Abstract
The anti-apoptotic protein FLIPS is a key suppressor of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) -induced apoptosis in human glioblastoma multiforme (GBM) cells. We previously reported that a novel phosphatase and tensin homolog (PTEN)-Akt-atrophin interacting protein 4 (AIP4) pathway regulates FLIPS ubiquitination and stability, although the means by which PTEN and Akt were linked to AIP4 activity were unclear. We here report that a second regulator of ubiquitin metabolism, the ubiquitin-specific protease (USP) 8, is a downstream target of Akt, and that USP8 links Akt to AIP4 and the regulation of FLIPS stability and TRAIL resistance. In human GBM xenografts, levels of USP8 correlated inversely with pAkt levels, and genetic or pharmacologic manipulation of Akt regulated USP8 levels in an inverse manner. Over-expression of WT USP8, but not catalytically inactive USP8, increased FLIPS ubiquitination, decreased FLIPS half-life, decreased FLIPS steady-state levels, and decreased TRAIL resistance, while siRNA-mediated suppression of USP8 levels had the opposite effects. Because high levels of the USP8 deubiquitinase correlated with high levels of FLIPS ubiquitination, USP8 appeared to control FLIPS ubiquitination through an intermediate target. Consistent with this idea, over-expression of WT USP8 decreased ubiquitination of the FLIPS E3 ubiquitin ligase AIP4, an event previously shown to increase AIP4-FLIPS interaction, while siRNA-mediated suppression of USP8 increased AIP4 ubiquitination. Furthermore, the suppression of FLIPS levels by USP8 over-expression was reversed by introduction of siRNA targeting AIP4. These results show that USP8, a downstream target of Akt, regulates the ability of AIP4 to control FLIPS stability and TRAIL sensitivity.
Keywords: glioblastoma, TRAIL, ubiquitin, PTEN, USP8
Introduction
Ubiquitination is a post-translational modification used by cells to alter protein stability and function (1, 2). Protein ubiquitination is accomplished by the co-ordinated action of a series of proteins referred to as E1, E2 and E3 enzymes. The E3 ubiquitin ligases (of which over 1000 are encoded in the human genome)(3) catalyze the rate-limiting step of the process and facilitate the transfer of the activated ubiquitin protein to lysine (K) in the target protein. The ligation of a single ubiquitin molecule at one or multiple lysines in the target protein (monoubiquitination) changes target protein activity and/or cellular location, while chain-like addition of multiple ubiquitin molecules to the sites of monoubiquitination (at K48 or K63 in ubiquitin itself)(polyubiquitination) leads to alterations in protein sorting and activity (K63 polyubiquitination), or more critically, to degradation of the targeted protein (K48 polyubiquitination)(2, 4). Ubiquitination, however, is a reversible process, and in addition to ubiquitin-binding, -activating, and -ligating enzymes, de-ubiquitinating enzymes also exist. These ubiquitin-removing proteases, referred to as de-ubiquitinating proteins, exist in several categories including ubiquitin C-terminal hydrolases, sumo-specific proteases, and perhaps most importantly, ubiquitin-specific proteases (USPs) (5). The USPs comprise the bulk of de-ubiquitinating enzymes in the genome (6, 7) and by their ability to cleave the isopeptide bond between ubiquitin and substrate proteins or other ubiquitin molecules, are involved in both the generation of ubiquitin as well as the tailoring of patterns of ubiquitination in target proteins (8). Although the USPs play a clear and complementary role in the ubiquitination process, substrates have been identified for only a few of the over 50 USPs that exist in the human genome, and processes known to be regulated by USPs are limited.
In a previous study, we reported that ubiquitination plays a key role in the sensitivity of GBM cells to the pro-apoptotic tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)(9). TRAIL binds to the death receptors 4 and 5 and induces a tumor-selective, type I extrinsic apoptotic cell death (10, 11). The sensitivity of glioblastoma multiforme (GBM), the most aggressive form of brain cancer, is variable, and in a previous report we showed that levels of FLIPS, a truncated splice variant of FLIP, were regulated by the PTEN-Akt-mTOR pathway, and that PTEN loss and Akt activation correlated in vitro, in human GBM xenografts, and in primary human GBM samples, with increased FLIPS mRNA translation, high levels of FLIPS expression, and TRAIL resistance (12). We also noted, however, that the PTEN pathway not only regulated FLIPS mRNA translation, but also the stability of the FLIPS protein. Specifically we found that PTEN, via an Akt-dependent but mTOR independent mechanism, regulated the ubiquitination, localization, and activity of the E3 ubiquitin ligase atrophin-interacting protein 4 (AIP4), and that in response to PTEN loss and AIP4 inactivation, FLIPS was retained in a non-ubiquitinated, stable form that accumulated and contributed to TRAIL resistance. AIP4, however, is not known to be a substrate of Akt, and the mechanism by which PTEN loss and Akt activation are linked to AIP4 activity was not defined. In this study we provide evidence that a deubiquitinase, USP8, provides the link between Akt activation and loss of AIP4 function, and that PTEN uses this USP8-AIP4 ubiquitin switch to regulate the process of ubiquitination, protein stability, and TRAIL sensitivity.
Materials and Methods
Cell culture and drug treatment
Transformed mouse astrocytes (TMA) derived from wild-type (WT) or PTEN conditional knockout (KO) mice [P2 animals with genotypes Pten+/+;GFAP-cre (WT) and PtenloxP/loxP;GFAP-cre (Pten cKO) provided by Suzanne Baker, St. Jude's Children's Research Hospital, Memphis, TN](13) and transformed with SV40 large T antigen and mutant V12 H-Ras (14, 15) were provided by Dr. Gabriele Bergers (UCSF, San Francisco, CA). Freshly resected human GBM xenografts were obtained from the UCSF Brain Tumor Research Tissue Bank, dissected into small (<1 mm diameter) pieces, passed through a 100-μm-pore-size tissue culture sieve and grown on reduced matrigel-coated dishes (Fisher Scientific). The PTEN status of GBM tissues was previously described (12). The TMA and GBM xenografts were incubated with either vehicle (DMSO, 24-72h), Akt inhibitor III (50 μM, 24h; Calbiochem), or rapamycin (100 nM, 24h; Cell Signaling Technology), after which cycloheximide (CHX, 100 μg/ml) or vehicle was added for an additional 24-72h. For studies of apoptosis, cells were incubated with TRAIL (800 ng/ml, 24 h) dissolved in a hypertonic solution containing Tris buffer pH 7.2 and supplied as a gift from Avi Ashkenazi (Genentech, South San Francisco, CA). Both TMAs and GBM xenograft cell lines were cultured in Dulbecco's modified Eagle's medium (H-21) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere.
Retroviral infection, transfection of plasmids, and siRNA
The retroviral construct pWzl-hygro-M(+)AktER was a generous gift from Martin McMahon (UCSF Comprehensive Cancer Center, San Francisco, CA). The construction of this vector has been previously described (12). Expression of the construct following infection of target cells was induced by the addition of 10 μM 4-hydroxytamoxifen (4-HT)(24h). The pcDNA mammalian expression construct encoding either a wild-type or catalytically inactive mutant (C478A) of USP8 was kindly provided by Kermit C. Carraway (University of California-Davis, Davis, CA). Cells were retrovirally infected as previously described (16-18), with pools of productively infected cells obtained by selection with neomycin (1 mg/ml, 7 days) or hygromycin B (400μg/ml, 7 days). Mammalian expression constructs were transfected into target cells using lipofectamine (Invitrogen) per manufacturer's instructions, after which stably transfected cells were selected using neomycin (1 mg/ml, 7 days), and verified for over-expression of the target protein by Western blot. For ubiquitination studies, cells were similarly transiently transfected with a construct encoding hemagglutinin (HA)-tagged ubiquitin for 48 hrs before the onset of further cell procedures. For siRNA studies, AIP4-targeted siRNA (300 nM, Ambion, identification no. 120674), USP8-targeted siRNA (300 nM, Ambion, identification no.105117), or scramble control (300 nM, silencer negative control #1, Ambion) were transfected into cells using lipofectamine and target protein levels were analyzed 1 to 4 days later by Western blot. In studies involving CHX, cells received the appropriate siRNA, scramble control, or vehicle (lipofectamine) control for 48h after which CHX was added (100 μg/ml) for an additional 24-72h. For Western blot studies, immunoprecipitation studies, and introduction of siRNA into stably infected/transfected cell lines, cells received the appropriate siRNA, scramble control, or vehicle for 48h prior to analysis. In studies using TRAIL, cells received either the target siRNA, scramble control, or vehicle for 48h, then TRAIL (800ng/mL) for an additional 24h, after which cell death was assessed by flow cytometry.
Immunoblot analysis and analysis of apoptosis by flow cytometry
Cells were washed with ice-cold phosphate-buffered saline, scraped from the culture dish, and incubated in tissue lysis buffer containing 10 mM KCl, 1mM sucrose, 2 mM MgCl2, 0.5% Igepal CA-630, 1 mM EDTA, 1 mM DTT, 10 mM β-glycerophosphate, 1 mM Na3VO4, 10mM sodium fluoride, 100 μg/ml phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin for 30 min on ice. The cell lysate was centrifuged, and the supernatant was stored at −80 °C until use. The protein concentration of extracts was measured using Protein Assay reagent (Bio-Rad Laboratories). Protein (30 μg) was subjected to SDS-polyacrylamide gel electrophoresis and electroblotted onto Immobilon-P membrane (Millipore). The membrane was blocked in 5% nonfat skim milk/TBST [20mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Tween 20] at 4 °C overnight and incubated with goat polyclonal antibody against FLIPS or FLIPL (Santa Cruz Biotechnology), or rabbit polyclonal antibody against USP8 (Abcam), AIP4, or HA (all from Cell Signaling Technology). Bound antibody was detected with anti-goat IgG or anti-rabbit IgG (Santa Cruz Biotechnology) using enhanced chemiluminescence Western blotting detection reagents (Amersham Pharmacia Biotech). Densitometric measurements of immunoreactive bands were acquired using an AlphaImager 2200 (Alpha Innotech Corporation). The extent of apoptosis in cultures (attached and floating cells) was determined by fluorescence-activated cell sorting analysis (sub-G1 DNA content), with measurements verified by annexin V staining (19).
Immunoprecipitation and analysis of ubiquitinated proteins
AIP4 or FLIPS was immunoprecipitated from cell lysates by a 1h incubation at 4°C with the appropriate antibody pre-conjugated to Protein G beads (Santa Cruz Biotechnology). The beads were subsequently pelleted by centrifugation at 2000 rpm for 5 min. and washed three times using tissue lysis buffer. The samples were then boiled at 95°C for 10 minutes to elute the immunocomplexed proteins. Levels of protein in the eluate were assessed by Western blot using the appropriate antibody. Levels of ubiquitinated protein were assessed by Western blot using an antibody targeting HA. Immunoprecipitations carried out using a nonspecific normal rabbit IgG antibody (for AIP4 immunoprecipitations) or a goat IgG antibody (for FLIPS immunoprecipitations, Santa Cruz Biotechnology) were included as negative controls.
Statistical analysis
All statistical analyses were performed using the Student's t test, with significance defined as P<0.05.
Results
We had previously shown that levels of the anti-apoptotic protein FLIPS were higher in TRAIL-resistant, PTEN-deficient GBM cells and TMA than in TRAIL-sensitive PTEN WT cells, and that these higher levels of FLIPS were associated with a longer FLIPS half life and lower levels of FLIPS ubiquitination (9). These data suggested an Akt-dependent, but mammalian target of rapamycin (mTOR)-independent link between PTEN and FLIPS ubiquitination which we subsequently showed was the result of Akt-mediated regulation of the activity of the FLIPS E3 ubiquitin ligase AIP4. Because AIP4 is not known to be a substrate of Akt, we initiated a search for pathways that might link Akt to AIP4 regulation. The de-ubiquitinating enzyme USP8 (ubiquitin specific protease 8) has been suggested to be regulated by Akt (20), and has also been reported to play a broad role in growth factor receptor trafficking and degradation, in part, by its ability to stabilize the E3 ligase neuregulin receptor degradation pathway protein 1(Nrdp1)(20, 21). We therefore considered the possibility that USP8 might be a link between the PTEN/Akt pathway and a ubiquitin E3 ligase involved in FLIPS protein stability and apoptotic sensitivity.
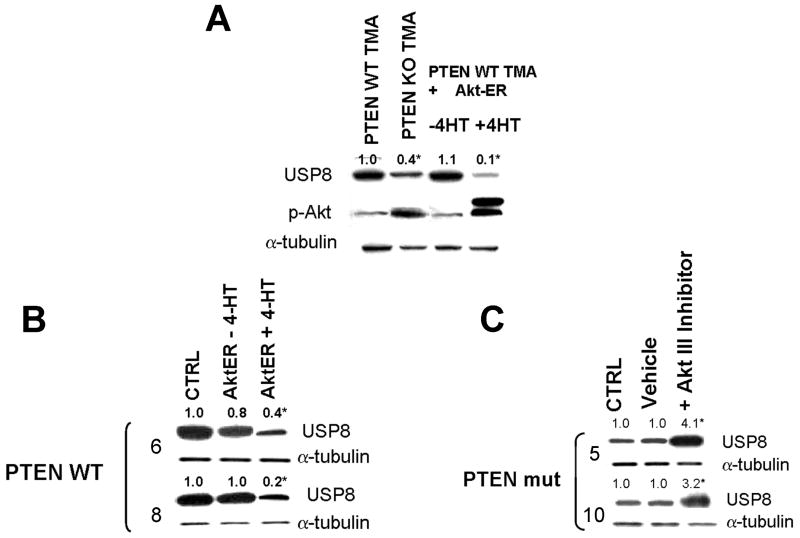
To begin to address this possibility, we first examined whether USP8 was regulated in a PTEN/Akt-dependent manner. Levels of USP8 were significantly higher in PTEN WT TMA than in PTEN KO TMA (lane 1 vs 2, Fig 1A), and also higher in PTEN WT human GBM cells than in PTEN mutant GBM cells (compare lane 1, Figs 1B and C). Furthermore, 4-hydroxytamoxifen (4HT)-mediated activation of a retrovirally-encoded exogenous Akt-estrogen receptor(ER) protein in PTEN WT TMA (last lane, Fig 1A) or PTEN WT human GBM cells (last lane, Fig 1B) resulted in a decrease of USP8 levels to those noted in corresponding PTEN-deficient cells. Conversely, exposure of PTEN mutant human GBM cells to an Akt inhibitor enhanced USP8 levels (last lane, Fig 1C). These results therefore show that PTEN loss and Akt activation are linked to suppression of USP8 levels, and that USP8 is a target for PTEN-mediated regulation.
Figure 1.
The PTEN-Akt pathway regulates levels of the deubiquitinase USP8. Mouse PTEN WT or KO TMA, human PTEN WT or mutant xenograft GBM cells, or the same cells infected with a construct encoding 4HT-activated Akt-ER were incubated with vehicle, 4-hydroxytamoxifen (4HT, 100 nM, 24 hrs)(A, B), or Akt III inhibitor (50 μM, 24 hrs)(C), after which cells were lysed and analyzed for levels of USP8 and α-tubulin.
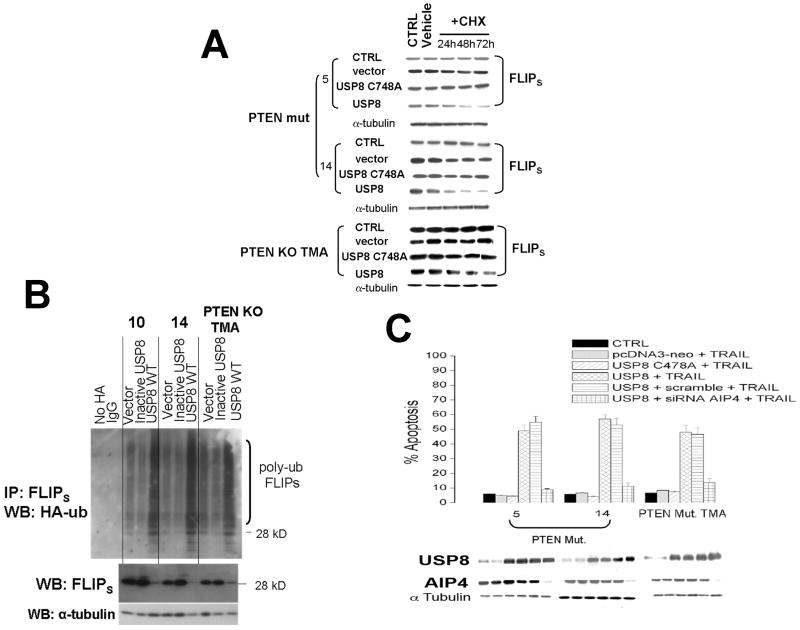
To address whether the PTEN/Akt-mediated control of USP8 is in turn directly linked to the control of FLIPS stability and/or apoptotic sensitivity, we manipulated USP8 levels in PTEN WT and PTEN-deficient cells, after which effects on FLIPS half-life, FLIPS steady-state levels, FLIPS ubiquitination, and apoptotic sensitivity to TRAIL were measured. In control PTEN mutant GBM and PTEN-KO TMA (which have relatively low levels of endogenous USP8) in which new protein synthesis was inhibited by CHX exposure, the pre-existing FLIPS protein exhibited a relatively long half-life (Fig. 2A), consistent with previous data. Retroviral introduction of a WT USP8 into these cells increased levels of USP8 (bottom panel, Fig 2C) but also significantly reduced FLIPS half-life relative to that noted in cells receiving either a blank (pcDNA3-neo) vector or a vector encoding catalytically inactive USP (21)(USP C748A, Fig. 2A). Consistent with this decrease in FLIPS stability, steady state levels of FLIPS also were significantly decreased (Fig 2B, bottom panel), and these cells were sensitized to TRAIL-induced apoptosis (Fig 2C, fourth bar vs first bar in each group). Because PTEN-dependent regulation of FLIPS stability is mediated by changes in FLIPS ubiquitination, we took the above USP8-modulated cells, transiently transfected a construct encoding HA-tagged ubiquitin, and following FLIPS immunoprecipitation used Western blot analysis to monitor the effect of USP8 alteration on the extent of HA-ubiquitin incorporated into FLIPS. Immunoprecipitates generated using IgG, or from cells not transfected with the HA-ubiquitin construct did not exhibit any HA-ubiquitinated FLIPS (Fig 2B, top panel). Levels of ubiquitinated FLIPS were also relatively low in control cells and cells expressing catalytically inactive USP8, consistent with the long half life of FLIPS in these cells. Expression of WT USP8, however significantly increased the amount of higher molecular weight, HA-labeled FLIPS (each ubiquitin added increases the apparent mass of the target protein by 7 kD). These results suggest that the PTEN-dependent regulation of USP8 is tied to the control of FLIPS ubiquitination and stability.
Figure 2.
Over-expression of USP8 decreases FLIPS half life, decreases FLIPS steady state levels, increases FLIPS ubiquitination, and increases TRAIL sensitivity. PTEN mutant human GBM xenograft cells or PTEN KO TMA were retrovirally infected with a constructs encoding either USP8 or a catalytically inactive mutant form of USP8 (C748A)(A-C), after which cells were transiently transfected with a blank vector, a construct encoding HA-ubiquitin (B), or siRNA targeting AIP4 (C). Cells were then incubated with vehicle, cycloheximide (CHX, 100 μg/ml)(A), or TRAIL (800 ng/ml, 24 hrs)(C), after which the cells were either lysed at the indicated time points and analyzed for levels of FLIPS, α-tubulin (A, B), USP8 and AIP4 (C), for the extent of HA-ubiquitination in FLIPS immunoprecipitates (B), or analyzed for the extent of TRAIL-induced apoptosis (C). The α-tubulin blots shown in A are representative of those for all experimental groups in each panel.
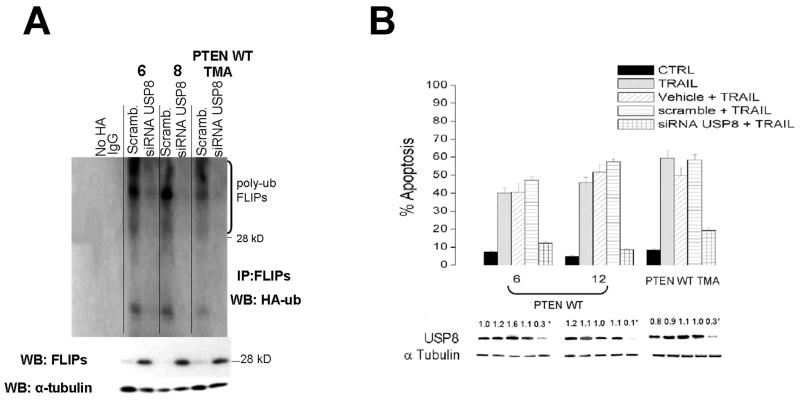
To confirm the apparent link between USP8, FLIPS stability, and TRAIL sensitivity, we also performed converse experiments in which PTEN WT GBM and TMA, which express relatively high levels of USP8, were transfected with scrambled siRNA or an siRNA targeting USP8, after which the effects on FLIPS steady state levels, ubiquitination, and TRAIL sensitivity were similarly monitored. The PTEN WT cells transfected with scrambled siRNA had relatively low levels of endogenous FLIPS (Fig 3A, bottom), consistent with the relatively high levels of ubiquitinated FLIPS noted in the FLIPS immunoprecipitates from the same cells engineered to express HA-ubiquitin (Fig 3A, top panel). Introduction of siRNA targeting USP8, however, not only decreased levels of USP8 (Fig 3, panel B, bottom), but also increased FLIPS steady-state levels (Fig 3A, bottom panel), decreased the extent of HA-ubiquitinated FLIPS (Fig 3A, top panel) and significantly reduced the extent of TRAIL-induced apoptosis in these cells (Fig 3B) relative to controls. Taken as a whole, these results show that USP8 levels are regulated in a PTEN-dependent manner, and that loss of PTEN function leads to decreased USP8 levels, decreased FLIPS ubiquitination, increased FLIPS stability, and increased TRAIL resistance.
Figure 3.
siRNA-mediated suppression of USP8 increases FLIPS steady state levels, decreases FLIPS ubiquitination, and decreases TRAIL sensitivity. PTEN WT human GBM xenograft cells or PTEN WT TMA were transiently transfected with a scrambled siRNA or an siRNA targeting USP8, after which cells were transiently transfected with a blank vector or a construct encoding HA-ubiquitin (A), or incubated with TRAIL (800 ng/ml, 24 hrs)(B). Cells were then either lysed and analyzed for levels of FLIPS, α-tubulin, and for the extent of HA-ubiquitination in FLIPS immunoprecipitates (A), or analyzed for levels of USP8 and the extent of TRAIL-induced apoptosis (B).
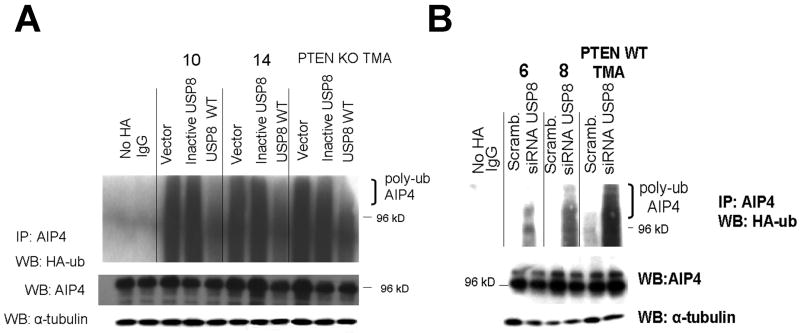
We previously reported that the E3 ligase AIP4, like USP8, was regulated in a PTEN-dependent manner, and that AIP4 contributed to the ubiquitination of FLIPS (9). Given the apparent connection between PTEN, USP8, and AIP4, we questioned whether these components might be parts of a single pathway that control FLIPS ubiquitination, and whether USP8 might act as a PTEN-dependent stimulator of AIP4 activity. To address this point we first asked whether the modulation of USP8 that resulted in changes in FLIPS stability and ubiquitination also influenced AIP4. Levels of AIP4 were similar in PTEN WT and PTEN-deficient cells, and neither introduction of WT USP8 into PTEN mutant cells nor siRNA-mediated suppression of USP8 in PTEN WT cells altered total AIP4 levels (Fig 4 A and B). We previously noted, however, that while the PTEN loss did not alter total levels of AIP4, it did increase the ubiquitination of AIP4 and interfere with the ability of AIP4 to interact with its targets (9), raising the possibility that the PTEN loss interferes with AIP4 action not by lowering AIP4 levels, but by decreasing USP8 levels, which may in turn leave AIP4 in a ubiquitinated, inactive state. Consistent with this possibility, AIP4 in the PTEN mutant cells (which have low levels of USP8 and high levels of FLIPS) was retained in a relatively highly ubiquitinated state, and introduction of WT USP8, but not catalytically inactive USP8, significantly decreased both AIP4 ubiquitination (Fig 4A) as well as FLIPS stability (Fig 2A). Conversely, AIP4 in the PTEN WT cells (which have high levels of USP8 and low levels of FLIPS) was retained in a relatively under-ubiquitinated state, and the introduction of an siRNA targeting USP8 that significantly decreased FLIPS ubiquitination also significantly increased AIP4 ubiquitination (Fig 4B). These data suggest that USP8, by modulating the ubiquitination status of AIP4, serves as link between Akt and the AIP4-mediated regulation of FLIPS stability.
Figure 4.
USP8 alterations are linked to changes in AIP4 ubiquitination. Cells (PTEN mutant human GBM xenograft cells or PTEN KO TMA, A, or PTEN WT human GBM xenograft cells or PTEN WT TMA, B) were either retrovirally infected with a constructs encoding either USP8 or a catalytically inactive mutant form of USP8 (C748A)(A), or transiently transfected with a scrambled siRNA or an siRNA targeting USP8 (B), after which all cells were transiently transfected with a blank vector or a construct encoding HA-ubiquitin. Cells were then lysed and analyzed for levels of AIP and α-tubulin, or for the extent of HA-ubiquitination in AIP4 immunoprecipitates (A).
As a final test of the proposed linkage between PTEN, Akt, USP8, AIP4, FLIPS stability, and TRAIL sensitivity, USP8 and AIP4 levels were co-modulated in PTEN-deficient cells, after which effects on TRAIL sensitivity were monitored. PTEN deficient cells, which have low levels of USP8 and high levels of FLIPS, were relatively TRAIL resistant, and as previously noted, introduction of WT USP8 (but not blank vector or catalytically inactive USP8) increased USP8 levels and significantly increased the extent of TRAIL-induced apoptosis (Fig 2C). This USP8-induced increase in TRAIL sensitivity, however, could be reversed by introduction of siRNA that targeted AIP4 but did not alter USP8 levels (Fig 2C). These results as a whole show that USP8 is a PTEN-regulated deubiquitinase which, by altering the ubiquitination status of the E3 ligase AIP4, helps control FLIPS stability and TRAIL sensitivity in GBM cells.
Discussion
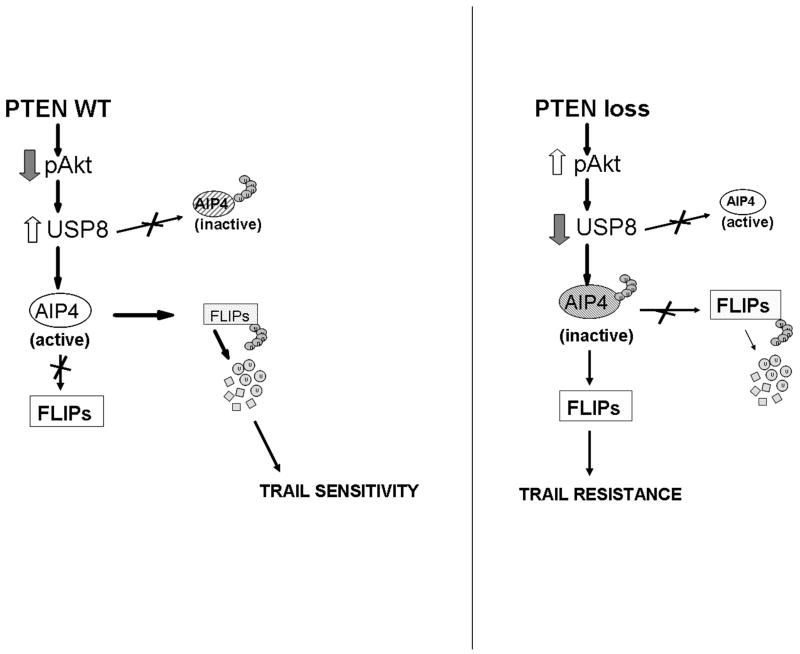
The pathway that links PTEN to the control of FLIPS ubiquitination and TRAIL sensitivity described in this work is presented in Fig. 5. In this model, PTEN suppresses levels of pAkt (left panel) which in turn leads to increased levels of USP8. USP8 interacts with AIP4 and retains this E3 ubiquitin ligase in a state in which it can interact with FLIPS. Under these conditions FLIPS undergoes ubiquitin-mediated degradation, leaving the cell susceptible to TRAIL-induced apoptosis. Loss of PTEN function (right panel, Fig. 5), in contrast increases pAkt levels, decreases USP8 levels, and turns off the USP8/AIP4 ubiquitin switch, allowing FLIPS to accumulate and suppress TRAIL-induced apoptosis. In this manner PTEN appears to use control of ubiquitination to help regulate TRAIL sensitivity in GBM cells.
Figure 5.
Schematic representation of the PTEN-Akt-USP8-AIP4 pathway that controls FLIPS ubiquitination and TRAIL sensitivity.
Although the linkage between PTEN status and Akt activation state has been well described, the linkage between Akt and the DUB USP8 has been less well studied. In breast tumor cells, Akt activation led to USP8 (thr907) phosphorylation, which in turn led to increased DUB activity toward the E3 ligase Nrdp1 (20). In the present study the linkage between PTEN loss, Akt activation, and USP8 levels and activity appeared to be somewhat different, and in our work Akt activation decreased, rather than enhanced, USP8 function by increasing USP8 ubiquitination and decreasing steady-state USP8 levels (data not shown). Furthermore, a preliminary immunohistochemical analysis of paired tissue from 12 newly diagnosed GBM revealed a statistically significant (p ≤ 0.05) inverse relationship between levels of expression of USP8 and PRAS40 (a downstream marker of Akt activity). In addition to suppressing levels of USP8, Akt may also stimulate the activity of USP8 toward select targets such as Nrdp1. Alternatively Akt activation may directly suppress Nrdp1 function in addition to having effects mediated via USP8-control of ubiquitination. In the present work and in that of Cao et al (20), Akt kinase inhibition blocked the effect of Akt on USP8 function, suggesting that events related to Akt-mediated phosphorylation of USP8 control the ability of USP8 to regulate AIP4 and the ubiquitination process. USP8 contains at least three consensus sites for Akt phosphorylation, although the requirements for these sites for phosphorylation, stability, and/or function have not been defined.
While the present studies show that USP8 regulates the E3 ligase function of AIP4, the exact means by which this occurs are only partially defined. Because the deubiquitination-deficient USP8 mutant used in this study was unable to alter AIP4 ubiquitination and FLIPS stability, the USP8-mediated regulation of AIP function clearly involves the ubiquitination process. The decreased AIP4 ubiquitination noted following introduction of WT USP8 combined with our previous work (9) showing mislocalization of ubiquitinated AIP4 strongly suggest that in the control PTEN WT setting, the deubiquitinating activity of USP8 prevents or reverses AIP4 K63 polyubiquitination, which in turn allows the AIP4 E3 ligase to interact with, and K48 polyubiquitinate FLIPS in the DISC, leading to FLIPS degradation and enhanced sensitivity to apoptotic stimuli. A direct interaction between USP8 and AIP4 has not been reported, and it is possible that instead of directly deubiquitinating AIP4, USP8 may alter the ability of other E3 ligases (or perhaps of AIP4 itself) to stimulate AIP4 K63 polyubiquitination (22). Regardless of the exact mechanism, the present work is the first to clearly define a mechanism by which a pathway related to tumorigenesis also regulates apoptotic sensitivity.
The ubiquitin control pathway described in this work has broad implications, not only for our understanding of TRAIL resistance, but also for our understanding of the control of ubiquitination and PTEN function. TRAIL resistance in GBM is multifactorial, and a number of alterations have been shown to contribute to TRAIL insensitivity (23-25). While Akt activation has been linked to several of these alterations, and to TRAIL resistance (26-28), this effect has been typically ascribed to Akt-mediated, phosphorylation-dependent effects on the activity and/or translational regulation of specific (often anti-apoptotic) proteins. The present study suggests that in addition to using phosphorylation, the PTEN-Akt pathway may use broader ubiquitin-based mechanisms to regulate protein stability, and in doing so may be able to rapidly change a variety of cell characteristics including apoptotic sensitivity. The linkage of PTEN to USP8 and AIP4 also suggests that the range of proteins whose stability is controlled by PTEN-regulated ubiquitination may be broader than expected. AIP4 is the E3 ligase for several proteins including FLIPL and Notch receptor 1 (29, 30), and our preliminary work suggests that several USPs in addition to USP8 are regulated in a PTEN-dependent manner. PTEN may therefore employ a modular approach to the regulation of protein ubiquitination and stability, recruiting different DUBs linked to different E3 ligases to co-ordinately stabilize or destabilize families of proteins that share a common function. The USP8-AIP4 ubiquitin switch described in this work may therefore represent the first of many different ubiquitin switches used by PTEN to control cellular behavior.
Acknowledgments
We thank L. Bin, K. Carraway, and M. McMahon for various construct, S. Baker and G. Bergers for the PTEN WT and KO TMA, A. Ashkenazi for TRAIL, D. James for the help with the human GBM xenografts, and Cynthia Cowdrey, King Chu, and Joanna Phillips for help with immunohistochemical analyses.
This work was supported by NIH Grants CA115638 and CA136774 to ROP, and CA097257 to ATP and ROP.
References
- 1.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nature Rev. 2006;6:776–88. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 3.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 4.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–9. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. Faseb J. 1997;11:1245–56. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 6.Nijman SM, Luna-Vargas MP, Velds, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, Lopez-Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Comm. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 8.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Panner A, Crane CA, Weng C, Feletti A, Parsa AT, Pieper RO. A novel PTEN-dependent link to ubiquitination controls FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res. 2009;69:7911–6. doi: 10.1158/0008-5472.CAN-09-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting proteinase, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–15. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 11.Muzio M, Chinnaiyan AM, Kischel FC, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–27. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 12.Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25:8809–23. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser MM, Zhu X, Kwon CH, Uhlmann EJ, Gutmann DH, Baker SJ. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–9. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- 14.Kanamori M, Vanden Berg SR, Bergers G, Berger MS, Pieper RO. Integrin beta 3 overexpression suppresses tumor growth in a human model of gliomagenesis: implications for the role of B3 overexpression in glioblastoma multiforme. Cancer Res. 2004;64:2751–8. doi: 10.1158/0008-5472.can-03-3354. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi T, Yamashita Y, Kanamori M, et al. The PTEN/Akt pathway dictates the direct alphaVbeta3-dependent growth-inhibitory action of an active fragment of tumstatin in glioma cells in vitro and in vivo. Cancer Res. 2006;66:11331–40. doi: 10.1158/0008-5472.CAN-06-1540. [DOI] [PubMed] [Google Scholar]
- 16.Hirose Y, Katayama M, Mirzoeva OK, Berger MS, Pieper RO. Akt activation suppresses Chk2-mediated, methylating agent-induced G2 arrest and protects from temozolomide-induced mitotic catastrophe and cellular senescence. Cancer Res. 2005;65:4861–9. doi: 10.1158/0008-5472.CAN-04-2633. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001a;61:6674–8. [PubMed] [Google Scholar]
- 18.Sonoda Y, Ozawa T, Hirose Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001b;61:4956–60. [PubMed] [Google Scholar]
- 19.Chang GH, Barbaro NM, Pieper RO. Phosphatidylserine-dependent phagocytosis of apoptotic glioma cells by normal human microglia, astrocytes, and glioma cells. Neuro-onc. 2000;2:174–183. doi: 10.1093/neuonc/2.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Z, Wu X, Yen L, Sweeney C, Carraway KL., 3rd Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol Cell Biol. 2007;27:2180–8. doi: 10.1128/MCB.01245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–57. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scialpi F, Malatesta M, Peschiaroli A, Rossi M, Melino G, Bernassola F. Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem Pharmacol. 2008;76:1515–21. doi: 10.1016/j.bcp.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Hopkins-Donaldson S, Ziegler A, Kurtz S, et al. Silencing of death receptors and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–64. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 24.Pai SI, Wu GS, Ozoren N, et al. Rare loss-of-function mutation of a death receptor gene in head and neck cancer. Cancer Res. 1998;58:3513–8. [PubMed] [Google Scholar]
- 25.Sheridan JP, Marsters SA, Pitti RM, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–21. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 26.Dida F, Li Y, Iwao A, Deguchi T, Azuma E, Komada Y. Resistance to TRAIL-induced apoptosis caused by constitutional phosphorylation of Akt and PTEN in acute lymphoblastic leukemia cells. Exp Hematol. 2008;36:1343–53. doi: 10.1016/j.exphem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Chen W, Zeng W, et al. Akt-mediated eminent expression of c-FLIP and Mcl-1 confers acquired resistance to TRAIL-induced cytotoxicity to lung cancer cells. Mol Cancer Ther. 2008;7:1156–63. doi: 10.1158/1535-7163.MCT-07-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poh TW, Huang S, Hirpara JL, Pervaiz S. LY303511 amplifies TRAIL-induced apoptosis in tumor cells by enhancing DR5 oligomerization, DISC assembly, and mitochondrial permeabilization. Cell Death Differ. 2007;14:1813–25. doi: 10.1038/sj.cdd.4402177. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–13. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Chastagner P, Israël A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]