previously reported strong linkage on chromosome 10q in pedigrees transmitting Alzheimer's disease through the mother, overlapping with many significant linkage reports including the largest reported study. Here, we report the most comprehensive fine mapping of this region to date. In a sample of 638 late-onset Alzheimer's disease (LOAD) cases and controls including 104 maternal LOAD cases, we genotyped 3,884 single nucleotide polymorphisms (SNPs) covering 15.2 Mb. We then used imputations and publicly available data to generate an extended dataset including 4,329 SNPs for 1,209 AD cases and 839 controls in the same region. Further, we screened eight genes in this region for rare alleles in 283 individuals by nucleotide sequencing, and we tested for possible monoallelic expression as it might underlie our maternal parent of origin linkage. We excluded the possibility of multiple rare coding risk variants for these genes and monoallelic expression when we could test for it. One SNP, rs10824310 in the PRKG1 gene, showed study-wide significant association without a parent of origin effect, but the effect size estimate is not of sufficient magnitude to explain the linkage, and no association is observed in an independent genome-wide association studies (GWAS) report. Further, no causative variants were identified though sequencing. Analysis of cases with maternal disease origin pointed to a few regions of interest that included the genes PRKG1 and PCDH15 and an intergenic interval of 200 Kb. It is likely that non-transcribed rare variants or other mechanisms involving these genomic regions underlie the observed linkage and parent of origin effect. Acquiring additional support and clarifying the mechanisms of such involvement is important for AD and other complex disorder genetics research.
Keywords: Alzheimer's disease, Genetic linkage, Genetic association, Human chromosome 10, Imprinting
Introduction
Alzheimer's disease (AD) is a complex neurodegenerative disorder of acquired impairment of intellect and memory. Late-onset AD (LOAD), the more common form of disease, affects an estimated 4.5 million people in the USA currently, with a projected increase in prevalence of up to 16 million in the next 40 to 50 years. Risk factors for LOAD include increased age and a family history of disease. Although aggregation of disease is observed within families and heritability of LOAD is estimated between 60% and 80%, studies in families with LOAD have not demonstrated a Mendelian mode of inheritance, and linkage and association studies in these families have not identified a particular sufficient causal variant. The APOE gene, encoding apolipoprotein E, is the first and strongest established susceptibility gene for LOAD, while several other promising candidates have been associated in multiple studies and populations [1], and new ones are emerging from genome-wide association studies (GWAS) [2–13]. Those however have small or at best moderate effects, never close to that of the APOE locus. This suggests that most of the disease heritability likely comes from either rare alleles or alleles with small effect sizes.
The small effect sizes that are typically observed might in part reflect the heterogeneity of the disease. Selection of affected individuals or families based on phenotypic or other characteristics that reflect the presence of specific genetic risk variants can increase the observed effect size and power. We and others have attempted to reduce heterogeneity by stratifying families based on features such as clinical presentation and parent of origin [14–17], borrowing a theme from successful linkage studies for early onset AD that identified amyloid precursor protein after accommodating genetic heterogeneity based on age of onset [18]. Recent linkage studies of LOAD have implicated at least three regions of the genome, including chromosome 10q, which has shown linkage to a LOAD locus in multiple independent studies [19, 20] and with varying methodologies [21, 22].
Our previous genome-wide linkage study showed evidence of linkage in this region, particularly when considering heterogeneity by parent of origin (43.7–61 Mb on National Center for Biotechnology Information (NCBI) Build 36.1) [14, 15]. Families with an affected mother showed peak logarithm of odds ratio (LOD) score of 3.7 at D10S1221 (chromosome 10 at ~57.2 Mbp), compared to a LOD score of 1.1 when parental affection status was not considered. Functional magnetic resonance imaging further supports evidence of heterogeneity, with offspring of cases from maternal families linked to chromosome 10 having markedly different brain activation patterns in response to word recall tasks compared to non-linked families [23]. Recently, a pooled linkage analyses that assembled the largest linkage collection of AD families to date strengthened the evidence for linkage on chromosome 10 [17]. Our samples were part of that study, but our maternal families only represented 10% of the total. The improvement of the score compared to previous smaller overlapping studies [16] and the shift of the peak toward our region (the peaks now overlap by two thirds) might indicate that our stratification strategy achieved similar increase in power with the larger sample size.
We report here single nucleotide polymorphism (SNP) fine mapping of this region using a case-control approach with the most comprehensive SNP coverage reported to date.
Materials and methods
Sample
-
(a)
Genotyped sample: Our cases include 255 unrelated LOAD patients from the three-site NIMH Genetics Initiative families and 94 additional AD patients from the National Cell Repository for AD (NCRAD), housed at Indiana University Medical Center (total= 349 cases, 104 with known maternal affection). Ascertainment criteria, sample procurement, and data gathering for the NIMH samples are further described in [24] and [14]. All cases met NINCDS/ADRDA criteria for diagnosis of probable or possible AD or had a definite diagnosis after autopsy confirmation. Controls included 197 unaffected individuals from the NCRAD and 92 unaffected individuals (spouses) from our ongoing longitudinal study of AD offspring [23]. All controls were screened to be cognitively healthy and were unrelated to each other in any of the cases. All participants included in the analysis reported European Caucasian ethnicity. Table 1 provides information on the sample demographics.
-
(b)
Extended dataset: In order to increase our sample size, we obtained data from the GWAS reported by Reiman et al. [13] on 1,410 cases and controls. These data are available at the Translational Genomics Research Institute (TGEN) website (http://www.tgen.org/research/) and include 312,316 SNPs across the genome with 1,943 SNPs in our region, for 860 cases and 550 controls. Of these, 643 cases and 404 controls were neuropathologically confirmed, while the remaining was clinically evaluated. The sample includes 41% males and is described in detail in the corresponding publication [13] and the TGEN website. Through data imputations using our dataset, the TGEN dataset, and the HapMap CEU data, we achieved an extended dataset that contained 2,048 individuals with complete data on 4,329 SNPs within our region of interest. Parent of origin information was not available for the extended dataset; thus, all such analyses as well as run of homozygosity (ROH) analyses were performed only in our genotyped dataset.
Table 1.
Genotyped sample demographics
| N | Agea | Male | Female | |
|---|---|---|---|---|
| AD cases | 349 | 73.5 | 150 | 199 |
| Maternal cases | 104 | 71.4 | 42 | 62 |
| Controls | 289 | 73.2 | 150 | 139 |
| Total | 638 | 73.4 | 300 | 338 |
Estimated age of onset for cases and age at screening for controls
SNP selection
SNPs for genotyping were chosen and genotyped in two steps, starting with selection of 3,072 SNPs in step 1, followed by an additional 1,499 in step 2. For step 1, HapMap (phase I)-validated SNPs were chosen based on location (chromosome 10: 43.7–61.0 Mb), linkage disequilibrium (LD; all phase I HapMap SNPs were included unless another SNP with r2≥ 0.8 was present), residence in coding sequence (all validated coding SNPs known to dbSNP were included), and conservation (based on University of California, Santa Cruz (UCSC) genome browser PhastCons >0.9), and Illumina design score >0.6 (reduced to 0.4 for irreplaceable tagging SNPs). In step 2, 1,499 SNPs were chosen to supplement step 1, based on newly available HapMap phase II genotyping data. All HapMap phase II SNPs with minor allele frequency (MAF) ≥5% were included unless another SNP with r2≥0.8 (based on phase II genotypes) was present in step 1 or was already selected.
We included five SNPs on chromosome X for quality control (QC) and assessment purposes, and another 152 SNPs outside our region for ancestry estimation. A portion of these were included in step 1 genotyping, chosen to be approximately equally spaced across the genome (one SNP per 3 Mb), of which, 64 passed design phase and were attempted. The remaining portion of QC SNPs was genotyped in step 2. These were chosen based on European-African divergence, from the online Table B of Smith et al. [25]. In total, after data cleaning, we retained 137 of these markers for QC analysis.
Genotyping methods
Genotyping was performed at the Broad/NCRR Center for Genotyping and Analysis using the Illumina Golden Gate custom array platform. We supplied 5 μg of DNA at 100 ng/μl including two blinded QC samples replicated on every 96-well microtiter plate sent to the BROAD for genotyping. The BROAD provided raw data (.idat intensity files) as well as initial genotype calls for analysis.
Raw intensity data for all 4571 SNPs were analyzed locally using the Illumina BeadStudio software to allow inspection and manual modification of clustering if necessary and the use of intensity-level information. We then considered particular genotypes missing if the Illumina quality score (GenCall score) was <0.6. We removed SNPs from final analyses if the proportion of missing genotypes per SNP exceeded 10%, if the MAF<0.01, or if Hardy–Weinberg equilibrium (HWE) in controls was rejected at p<10−4.
Statistical methods
Data cleaning, descriptive statistics, tests of departures from Hardy–Weinberg proportions, and genetic association analyses were performed in R version 2.6. Estimates and plots of LD were also carried out in Haploview (www.broad.mit.edu/mpg/haploview). To assess potential genetic outliers among our genotyped set of self-reported Caucasian samples, we performed STRUCTURE analyses [26] using the 137 markers genotyped for this purpose that were not in our candidate region. Assuming one, two, and three ancestral populations, we did not see evidence for population substructure nor did we detect individual outliers among our samples.
Similar ancestry analyses have been performed on the TGEN dataset [13] that we included in our extended dataset (see below), also with no evidence of substructure. We therefore did not further correct for ancestry in subsequent analyses, yet we did include a “dataset” covariate in the analyses of the extended dataset to account both for sample and platform differences.
Logistic regression models were used to estimate odds ratios (OR) per SNP assuming an additive genetic model, adjusting for age of onset or age at cognitive screen for controls. Nominal p values were estimated via 5,000 permutations of case–control status as the number of permutations with likelihood ratio test (LRT) values are greater than or equal to the observed LRT value per SNP. Given the stronger linkage signal in this region among maternally affected families, we also performed case–control analyses as above restricting cases to only those with known maternal affection. Our empirical p values were highly correlated with asymptotic results. Reported p values are SNP specific and not corrected for the testing of multiple SNPs in the region.
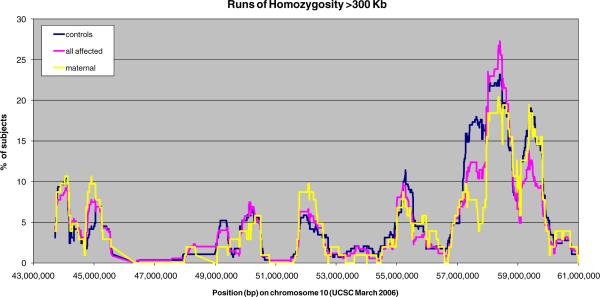
In order to explore the presence of disease predisposing deletions or long homozygous segments that might suggest the involvement of recessive alleles on long haplotypes, we used PLINK [27] (http://pngu.mgh.harvard.edu/purcell/plink/), with an ROH defined as 300 Kb of homozygous SNPs, allowing heterozygosity of ≤1 SNP within that distance, to allow for genotyping errors. None of the identified ROH provided evidence of a deletion after inspection of the signal intensity data in the BeadStudio software. We then plotted the frequency of homozygosity runs per case and controls by location (Fig. 1) to identify any disease-specific ROH.
Fig. 1.
Runs of homozygosity (ROH) within our region of interest. The fraction of individuals carrying an ROH greater than 300 Kb is plotted against the mid-point location of the ROH. Maternal cases, all cases, and controls are plotted separately
Extended dataset
We created a combined data set including genotypes from our case–control sample and GWAS genotypes available from the TGEN case–control data generated on the Affymetrix 500 K GeneChip platform. The TGEN data included 1,943 SNPs with genotypes in our region, of which, 424 overlapped with our genotyped SNPs. In order to accurately impute the largest possible set of SNPs in the combined dataset, we used the imputation platform Beagle v.3.0.4 [28] (http://www.stat.auckland.ac.nz/~bbrowning/beagle/beagle.html) and added the HapMap I and II CEU data which included 5,258 SNPs in common with either our or the TGEN sample. This provides probabilities for each of the three possible genotypes for an SNP when no observed genotype is available. The allele identities and strands (plus or minus) when ambiguous were matched using Chi-square tests between allele counts in the two samples. We required that the selected match is at least 1,000 times more likely than the alternative based on p values. When this level of confidence was not achieved, for example, in SNPs with complementary alleles (C/G or A/T) and with frequencies close to 50%, we used information from neighboring SNPs that were in complete (D′=1) but not perfect (r2<0.8) LD and, therefore, formed only three observed haplotypes with the SNP of interest.
After acquiring the imputed genotype probabilities, we removed SNPs that had an imputation r2<0.5 reflecting insufficient information for imputation, and SNPs that were not in HWE (p<0.001) in our sample, the TGEN sample, or the two combined.
Association tests in this extended dataset were performed via logistic regression analogous to those in our primary data analysis, using the genotype probabilities calculated by Beagle and the analytic package PLINK. We included a “dataset” covariate to correct for possible systematic differences between our and the TGEN datasets. In Supplementary Table 1, we show the statistical power of our extended dataset, the genotyped set, and the maternal subset.
Finally, we compared our strongest signals with those from the study of Li et al. [4]. Those results are publicly available on the GlaxoSmithKline Clinical Trials Registry (http://www.GSK.com).
Nucleotide sequencing
Sequencing by the Sanger method [29] was performed via standard protocols either in our laboratory or commercially by Polymorphic DNA technologies Inc. (Alameda, CA, USA). Primers were designed to allow high quality sequence of each exon as well as at least 10 bp of flanking sequence, including the 5′ and 3′ untranslated regions. Putative promoter sequences were not included. We sequenced the following genes selected because they showed evidence of association and improvement in the maternal stratum: PCDH15, CXCL12, ZNF488, ARHGAP22, CHAT, SLC18A3, PRKG1, MBL2, and GDF10. The PCDH15 gene was sequenced in our laboratory on 14 cases with maternal disease origin and ten unrelated controls. The remaining eight genes were sequenced by Polymorphic DNA technologies on 103 maternal cases and 37 additional cases from families showing linkage to this region. Additional two cases were included because they had available brain tissue for our imprinting analyses. We chose 143 of the oldest controls for comparison. Chromatographs were aligned using CodonCode Aligner (CodonCode Corporation, Dedham, MA, USA), and variations were visually inspected and annotated when they appeared valid and were observed on both strands. All variations were mapped in the genome and were characterized as coding synonymous, non-synonymous, nonsense, non-coding with potential functionality, and non-coding with no evidence of functionality. The latter two categories were assigned based on phylogenetic conservation and the bioinformatics tool RegRNA (http://regrna.mbc.nctu.edu.tw/).
Allele-specific expression
We investigated the possibility of monoallelic expression by genotyping transcribed SNPs on genomic DNA and then temporal lobe cDNA from heterozygous individuals. Polymerase chain reaction (PCR) amplifications specific for genomic DNA or cDNA were performed by use of primers within introns or spanning across exons. The corresponding PCR products were used for genotyping by nucleotide sequencing. We first genotyped genomic DNA from all samples with available brain tissue, 52 brains without gross pathology and 55 brains with AD pathology. We then followed up by genotyping the cDNA of any observed heterozygotes, expecting to fail to observe only one of the two alleles in the event of allele-specific expression.
DNA and RNA extractions and reverse transcription methods are described in detail in Szymanski et al. 2009 [30]. Briefly, our sample included 21 male cases, 34 female cases, 28 male controls, and 24 female controls. The average age for the cases was 79, and the average postmortem delay to brain harvest was 12.4 h. These were 74 years and 15.1 h, respectively, in controls. DNA was extracted using a Puregene kit (Gentra systems–Qiagen®), RNA was extracted using TRIzol (Invitrogen®), and first strand complementary DNA was generated using the TaqMan reverse transcription kit by Applied Biosystems (cat#N8080234) using random primers and the manufacturer-suggested protocols. The tissue samples have been used extensively in our laboratory and show no evidence of significant degradation.
Genotyping and analysis of rs5984894 in PCDH11X
For reasons explained in the discussion, we genotyped rs5984894 in our samples using TaqMan® to test for interactions with SNPs in the PCDH15 gene. We observed a 96.4% call rate, all males were hemizygotes as expected for an X linked marker, and the genotypes of females were in HW equilibrium. Interaction with SNPs showing a parent of origin effect was tested by using the rs5984894 genotypes as a covariate in the regression model alone or together with sex and by testing associations of pseudohaplotypes between rs5984894 and each chromosome 10 marker of interest using Haploview.
Results
Descriptive analyses
-
(a)
Genotyped sample: After missing data filters, 3,997 SNPs remained, of which, 101 had a MAF<1%, and an additional 12 had significant departure from HWE proportions among controls at p<10−4, resulting in 3,884 SNPs for our primary association analyses (Supplementary Table 2). Of the 152 ancestry informative SNPs, 16 were removed due to missing data, but we kept those with MAF<0.01, since they were chosen to have extreme MAF differences between populations, resulting in 136 for ancestry analyses. Four of the five X chromosome SNPs clustered well, and the data were consistent with the subjects' reported sex. Our region of interest lies between 43.7 and 61 Mb on chromosome 10, and after subtracting 2.1 Mb that are within segmental duplications and could not be covered with SNPs, we have an effective region size of 15.2 Mb covered by 3,884 SNPs or a density of one SNP/3.9 Kb for our genotyped dataset. The same region is covered by 3,018 and 2,788 on the Illumina 550 and the Affymetrix v 5.0 arrays, respectively. Our SNP coverage of this region is therefore about 30% greater than either of these platforms that have been commonly used for GWAS.
-
(b)
Extended dataset: We were able to match the alleles of 5,258 SNPs that had data in HapMap and in at least one of the two case–control samples and used them for imputations. We removed 803 SNPs that had an imputation r2<0.5 and 126 SNPs for deviations from HWE, leaving 4,329 SNPs with genotype probability calls on all individuals.
The publicly available result files from the Li et al. study [4] included p values for 3,018 SNPs in our region: 98% of the TGEN SNPs, 16% of our genotyped SNPs, and 39% of SNPs in the extended dataset.
Case–control analyses
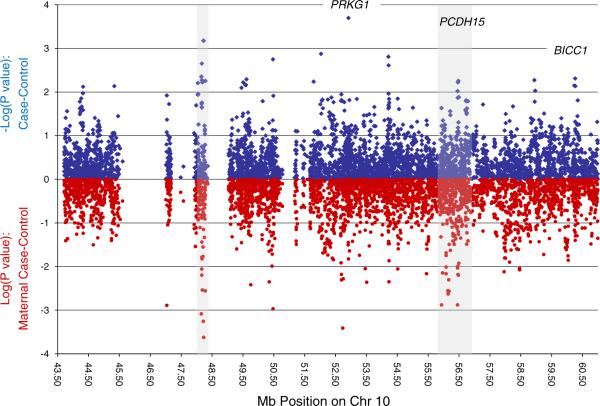
p values for our case–control analyses of each SNP across the region are shown in Fig. 2, along with analogous results when considering only maternal cases. Of 3,884 SNPs, 143 (3.7%) had nominal p values <0.05, and 29 (0.75%) had p values <0.01 for the case–control analysis. The SNPs in the top 1% of p values from case–control analyses are provided in Table 2, along with results for stratified analyses based on maternal affection status. The appropriate significance threshold based on p values is unclear for a set of correlated SNPs such as these, but the threshold likely to protect family-wise type 1 error can be estimated based on the correlation structure of SNPs in this region. We calculated all pairwise r2 values across SNPs and subtracted the maximum r2 for each SNP to any other from the total count of SNPs, without replacement. This provides an estimate of the independent association tests in our analysis (1,673.5). Bonferroni correction for the effective number of tests suggests a threshold of p<3×10−5 to consider study-wide significance. None of our SNPs achieved this level of significance.
Fig. 2.
p values for single nucleotide polymorphism associations across the linkage region among all cases and controls (blue) and when restricted to maternal cases versus controls (red)
Table 2.
Top 1% of associated single nucleotide polymorphisms across fine-mapping region
| Name | Position | Highlighted in Fig.1a | Minor allele | MAF | Case-control comparison |
Maternal case-control comparison |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | Cl_low | Cl_up | LRTpval | PERMp | ORmatb | Cl_low | Cl_up | LRTpval | PERMp | |||||
| rs1325251 | 52,918,572 | PRKG1 | A | 0.12 | 0.53 | 0.37 | 0.77 | 6.19E-04 | 2.00E-04 | 0.53 | 0.32 | 0.88 | 1.49E-02 | 1.50E-02 |
| rs6588761 | 48,219,815 | R1 | A | 0.03 | 0.28 | 0.11 | 0.62 | 1.10E-03 | 6.67E-04 | 0.19 | 0.07 | 0.48 | 5.57E-04 | <0.001 |
| rs2382748 | 48,228,985 | R1 | C | 0.03 | 3.86 | 0.67 | 10.50 | 1.05E-03 | 6.67E-04 | 6.24 | 2.34 | 18.42 | 2.38E-04 | <0.001 |
| rs17581623 | 52,030,619 | A | 0.10 | 0.56 | 0.37 | 0.83 | 3.58E-03 | 1.33E-03 | 0.67 | 0.39 | 1.16 | 1.50E-01 | 1.89E-01 | |
| rs7477069 | 54,212,144 | A | 0.34 | 0.69 | 0.54 | 0.87 | 2.03E-03 | 1.56E-03 | 0.69 | 0.49 | 0.97 | 3.31E-02 | 3.50E-02 | |
| rs7901053 | 50,473,548 | A | 0.41 | 1.43 | 1.14 | 1.80 | 2.11E-03 | 1.78E-03 | 1.74 | 1.24 | 2.46 | 1.08E-03 | <0.001 | |
| rs4922536 | 48,168,383 | R1 | A | 0.47 | 1.39 | 1.12 | 1.74 | 3.13E-03 | 2.22E-03 | 1.56 | 1.12 | 2.20 | 8.78E-03 | 7.00E-03 |
| rs7899656 | 54,219,374 | A | 0.28 | 1.47 | 1.15 | 1.90 | 2.33E-03 | 2.44E-03 | 1.48 | 1.02 | 2.18 | 3.76E-02 | 3.20E-02 | |
| rs9421758 | 48,153,610 | R1 | A | 0.29 | 1.43 | 1.12 | 1.82 | 3.97E-03 | 4.44E-03 | 1.89 | 1.29 | 2.81 | 8.21E-04 | 2.00E-03 |
| rs6481440 | 60,262,929 | G | 0.05 | 0.46 | 0.26 | 0.78 | 4.06E-03 | 4.89E-03 | 0.43 | 0.16 | 0.96 | 3.99E-02 | 3.70E-02 | |
| rs17698900 | 49,616,272 | G | 0.13 | 0.64 | 0.46 | 0.89 | 7.17E-03 | 5.11E-03 | 0.74 | 0.46 | 1.17 | 2.06E-01 | 2.15E-01 | |
| rs4922522 | 48,173,807 | R1 | C | 0.03 | 2.98 | 1.39 | 7.14 | 4.16E-03 | 5.33E-03 | 3.45 | 1.28 | 9.49 | 1.50E-02 | 1.10E-02 |
| rs2393352 | 58,941,770 | A | 0.49 | 1.40 | 1.12 | 1.75 | 2.88E-03 | 5.33E-03 | 1.20 | 0.86 | 1.66 | 2.79E-01 | 2.69E-01 | |
| rs4326728 | 48,283,860 | R1 | G | 0.02 | 3.92 | 1.44 | 13.69 | 6.03E-03 | 5.56E-03 | 6.11 | 1.87 | 23.45 | 2.76E-03 | 1.00E-03 |
| rs10763176 | 56,478,812 | R2/PCDH15 | A | 0.41 | 0.73 | 0.58 | 0.91 | 5.59E-03 | 5.56E-03 | 0.63 | 0.45 | 0.88 | 6.31E-03 | 6.00E-03 |
| rs12356013 | 51,791,236 | A | 0.07 | 0.55 | 0.34 | 0.84 | 5.92E-03 | 5.78E-03 | 0.88 | 0.45 | 1.86 | 7.33E-01 | 7.43E-01 | |
| rs10888205 | 48,186,564 | R1 | A | 0.24 | 1.43 | 1.11 | 1.83 | 5.06E-03 | 6.00E-03 | 1.77 | 1.21 | 2.67 | 2.90E-03 | 3.00E-03 |
| rs11101409 | 49,507,785 | G | 0.01 | 0.20 | 0.05 | 0.65 | 5.71E-03 | 6.00E-03 | 0.25 | 0.01 | 1.29 | 1.09E-01 | 1.17E-01 | |
| rs1922166 | 56,465,084 | R2/PCDH15 | C | 0.41 | 1.38 | 1.10 | 1.73 | 5.70E-03 | 6.00E-03 | 1.58 | 0.14 | 2.22 | 6.57E-03 | 8.00E-03 |
| rs4922505 | 48,042,154 | G | 0.02 | 0.27 | 0.09 | 0.70 | 6.41E-03 | 6.22E-03 | 0.36 | 0.06 | 1.30 | 1.29E-01 | 1.37E-01 | |
| rs10857619 | 49,577,319 | A | 0.43 | 0.74 | 0.59 | 0.93 | 8.61E-03 | 6.89E-03 | 0.82 | 0.60 | 1.13 | 2.21E-01 | 2.09E-01 | |
| rs10508906 | 49,601,155 | G | 0.28 | 1.42 | 1.10 | 1.83 | 6.45E-03 | 6.89E-03 | 1.16 | 0.80 | 1.67 | 4.36E-01 | 4.27E-01 | |
| rs11006267 | 60,233,640 | G | 0.04 | 0.46 | 0.25 | 0.80 | 5.57E-03 | 7.11E-03 | 0.46 | 0.17 | 1.05 | 6.82E-02 | 6.40E-02 | |
| rs12257237 | 45,334,677 | G | 0.09 | 1.79 | 1.17 | 2.76 | 6.52E-03 | 7.33E-03 | 1.61 | 0.87 | 2.92 | 1.26E-01 | 1.27E-01 | |
| rs1838539 | 60,269,468 | G | 0.05 | 0.50 | 0.30 | 0.83 | 7.32E-03 | 7.33E-03 | 0.46 | 0.19 | 1.00 | 5.03E-02 | 5.60E-02 | |
| rs1147967 | 44,322,938 | A | 0.15 | 1.53 | 1.13 | 2.09 | 6.54E-03 | 7.56E-03 | 1.42 | 0.91 | 2.29 | 1.21E-01 | 1.19E-01 | |
| rs7896935 | 49,453,503 | A | 0.12 | 1.58 | 1.12 | 2.23 | 8.78E-03 | 8.00E-03 | 1.72 | 1.04 | 3.02 | 3.53E-02 | 3.30E-02 | |
| rs11005844 | 58,969,302 | A | 0.47 | 0.74 | 0.59 | 0.92 | 7.95E-03 | 9.33E-03 | 0.84 | 0.60 | 1.17 | 2.93E-01 | 3.11E-01 | |
| rs11004683 | 56,454,702 | R2/PCDH15 | C | 0.41 | 1.35 | 1.07 | 1.70 | 9.76E-03 | 9.56E-03 | 1.56 | 1.12 | 2.19 | 9.29E-03 | 8.00E-03 |
| rs7919766 | 44,317,317 | G | 0.17 | 0.68 | 0.50 | 0.91 | 9.46E-03 | 1.07E-02 | 0.74 | 0.47 | 1.13 | 1.72E-01 | 1.93E-01 | |
| rs12570371 | 54,217,463 | A | 0.28 | 1.39 | 1.09 | 1.78 | 8.92E-03 | 1.09E-02 | 1.32 | 0.92 | 1.93 | 1.31E-01 | 1.50E-01 | |
| rs2607872 | 48,051,698 | G | 0.04 | 0.48 | 0.27 | 0.85 | 1.09E-02 | 1.11E-02 | 0.53 | 0.21 | 1.16 | 1.17E-01 | 1.28E-01 | |
| rs10996377 | 52,690,748 | PRKG1 | A | 0.07 | 0.57 | 0.36 | 0.89 | 1.39E-02 | 1.13E-02 | 0.44 | 0.25 | 0.79 | 6.60E-03 | 3.00E-03 |
| rs9421801 | 48,042,861 | A | 0.04 | 2.03 | 1.15 | 3.64 | 1.40E-02 | 1.16E-02 | 1.88 | 0.84 | 4.77 | 1.27E-01 | 1.18E-01 | |
| rs11595808 | 47,034,484 | A | 0.02 | 3.03 | 1.29 | 7.95 | 1.06E-02 | 1.20E-02 | NA | NA | NA | 1.29E-03 | 5.00E-03 | |
| rs885834 | 50,485,518 | G | 0.38 | 1.35 | 1.07 | 1.70 | 1.15E-02 | 1.22E-02 | 1.37 | 0.99 | 1.90 | 6.04E-02 | 5.80E-02 | |
| rs1922158 | 56,494,637 | R2/PCDH15 | A | 0.41 | 0.75 | 0.59 | 0.94 | 1.21E-02 | 1.31E-02 | 0.65 | 0.47 | 0.91 | 1.19E-02 | 1.30E-02 |
| rs10761612 | 52,368,092 | C | 0.44 | 0.76 | 0.60 | 0.94 | 1.39E-02 | 1.33E-02 | 0.86 | 0.62 | 1.18 | 3.50E-01 | 3.76E-01 | |
| rs1922155 | 56,458,141 | R2/PCDH15 | G | 0.41 | 1.33 | 1.06 | 1.68 | 1.31E-02 | 1.36E-02 | 1.55 | 1.11 | 2.18 | 1.00E-02 | 1.40E-02 |
CO controls, CA cases
Rl, R2 refer to region 1 or 2 in gray in Fig. 1
Italic indicates stronger effect in maternal subsets
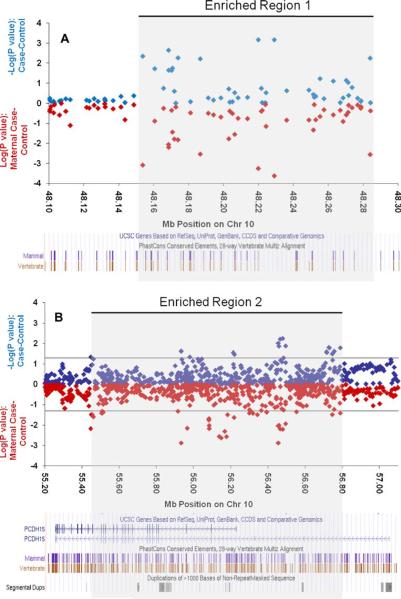
From our genotyped dataset results, two areas across our fine-mapping region (gray shading in Fig. 2) are worth noting for an observed clustering of nominally significant SNPs: a large intergenic region near position 48.2 Mb and a second region, at 56.4 Mb, containing the gene for protocadherin 15 (PCDH15). The first area, between 48.1 and 48.3 Mb on chromosome 10 (UCSC March 2006), contains 1.7% of all tested SNPs, but 18% (7/39) of the top 1% most significantly associated SNPs (Fig. 3a and Table 2). These SNPs are not strongly correlated with each other (only one pair has an r2 of 1, all others are below 0.6 with an average r2=0.2 for all seven). All seven SNPs show stronger association in the maternal subset, and 20% (8/39) of the top 1% in the maternal stratum is found in this small region. This region is devoid of known genes, harboring only two gene predictions that show no homologies with known proteins and no RNA evidence. It is flanked on the telomeric side by a “non-SNPable” segmental duplication which contains one of two almost identical copies of the gene PTPN20, the other copy located at ~46 Mb also within our region of interest. On the centromeric side, the region is flanked by the gene GDF10.
Fig. 3.
Regions with enrichment for nominally significant single nucleotide polymorphisms. Full case-control comparisons in blue, maternal case-control comparisons in red. Genes and conservation information from University of California, Santa Cruz shown at bottom of each panel
The second area showing an enrichment of nominally significant SNPs among the maternal cases was observed between 55.4 and 56.8 Mb, which overlaps with the PCDH15 gene locus. Thirteen percent (5/39) of our top 1% most significantly associated SNPs is located in this gene, which includes 11.6% of all tested SNPs, and is thus not greater than expected. However, 13 of 19 nominally significant SNPs in this gene improve association (and ORs) when restricting cases to only those with maternal affection (see Fig. 3b and Table 2). If one considers the distribution of p values among the maternal subset analyses across the entire fine-mapping region, 36% of the top 1% most significantly associated SNPs is located in this gene, more than threefold higher than the expected 11.6%.
Of additional interest among the top results of the maternal subset was an area which corresponds to the cyclic GMP-dependent protein kinase gene (PRKG1), spanning positions 52.4 to 53.7 Mb (11.8% of all SNPs in our fine-mapping panels). This gene includes the strongest association in the overall sample, while seven (18%) of the SNPs in the top 1% of the maternal subset analysis also mapped to this gene region. p values among those SNPs ranged from of 0.001 to 0.014.
Neither haplotype analyses nor stratification by APOE status or sex revealed additional areas of the fine-mapping region with significance levels above those expected by chance. These results are therefore not presented in detail. Finally, although at least one notable ROH was identified located at around 58 Mb, no significant differences were observed between cases and controls (Fig. 1).
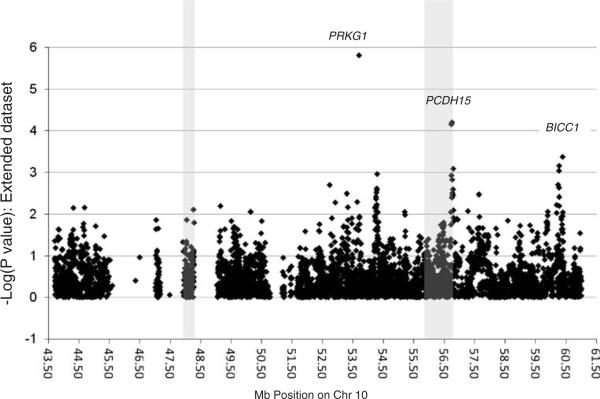
Results for analyses of the extended dataset are shown in Fig. 4 and Table 3. One SNP, rs10824310, showed statistical significance (p = 1.5 × 10−6) that can withstand correction for multiple testing. This significance was due entirely to the TGEN dataset which alone provided a p = 1.0 × 10−6, while our genotyped dataset showed the same direction but no support alone (p=0.24). SNP rs10824310 was genotyped by TGEN and imputed in our dataset; therefore, imputation errors would not generate a spurious association. This SNP is located in the PRKG1 gene, the same as rs1326251, which was the best signal in our genotyped sample. The TGEN sample showed only weak excess of risk alleles for the latter SNP (p=0.33 in TGEN alone, p=0.005 in the extended dataset). We examined the results of Li et al. [4] and found that rs10824310 was present but did not show evidence of association (p=0.9), while rs1326251 was not reported.
Fig. 4.
p values for single nucleotide polymorphism associations across the linkage region in the extended dataset. The shaded areas are the same as in Fig. 2
Table 3.
Top 1% associated single nucleotide polymorphisms in the extended dataset
| Name | Position | Region | Minor allele (+ strand) | MAF | OR | Cl_low | Cl_up | Pval |
|---|---|---|---|---|---|---|---|---|
| rs10824310 | 53,698,470 | PRKG1 | T | 0.07 | 0.52 | 0.25 | 0.79 | 1.5E-06 |
| rs2441760 | 56,747,641 | R2/PCDH15 | C | 0.27 | 1.40 | 1.23 | 1.57 | 6.2E-05 |
| rs7089764 | 60,382,682 | C | 0.17 | 0.35 | 1.18 | 1.52 | 0.00042 | |
| rs1838539 | 60,269,468 | BICC1 | G | 0.07 | 0.63 | 0.36 | 0.90 | 0.0007 |
| rs6481440 | 60,262,929 | BICC1 | C | 0.06 | 0.64 | 0.37 | 0.91 | 0.00092 |
| rs11006267 | 60,233,640 | BICC1 | G | 0.06 | 0.65 | 0.36 | 0.93 | 0.00198 |
| rs7085810 | 60,263,748 | BICC1 | C | 0.09 | 0.67 | 0.41 | 0.93 | 0.00231 |
| rs7098555 | 60,262,814 | BICC1 | A | 0.08 | 0.67 | 0.41 | 0.93 | 0.00234 |
| rs1900493 | 56,783,626 | R2/PCDH15 | T | 0.48 | 1.22 | 1.09 | 1.35 | 0.00255 |
| rs10825499 | 56,779,323 | R2/PCDH15 | A | 0.48 | 1.22 | 1.09 | 1.35 | 0.00257 |
| rs1935927 | 54,282,884 | C | 0.23 | 1.27 | 1.11 | 1.43 | 0.00264 | |
| rs10824815 | 54,269,972 | G | 0.22 | 1.26 | 1.10 | 1.41 | 0.00304 | |
| rs1900472 | 56,765,259 | R2/PCDH15 | C | 0.48 | 1.21 | 1.08 | 1.35 | 0.00313 |
| rs6480524 | 53,297,004 | PRKG1 | G | 0.26 | 0.80 | 0.65 | 0.95 | 0.00321 |
| rs2218582 | 56,773,366 | R2/PCDH15 | A | 0.48 | 1.21 | 1.08 | 1.34 | 0.00345 |
| rs1900482 | 56,778,201 | R2/PCDH15 | C | 0.49 | 1.21 | 1.08 | 1.34 | 0.00355 |
| rs11819131 | 54,253,439 | T | 0.23 | 1.25 | 1.09 | 1.40 | 0.00378 | |
| rs4098805 | 54,256,054 | C | 0.23 | 1.24 | 1.09 | 1.40 | 0.00428 | |
| rs10508965 | 53,639,284 | PRKG1 | A | 0.03 | 1.83 | 1.40 | 2.26 | 0.00505 |
| rs1326251 | 52,918,572 | PRKG1 | T | 0.12 | 1.36 | 1.14 | 1.58 | 0.00522 |
| rs7902899 | 54,280,438 | G | 0.42 | 1.19 | 1.06 | 1.32 | 0.00587 | |
| rs7094271 | 60,185,028 | BICC1 | T | 0.46 | 0.82 | 0.67 | 0.96 | 0.00594 |
| rs11003193 | 54,260,405 | C | 0.42 | 1.19 | 1.06 | 1.32 | 0.00611 | |
| rs17009540 | 49,149,175 | T | 0.05 | 0.67 | 0.38 | 0.96 | 0.00633 | |
| rs11819277 | 54,248,264 | T | 0.21 | 1.24 | 1.08 | 1.40 | 0.00656 | |
| rs1937663 | 53,330,084 | PRKG1 | A | 0.29 | 0.83 | 0.68 | 0.97 | 0.00674 |
| rs7899656 | 54,219,374 | T | 0.29 | 0.82 | 0.66 | 0.97 | 0.00694 | |
| rs4098804 | 54,256,334 | A | 0.22 | 1.24 | 1.08 | 1.40 | 0.00694 | |
| rs915085 | 44,323,185 | A | 0.08 | 0.73 | 0.50 | 0.96 | 0.00702 | |
| rs7917334 | 54,240,857 | G | 0.37 | 1.19 | 1.06 | 1.32 | 0.00705 | |
| rs1935924 | 54,258,086 | G | 0.42 | 1.18 | 1.06 | 1.31 | 0.00842 | |
| rs2114561 | 59,895,941 | A | 0.22 | 1.23 | 1.07 | 1.39 | 0.00868 | |
| rs4385797 | 54,291,036 | A | 0.34 | 0.83 | 0.70 | 0.97 | 0.00868 | |
| rs11812365 | 50,139,287 | C | 0.16 | 1.27 | 1.09 | 1.45 | 0.0087 | |
| rs10857452 | 50,139,698 | C | 0.07 | 1.43 | 1.16 | 1.71 | 0.0087 | |
| rs11003238 | 54,309,106 | C | 0.33 | 0.83 | 0.69 | 0.97 | 0.00883 | |
| rs11006325 | 60,390,246 | T | 0.16 | 1.26 | 1.08 | 1.44 | 0.00897 | |
| rs1343043 | 54,306,825 | A | 0.34 | 0.84 | 0.70 | 0.97 | 0.00903 | |
| rs11815410 | 60,186,341 | BICC1 | T | 0.44 | 1.19 | 1.05 | 1.32 | 0.00927 |
| rs11003831 | 55,208,481 | A | 0.43 | 0.84 | 0.71 | 0.98 | 0.01032 | |
| rs1125139 | 59,889,658 | G | 0.20 | 1.23 | 1.07 | 1.39 | 0.01075 | |
| rs10763591 | 60,375,162 | A | 0.16 | 1.25 | 1.07 | 1.42 | 0.01092 | |
| rs11003138 | 54,217,179 | C | 0.46 | 1.18 | 1.05 | 1.31 | 0.011 |
The PCDH15 locus remained of interest in the extended dataset despite the lack of parent of origin information. Although the set of SNPs toward the middle of the gene that had been previously identified for a parent of origin effect did not show significance, another set of six SNPs toward the 5′ (telomeric) end of the gene showed p values between 6.2×10−5 and 3.5×10−3. The strongest signal was for rs2441760, which was also nominally significant in the genotyped sample alone (p=0.03).
The intergenic region at 48.2 Mb showed no evidence of association in the extended dataset. Since that region was highlighted mainly because of the parent of origin effect, its importance cannot be assessed in the extended dataset due to the lack of parental data.
Finally, the extended dataset analysis highlighted a new region toward the telomeric end of our linkage at 60.18 to 60.39 Mb on chromosome 10 containing the gene BICC1. In this region, the extended dataset showed ten SNPs with p values between 4×10−4 and 9×10−3. Four of these SNPs (rs11006267, rs7098555, rs6481440, and rs1838539) were nominally significant in both our genotype and TGEN dataset. One SNP, rs7089764, showed the strongest significance in the stratum of patients not carrying any APOE e4 alleles (p=7×10−5 compared to p=6×10−3;OR 1.55 compared to 1.23), a trend that was in the same direction when examining the genotyped sample alone (p=0.044 from 0.07; OR 1.5 from 1.3).
Sequencing
We followed up our association analysis for common variation contributing to disease with a nucleotide sequencing analysis to test the hypothesis that multiple individually rare alleles in one or many genes contribute to risk. We selected what we considered the most likely candidate genes for sequence analysis weighing strongly toward genes that fit our primary hypothesis showing a possible parent of origin effect, but including our strongest result overall in the PRKG1 gene. Our rational was that the observed relatively weak associations might be the result of concentrations of such rare alleles on a haplotype or that the same gene might include both common and rare risk variants. PCDH15 was sequenced in 14 cases with maternal disease origin and ten unrelated controls. We identified 13 common previously known variants and six previously unknown variants; however, allele distributions were similar cases and controls. Eight additional genes (CXCL12, ZNF488, ARHGAP, CHAT, SLC18A3, PRKG1, MBL2, and GDF10) were then sequenced in 283 individuals (140 cases and 143 controls). These genes were selected because SNPs located in their vicinity showed significance, which improved in the maternal subset. A total of 134 DNA variants were identified within or close to exons including 74 previously unknown variants (NCBI submission numbers ss124987499 through ss124987572). We examined all common variants for frequency differences between cases and controls. One novel variant (ss124987507 genome coordinates chr10:49,333,190, build March 2006) showed a significant allele frequency difference between cases and controls (p=0.0035) and no strong LD with any genotyped SNP. This difference however was no longer observed when additional cases and controls were genotyped. We then grouped variants together and globally compared frequency of the alternative (non-reference) allele between cases and controls in the following nested categories: (1) coding non-synonymous SNPs, (2) all coding SNPs, (3) the previous SNPs plus any SNPs in conserved regions and with functional potential based on RegRNA predictions, and finally, (4) the previous SNPs plus any with functional potential even in non-conserved regions. In all cases, we further sub-grouped SNPs with MAF<0.01, MAF<0.05, and all MAF. None of these comparisons provided any evidence of difference between cases and controls for any one gene or all genes combined, failing to support the multiple rare allele hypothesis for the genes in question.
Allele-specific expression
We were able to find heterozygotes with available tissue for RNA extraction for six genes. The list of genes follows with the number of controls and cases that were informative for monoallelic expression in parenthesis: ARHGAP22 (3, 4), GDF10 (6, 1), PCDH15 (3, 2), PRKG1 (0, 1), SLC16A9 (0, 3), and ZNF488 (2, 5). In all instances, sequencing detected both allelic variants on the transcript ruling out the possibility of exclusively monoallelic expression unless it is tissue specific, splice variant specific, temporally variant, or polymorphic. Our results suggest no consistent genomic imprinting in the temporal lobe for the tested genes.
Discussion
We have performed a dense association study of the chromosome 10 region implicated in AD by many groups. We have covered the one-LOD interval of our linkage peak, which largely coincides with the region identified in a recent combined linkage analysis [17]. The peak marker (D10S464 at 60.9 Mb) in that report is less than 4 Mb from our peak linkage marker, D10S1221, and our fine-mapping region includes two thirds (12 Mb) of their one-LOD interval [17]. Our study of 3,884 SNPs and the extended dataset of 4,329 markers on 2,048 samples represents the mostcomplete coverage of this genomic region to date. One SNP in the PRKG1 gene achieved study-wide statistical significance; however, the same SNP shows no trend for association in the independent dataset of Li et al. [4]. Other interesting regions emerged with over-representation of nominally significant SNPs and support from both the genotyped and the extended dataset, or from strong parent of origin effects. These include the genes BICC1 and PCDH15, as well as an intergenic region of 240 kb. Signals in these regions often improve when we restrict analysis to maternal cases, consistent with our linkage findings.
The cyclic GMP-dependent protein kinase gene (PRKG1) product, protein kinase G (PKG), is a serine/threonine-specific protein kinase that is activated by cyclic GMP. This gene showed the strongest signal in the genotyped sample, a high concentration of other strong signals in maternal cases, and the strongest, albeit not the same, signal in the extended dataset. PKG is known to be involved in the regulation of smooth muscle relaxation and platelet function but, most importantly, in neural cell survival [31]. Due to its function, it has been proposed as a therapeutic target for neurodegenerative disorders including AD [31]. Its involvement in neuroprotection makes PRKG1 an exceptional candidate for AD as any perturbation of this function would presumably make the brain vulnerable to increased loss of neurons and lower the biological liability threshold for developing dementia. The lack of consistency in the results from different datasets, the lack of significance in the results of Li et al., and the lack of candidate rare coding variants reduce the enthusiasm; however, these results could reflect multiple non-coding variants with unequal representation in different haplotypes and in different datasets. This is a difficult hypothesis to test, although recent high throughput sequencing methods might make it possible.
Protocadherin15 (PCDH15) is a member of the cadherin superfamily, which codes for integral membrane proteins that mediate calcium-dependent cell-cell adhesion. The high concentration of association signals in the maternal case analysis, especially in the absence of signals before the stratification, is reminiscent of our linkage results, and it makes this gene region particularly interesting. Adding to this, another protocadherin, PCDH11X, was recently reported to be strongly associated with AD [6]. Adhesion molecules such as PCDH15 and PCDH11X regulate cellular adhesion through specific interactions on the opposing cell or surface. Beyond its role as a cell adherence protein, the PCDH15 gene has been identified as essential to the maintenance of normal retinal and cochlear function. PCDH15 is best known for its role in the development and maintenance of hairlike projections called stereocilia in the inner ear and in the development and maintenance of retinal photoreceptors. Mutations in the PCDH15 gene are associated with deafness and hearing loss as well as vision problems [32]. PCDH15 is located exactly under the peak of our linkage signal. Since protocadherins are by definition involved in interactions, we hypothesized that PCDH15–PCDH11X interactions might underlie AD risk. The sexual dimorphism that has been observed for PCDH11X which is located on chromosome X but escapes X-inactivation [33] and the dose-dependent risk observed for the risk allele of PCDH11X [6] provides an interesting possibility that might explain the maternal origin effect that we observe around PCDH15. However, after genotyping the AD-associated variant in PCDH11X (rs5984894), we found no association with AD and no evidence of interaction with any of the SNPs in PCDH15 that showed association in the maternal cases (PCDH11X-rs5984894 allele counts are provided in Supplementary Table 3).
We also noted a cluster of nominally significant associations in the genotyped dataset within a gene devoid region at 48.1–48.3 Mb. This region contains only 1.7% of all tested SNPs but concentrates 18% (7/39) of the top 1% signals in the analysis of all genotyped cases and 20% (8/39, a 15-fold enrichment) of the top 1% signals in maternal cases including the best maternal signal (p<0.001, see Supplementary Table 4). The associations were not observed in the extended dataset and might be specific to the maternal cases. Intergenic regions have been shown to often contain regulatory sequences that may act from a distance to control the expression of neighboring genes [34–36]. The genes which flank this region are GDF10 (growth differentiation factor 10) upstream and protein tyrosine phosphatase (PTP), non-receptor type 20B (PTPN20B) downstream. GDF10 codes for a protein in the transforming growth factor beta superfamily that is closely related to bone morphogenetic protein family. Proteins in this family regulate cell growth and differentiation in both embryonic and adult tissues. The PTPN20B gene product is PTPs. These proteins are a group of enzymes that remove phosphate groups from phosphorylated tyrosine residues on proteins. Together with tyrosine kinases, PTPs regulate the phosphorylation state of many important signaling molecules, such as the MAP kinase family. PTPs are associated with the regulation of several fundamental cellular processes which may include cell growth and differentiation, mitotic cycles, and oncogenic transformation. Of note, the PTPN20B gene is the result of a segmental duplication on chromosome 10 and is the near identical copy of PTPN20A. A third copy is only partially duplicated and contains a pseudogene, designated as PTPN20C. As a result of the duplication, the PTPN20A gene is in the non-SNPable part of our region.
The BICC1 gene located at 60.2 Mb attracted our attention only in the extended dataset due to a concentration of association signals, which we also found to be present but weaker in the genotyped dataset. The gene encodes for the bicaudal C homolog 1, a protein that was originally recognized to be important in Drosophila oogenesis [37, 38] and later found to control cilia orientation [39], to cause polycystic kidney disease in mouse models [40] and to be a substrate for caspases and essential for cell survival [41]. The gene has been previously explored for association with AD in a relatively small sample with negative results [42].
Our previous expression studies on genes in the chromosome 10 linkage region revealed a novel gene at 10q11.23, ASAH2L [43]. This partial paralog of the alkaline ceramidase gene displayed reduced expression with increasing age, reduced expression in females across ages, and further reduction in LOAD patients. In concordance with the observed parent of origin effect on the linkage, this reduction was more pronounced in patients with an affected mother. In the present study, only nominally significant associations were observed near this gene region which, however, is not adequately tested due to the segmental duplication it lies within. Stratification by maternal parent of origin marginally improved significance.
Additional candidate genes have been investigated for association with development of AD in the chromosome 10 linkage region by other research groups. These genes include RASSF4 (Ras association (RalGDS/AF-6) domain family 4), ALOX5 (arachidonate 5-lipoxygenase), ANXA8 (annexin A8), SLC18A3 (solute carrier family 18 (vesicular acetylcholine), member 3), ACF (apobec-1 complementation factor), DKK1 (dickkopf homolog 1), and ZWINT (human ZW10 interacting protein-1). Our analysis did not provide significant support for any of these genes.
Our fine-mapping effort provides the greatest density of coverage for this region to date. Nevertheless, we remain underpowered to detect associations of alleles with very small effects (i.e., ORs at or below ~1.3, see Supplementary Table 1). While prior linkage results argue for a relatively strong effect, that signal may be divided across multiple alleles and/or genes with smaller individual effects in this region, leading to the lack of region-wide statistical significance among our association results. Our efforts to identify such alleles by sequencing the coding regions of selected genes did not provide a positive result. We also did not detect genomic imprinting in the temporal lobe. Our results provide a few interesting candidate regions that often show the same parent of origin effect observed by linkage, yet we cannot conclusively point to the AD risk variants underlying the reported linkage signals. It is likely that as more genome-wide association data become available for AD, perhaps some with parent of origin information, our data, together with new knowledge about the disease, will contribute to the identification of the elusive chromosome 10 gene(s).
Supplementary Material
Acknowledgements
This work was supported by National Institute on Aging (NIA) grants to DA and SSB (RO1AG022099 and RO1AG021804) and an award from the Neurosciences Education and Research Foundation to DA. We thank the Harvard Brain Tissue Resource Center (MH/NS077550) and the Johns Hopkins Brain resource center for providing us with the postmortem brain tissue used for these studies. Genotyping was in part subsidized by the Broad Institute Center for Genotyping and Analysis, which is supported by grant U54 RR020278-01 from the National Center for Research Resources. Samples from the NCRAD, which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the NIA, were used in this study. We also thank Kelly Benke, Katherine Miller, and Delphine Fradin for help with data cleaning and descriptive analysis scripts. We thank contributors, including the Alzheimer's Disease Centers who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10048-010-0234-9) contains supplementary material, which is available to authorized users.
Ethical standards The experiments reported in this manuscript comply with the current laws of the country in which they were performed (USA).
References
- 1.Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- 2.Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, et al. A genome-wide association study for late-onszheimer's disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onszheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Wetten S, Li L, Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 5.Poduslo SE, Huang R, Huang J, Smith S. Genome screen of late-onszheimer's extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:50–55. doi: 10.1002/ajmg.b.30767. [DOI] [PubMed] [Google Scholar]
- 6.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onszheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA. Genome-wide association study implicates a chromosome 12 risk locus for late-onszheimer disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, et al. Evidence for novel susceptibility genes for late-onszheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 10.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 12.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS ONE. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late-onszheimer disease. Am J Med Genet. 2002;114:679–686. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- 15.Bassett SS, Avramopoulos D, Perry RT, Wiener H, Watson B, Jr, Go RC, Fallin MD. Further evidence of a maternal parent-of-origin effect on chromosome 10 in late-onszheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:537–540. doi: 10.1002/ajmg.b.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, et al. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 17.Hamshere ML, Holmans PA, Avramopoulos D, Bassett SS, Blacker D, Bertram L, Wiener H, Rochberg N, Tanzi RE, Myers A, et al. Genome-wide linkage analysis of 723 affected relative pairs with late-onszheimer's disease. Hum Mol Genet. 2007;16:2703–2712. doi: 10.1093/hmg/ddm224. [DOI] [PubMed] [Google Scholar]
- 18.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 19.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, et al. Susceptibility locus for Alzheimer's disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 20.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 21.Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onszheimer's disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 22.Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD. Evidence for a novel late-onszheimer disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75:398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassett SS, Kusevic I, Cristinzio C, Yassa MA, Avramopoulos D, Yousem DM, Fallin MD. Brain activation in offspring of AD cases corresponds to 10q linkage. Ann Neurol. 2005;58:142–146. doi: 10.1002/ana.20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D, et al. ApoE-4 and age at onset of Alzheimer's disease: the NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 25.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szymanski M, Wang R, Fallin MD, Bassett SS, Avramopoulos D. Neuroglobin and Alzheimer's dementia: Genetic association and gene expression changes. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.10.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiscus RR. Involvement of cyclic GMP and protein kinase G in the regulation of apoptosis and survival in neural cells. Neurosignals. 2002;11:175–190. doi: 10.1159/000065431. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopes AM, Ross N, Close J, Dagnall A, Amorim A, Crow TJ. Inactivation status of PCDH11X: sexual dimorphisms in gene expression levels in brain. Hum Genet. 2006;119:267–275. doi: 10.1007/s00439-006-0134-0. [DOI] [PubMed] [Google Scholar]
- 34.Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- 35.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 36.Uchikawa M, Takemoto T, Kamachi Y, Kondoh H. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev. 2004;121:1145–1158. doi: 10.1016/j.mod.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Mahone M, Saffman EE, Lasko PF. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. Embo J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol. 1998;18:4855–4862. doi: 10.1128/mcb.18.8.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development. 2009;136:3019–3030. doi: 10.1242/dev.038174. [DOI] [PubMed] [Google Scholar]
- 40.Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, Bryda EC. Positional cloning of jcpk/bpk locus of the mouse. Mamm Genome. 2003;14:242–249. doi: 10.1007/s00335-002-2241-0. [DOI] [PubMed] [Google Scholar]
- 41.Creagh EM, Brumatti G, Sheridan C, Duriez PJ, Taylor RC, Cullen SP, Adrain C, Martin SJ. Bicaudal is a conserved substrate for Drosophila and mammalian caspases and is essential for cell survival. PLoS ONE. 2009;4:e5055. doi: 10.1371/journal.pone.0005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riemenschneider M, Mahmoodzadeh S, Eisele T, Klopp N, Schwarz S, Wagenpfeil S, Diehl J, Mueller U, Foerstl H, Illig T, et al. Association analysis of genes involved in cholesterol metabolism located within the linkage region on chromosome 10 and Alzheimer's disease. Neurobiol Aging. 2004;25:1305–1308. doi: 10.1016/j.neurobiolaging.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Avramopoulos D, Wang R, Valle D, Fallin MD, Bassett SS. A novel gene derived from a segmental duplication shows perturbed expression in Alzheimer's disease. Neurogenetics. 2007;8:111–120. doi: 10.1007/s10048-007-0081-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.