Abstract
Sunlight UV exposure produces DNA photoproducts in skin that are repaired solely by nucleotide excision repair in humans. A significant fraction of melanomas are thought to result from UV-induced DNA damage that escapes repair, however, little evidence is available regarding the functional capacity of normal human melanocytes, malignant melanoma cells and metastatic melanoma cells to repair UV-induced photoproducts in DNA. In this study, we measured nucleotide excision repair in both normal melanocytes and a panel of melanoma cell lines. Our results show that in 11 of 12 melanoma cell lines tested, UV photoproduct repair occurred as efficiently as in primary melanocytes. Importantly, repair capacity was not affected by mutation in the N-Ras or B-Raf oncogenes, nor was a difference observed between a highly metastatic melanoma cell line (A375SM) or its parental line (A375P). Lastly, we found that while p53 status contributed to photoproduct removal efficiency its role did not appear to be mediated by enhanced expression or activity of DNA binding protein DDB2. We concluded that melanoma cells retain capacity for nucleotide excision repair, the loss of which probably does not commonly contribute to melanoma progression.
Keywords: melanoma, DNA repair, UV light, p53, skin cancer
Introduction
Ultraviolet (UV) light from the sun causes a variety of lesions in the genome that distort the structure of DNA, resulting in blocks to gene transcription and DNA replication. Epidemiological evidence strongly indicates that UV-induced DNA damage is a primary cause of skin cancer, including melanoma (1, 2), an aggressive form of skin cancer which arises from specialized pigmented cells called melanocytes. Though comprising only 5-10% of human skin, melanocytes synthesize the pigment melanin, which provides skin tone, hair color, and protection from UV radiation. The significant increase in melanoma cases in the past half-century and the poor survival rates among patients with metastatic melanoma (3, 4) therefore merits thorough investigations of the underlying causes of melanoma initiation and progression.
Among the lesions induced by UV, cyclobutane pyrimidine dimers (CPDs; 80-90%) and [6-4] pyrimidine-pyrimidone photoproducts ([6-4] PPs; 10-20%) are most abundant, though both can be accurately removed from the genome by nucleotide excision repair (henceforth termed “excision repair”). This well-characterized repair system responds to a variety of environmental and chemotherapeutic agents that form bulky adducts in DNA, and in humans is the sole mechanism for removal of CPDs and [6-4] PPs from DNA (5). Importantly, reconstitution of excision repair in vitro with the six essential factors (XPA, RPA, XPC, TFIIH, XPG, and XPF-ERCC1) has provided a significant understanding of the individual steps of repair and allowed a clear determination of the minimal set of factors necessary and sufficient for the complete removal of UV photoproducts from DNA (6, 7). The importance of the excision repair system to human health is most obvious in xeroderma pigmentosum (XP), a disease in which most patients lack one of the essential excision repair factors (8). One consequence of this loss is a 2000-fold higher incidence of metastatic melanoma compared to normal individuals (9).
Melanomas display complex genetic profiles but often show activating mutations in the oncogenes B-Raf (50-75%) and N-Ras (10-15%) (10, 11), resulting in enhanced cell growth through signaling of the mitogenic ERK1/2 pathway (12, 13). Similarly, though the tumor suppressor p53 is disrupted in nearly half of all cancers and is known to promote repair of UV photoproducts in many cell types (14), studies indicate that p53 mutations are rare in primary melanoma (less than 1%) (15) but do increase in frequency in metastatic melanoma (5%) (16). Though the p53 gene is not commonly altered in melanoma, disruption of the tumor suppressor ARF, which regulates p53 protein stability, is a common occurrence in metastatic melanoma through genetic deletion of the CDKN2A locus (17-19). Therefore alterations of p53-dependent pathways have the potential to influence melanoma progression.
In contrast, though there is some evidence linking a polymorphism in the excision repair gene XPD and susceptibility to cutaneous melanoma (20), there is little available data indicating that altered expression of excision repair genes contributes to melanoma, and indeed a recent microarray analysis of mRNA expression profiles in metastatic melanomas did not find changes in excision repair genes (21). Though analyses of mRNA transcript and protein expression profiles have the potential to be informative, it may be more relevant to test for functional excision repair capacity in order to make proper correlations of DNA repair and carcinogenesis. Along these lines, though early work initially indicated that melanoma cells did not show enhanced repair rates (22), other work suggested that sub-clones of a metastatic melanoma line did indeed show elevated repair rates in comparison to non-melanoma cells, and this repair correlated with increased survival after UV (23). An additional study similarly concluded that DNA repair capacity in mouse melanoma cell lines correlated with metastatic potential (24). However, more recent in situ work indicated that cutaneous melanoma patients show normal repair kinetics (25). It is therefore unclear whether excision repair capacity is altered in melanoma cells relative to normal melanocytes, or whether genetic background (B-Raf/N-Ras/p53 status) or metastatic potential are directly correlated with excision repair capacity.
In this study, we used normal human melanocytes (NHMs) and a variety of melanoma cell lines to characterize excision repair capacity as a function of genetic and metastatic states. Our results show that in nearly all melanoma cell lines tested, excision repair occurred as efficiently as in NHMs, irrespective of mutations in the N-Ras and B-Raf oncogenes. In addition, we found no change in excision repair capacity in a highly metastatic melanoma cell line (A375SM) compared to its parental melanoma cell line (A375P), which has a low metastatic potential. Lastly, we observed that melanoma cell lines containing functional p53 repair UV photoproducts more efficiently than lines with inactive p53, but that this difference appears to be not due to enhanced levels of the UV photoproduct binding protein DDB2.
Materials and Methods
Cell lines
Description of the sources, culture method, and UV irradiation of the melanoma cell lines are provided in Supplementary Table S1. All of the cell lines used in our study are authenticated by microarray analysis as described previously (13).Secondary cultures of normal human melanocytes were derived as reported previously (26). Briefly, these cells (NHM-16 and NHM-21) were grown in Medium 254 (Gibco, Carlsbad, CA) containing Human Melanocyte Growth Supplement (HMGS-1; Gibco) at 37°C in a cell culture incubator with 5% CO2. A375P and A375SM cell lines were a gift from Dr. Richard O. Hynes (MIT, Cambridge, MA).
UV irradiation
Culture medium was removed from exponentially growing cells, set aside, and then cells were washed once with warm PBS before placement under a GE germicidal lamp emitting primarily 254 nm UV light (UV-C) connected with a digital timer. After receiving the indicated dose of UV (typically 5-20 J/m2, as indicated), culture medium was added back to the cells, which were subsequently placed back into the cell culture incubator for the indicated length of time. A UV-C sensor (UV Products, San Gabriel, CA) was used to calibrate the fluence rate of the incident light.
Immunoslot blot assay for measurement of CPD and [6-4] PP repair in vivo
Repair of CPDs and [6-4] PPs by immunoslot blot was performed essentially as reported previously (27).
Fluorescence microscopy
Immunofluorescence microscopy was performed as described previously (28). Briefly, cells were cultured in 35-mm glass-bottom dishes (MatTek, Ashland, MA) for 24 hrs before UV irradiation (10 J/m2). Cells harvested at various time points post-irradiation were fixed with 4% formalin and further treated with ice-cold detergent (0.5% Triton X-100) for 5 min. After denaturation of DNA with 2 M HCl for 30 min at room temperature, CPDs were detected with the mouse monoclonal anti-CPD antibody (Kamiya Biomedical) and Alexa Fluor 488 goat-anti mouse IgG conjugate (Invitrogen). Nuclear DNA was counterstained with propidium iodide and CPD signals observed with a Leica inverted DMIRB fluorescence microscope.
Measurement of [6-4] PP repair activity in cell-free extracts
Preparation of radiolabeled substrate and the in vitro excision repair assay was essentially as previously described (27, 29). Cell-free extracts for use in the excision assay were prepared as reported (30).
Immunoblot analyses
Protein lysates from exponentially growing cultures of cells were harvested and analyzed by SDS-PAGE and immunoblotting as described elsewhere (26). The following antibodies were used to detect the respective proteins: XPA, XPC, RPA70, TFIIH (p62 subunit, XPB), p53, Actin (Santa Cruz Biotechnology, Santa Cruz, CA); p21, DDB2, GAPDH (Cell Signaling Technology, Inc., Danvers, MA), RPA34 (Calbiochem, Gibbstown, NJ), XPF, and XPG (Abcam, Cambridge, MA).
siRNA transfection
Exponentially growing cells of SK-Mel-103 or SK-Mel-187 were transfected with either p53 siRNA (Santa Cruz) or non-targeting siRNA (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen) transfection reagent. Cells were UV irradiated 48 hours after transfection and then harvested at the indicated time points to assay for CPD repair.
Electrophoretic mobility shift assay for UV-DDB binding activity
UV-DDB binding activity was performed using cell-free extracts from both SK-Mel-103 and SK-Mel-187 cell lines as reported previously (30-33). Briefly, 5 fmol of 136-bp double-stranded DNA containing a [6-4] PP was incubated with the indicated amount of proteins or cell-free extract in 12.5 μl reaction mixtures. After a 30-min incubation at 30°C, glycerol was added to ∼8%, and reaction mixtures were resolved using 5% native PAGE at room-temperature with a constant current (25 mA). DNA binding was visualized by autoradiography and quantified using ImageQuant 5.2 software (Molecular Dynamics). Recombinant UV-DDB protein was used as a positive control for DDB binding activity (31).
Matrigel invasion assay
Cellular invasion assays were performed using 8.0-μm pore size Biocoat Matrigel Invasion chambers (BD Biosciences) as described by the manufacturer. Each data point represents the average of three independent experiments and error bars represent the standard deviation of the mean.
Results
Measurement of [6-4] photoproduct repair in vitro
Epidemiological evidence suggests a strong correlation between exposure to UV from the sun and development of melanoma in humans (22, 34, 35), and we therefore sought to investigate whether the capacity to repair UV photoproducts differed between normal human melanocytes and melanoma cell lines. Our initial approach employed an in vitro, cell-free excision assay to measure excision repair. Our laboratory has used this assay extensively to study the mechanism of nucleotide excision repair and to measure repair capacity in mammalian cell lines and tissues (29, 36). The assay involves incubation of an internally 32P-labeled oligonucleotide containing a site-specific [6-4] PP in cell-free extract and then electrophoresis of the purified DNA on a denaturing gel, which allows visualization and quantitation of the small 24-32-nt oligomers that are generated during the repair reaction (Figure 1A). Though the assay has been used with extracts from a variety of mammalian cell types and tissues, it has not previously been applied to measure repair in melanocytes or melanoma cells, a physiologically relevant and medically important target of UV-induced DNA damage.
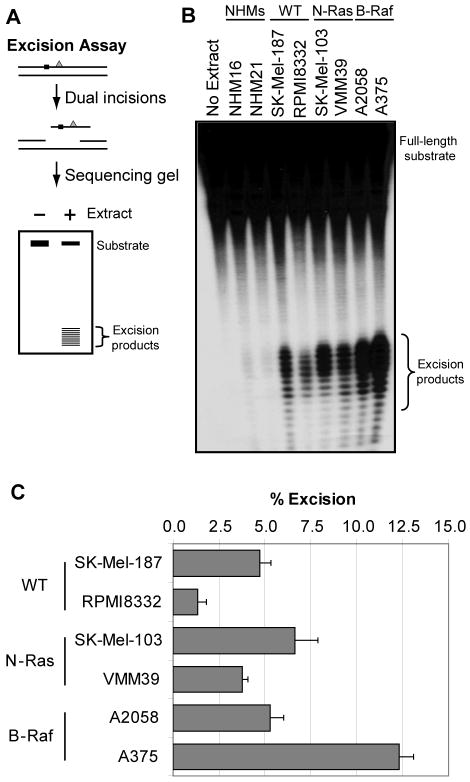
Figure 1. [6-4] photoproduct repair in cell-free extracts.
(A) Schematic of the in vitro excision assay. An internally 32P-labeled (black square), 136-bp double-stranded DNA containing a centrally located [6-4] PP (gray triangle) is incubated with cell-free extract for 90 min. The damaged DNA is removed through dual incisions of the DNA at sites bracketing the lesion, resulting in release of 24- to 32-nt-long oligomers (excised products) that are visualized by denaturing PAGE and phosphorimager analysis. (B) Excision repair in cell-free extracts. Cell lines examined included two normal human melanocyte lines (NHMs) and two cell lines each that lack (wild-type: WT) or contain mutations in either N-Ras or B-Raf. (C) Quantitative analyses of excision repair in cell-free extracts. The percent of total radiolabeled material released as excision products represents the amount of excision repair (% excision). Excision assays were performed three times with three independent preparations of cell-free extract for each cell line, and the data indicate the average and standard deviation from these assays.
Our initial studies focused on two normal human melanocyte lines (NHM: NHM16 and NHM21) and two cell lines each from melanoma cells containing normal or mutant forms of the B-Raf and N-Ras oncogenes (WT: SK-Mel-187 and RPMI8322; N-Ras: SK-Mel-103 and VMM39; B-Raf: A2058 and A375). Using cell-free extracts prepared from the indicated cell lines, we observed variable amounts of [6-4] PP removal among the different melanoma lines (Figure 1B), ranging from ∼1% in the RPMI8332 line and up to ∼12.5% in A375 cells (Figure 1C). This variation is consistent with previous reports showing that human melanomas show a high degree of inter-individual variability in excision repair in situ (25). Importantly, immunoblot analyses of these extracts showed no clear correlation between excision repair capacity and expression level of any specific excision repair protein (Supplementary Figure 1).
Interestingly, we observed very little excision activity in the extracts from the normal human melanocytes. To better understand this lack of excision repair activity with this assay, we supplemented extracts from excision-competent A375 and CHO (Chinese hamster ovary) cells, which do not contain detectable level of melanin (Supplementary Table 2), with either NHM16 or NHM21 extract. As shown in Supplementary Figure 2A, both NHM extracts inhibited the excision activity of A375 and CHO extracts, indicating the presence of an inhibitory factor in the NHM extract. We considered that the presence of melanin in the NHM extracts might interfere with the in vitro excision activity. To test for the effect of melanin we titrated CHO cell-free extract with increasing amounts of synthetic melanin and observed a concentration-dependent inhibition of excision repair (Supplementary Figure 2B). This inhibition was not specific to excision repair, however, as the ability of restriction enzymes to digest the substrate was also inhibited by melanin (Supplementary Figure 2C), indicating that melanin may non-specifically bind to DNA and inhibit the action of multiple nucleases. We conclude, however, that though the excision assay is a convenient and reliable tool for measuring repair capacity in many cell lines, including melanoma cells, it cannot be used for all cell types, such as melanocytes.
Measurement of [6-4] PP repair in vivo
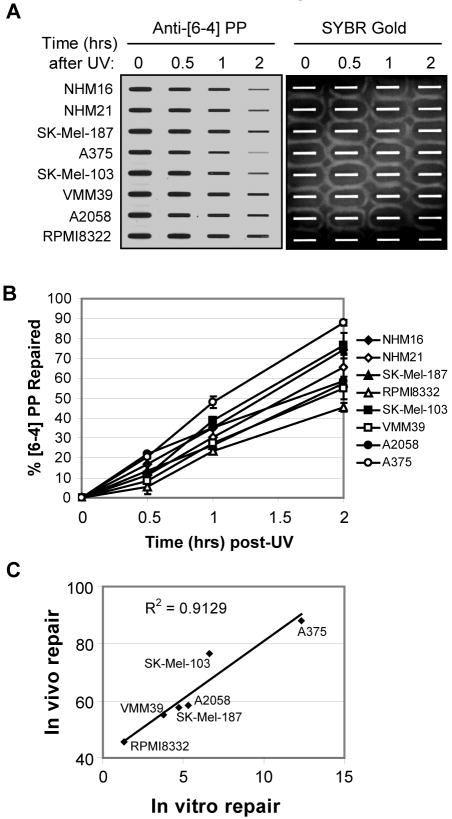
We next uded an immunoslot blot assay to monitor removal of [6-4] PPs in NHMs and melanoma cell lines. This assay involves the immobilization of genomic DNA from cells at various times after UV irradiation onto a nitrocellulose membrane and then immunoblotting with antibodies that specifically recognize either CPDs or [6-4] PPs (37). As shown in Figure 2A, we observed a time-dependent reduction in anti-[6-4] PP antibody reactivity in genomic DNA from both the normal human melanocytes and the melanoma cell lines, at rates near those previously reported for other cell types (38), with typically 50-80% of [6-4] PPs removed within 2 hours in the various cell lines (Figure 2B). Importantly, we observed very similar [6-4] PP signals in the UV-irradiated melanocytes and melanoma cells prior to repair, indicating that similar numbers of photoproducts were induced by UV. Cell lines lacking or containing mutations in the B-Raf and N-Ras oncogenes repaired [6-4] PPs at similar rates, indicating that mutation of these oncogenes does not significantly affect excision repair. Interestingly, when we compared the relative amount of [6-4] PP repair in the immunoslot blot assay (Figure 2A, B) with repair in the in vitro excision assay (Figure 1) among the six melanoma cell lines tested, we observed a strong correlation (r2=0.91) between the two assays (Figure 2C), indicating that the two approaches are reliable measures of [6-4] PP repair capacity.
Figure 2. Immunoslot blot assay for [6-4] photoproduct repair.
(A) An immunoslot blot assay was used to measure [6-4] PP repair at various time points post-irradiation in NHMs and melanoma cell lines exposed to 10 J/m2 of UVC. The image shows the [6-4] PP signal detected with an anti-[6-4] PP antibody and SYBR Gold staining to show equal loading of total genomic DNA. (B) Quantitative analysis of the repair assay. The averages and standard deviations are from two independent experiments. (C) Correlation between [6-4] PP repair levels using the immunoslot blot and in vitro cell-free excision assays. For each of the six melanoma cell lines examined, the level of [6-4] PP repair as measured at the two hour time point by immunoslot blot assay (in vivo repair) was plotted along with the amount of [6-4] PP substrate repaired for 90 min in the excision assay in Figure 1 (in vitro repair).
Measurement of CPD repair in vivo
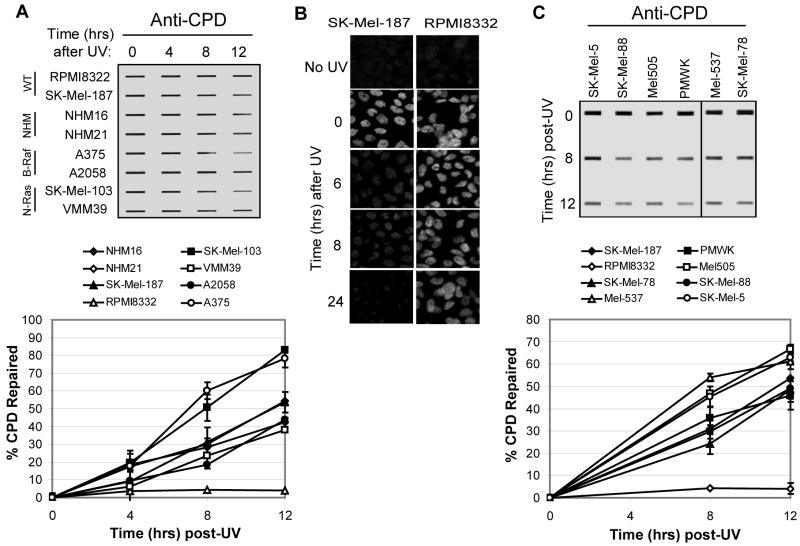
We next measured CPD repair in the same set of cell lines. CPDs constitute the majority of UV photoproducts in DNA and are recognized and repaired by the excision repair machinery more slowly than for [6-4] PPs and inefficiently in the in vitro excision assay making of quantitative comparison of CPD repair with the excision assay difficult. Therefore, we solely relied on the slotblot assay for in vivo repair. As displayed and quantified in Figure 3A, with the exception of RPMI8332 cells, which repaired CPDs very inefficiently, all the other cell lines removed 40-80% of CPDs within 12 hours. Importantly, the two NHM lines showed very similar CPD repair kinetics with three of these melanoma lines, indicating that melanocytes and melanoma cells repair UV photoproducts with similar kinetics. The RPMI8332 melanoma line, which repaired [6-4] PPs least efficiently in both the in vitro excision assay (Figure 1) and in vivo slot blot assay (Figure 2), appeared to lack any CPD repair capacity, with only ∼4% of CPDs removed within 12 hours (Figure 3A). To confirm this inefficient repair phenotype, we used immunofluorescence microscopy to detect CPD formation and removal in the repair-deficient RPMI8332 line and the repair-proficient line SK-Mel-187. Very little CPD removal was observed in RPMI8332 cells by immunofluorescence (Figure 3B), consistent with the results of the immunoslot blot assay.
Figure 3. CPD repair in melanocytes and melanoma cell lines.
(A) Cells were exposed to UVC (10 J/m2) and harvested at the indicated time points for immunoslot blot analysis of CPD repair. The top image shows a representative experiment. CPD repair assays were performed three times for each cell line and average CPD repair (and standard deviation) graphed below. (B) Immunofluorescence analyses of CPD repair in SK-Mel-187 and RPMI8332 cells. (C) CPD repair in six additional melanoma cell lines with wild-type N-Ras and B-Raf. The graph (right panel) includes SK-Mel-187 and RPMI cells analyzed in (A).
Since we observed a significant difference in CPD repair in RPMI8332 cells and SK-Mel-187 cells, both of which are WT for both B-Raf and N-Ras, we decided to measure CPD removal in six additional melanoma lines with wild-type forms of these genes. As shown in Figure 3C, all six lines displayed CPD removal rates comparable to SK-Mel-187 and the other cell lines and were unlike that seen in the RPMI8332 cells. Though the cause of the inefficient CPD and [6-4] PP removal in RPMI8332 cells is not known, we conclude that the repair deficiency is not characteristic of melanoma cells that are WT for both B-Raf and N-Ras. Interestingly, RPMI8332 cells were also significantly more sensitive to UV than the other cell lines in a colony formation assay (Supplementary Figure 3). This line has high level of chromosomal instability and some unusual growth properties as the cells undergo massive cell death upon reaching confluency, suggesting that inefficient repair is secondary to gross dysregulation of many pathways.
p53 contributes to CPD repair efficiency in melanoma cells
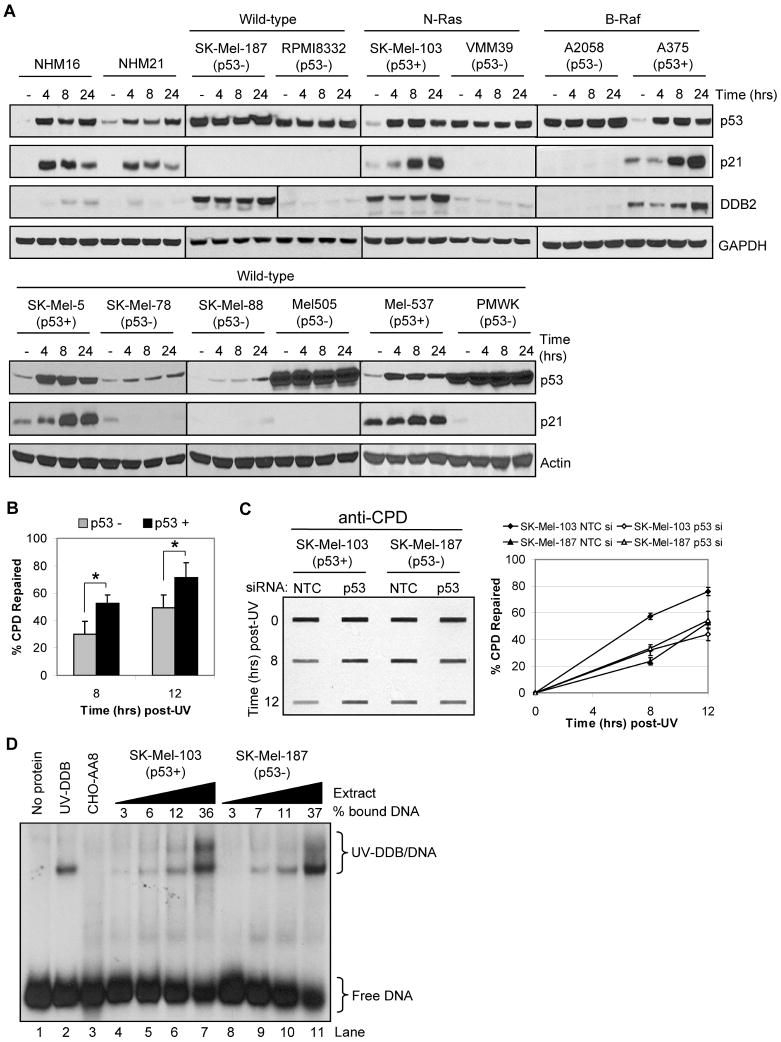
Though the tumor suppressor p53 has been shown to contribute to CPD repair rates and UV survival in human skin fibroblasts and other cell types (14, 38), other work has indicated no effect of p53 deficiency on UV photoproduct repair in keratinocytes (39). We therefore sought to examine p53 functionality in normal melanocytes and melanoma cell lines to determine whether p53 status may influence CPD repair in melanoma. Since p53 mutation status has only been characterized in a subset of these lines (13, 40), we tested p53 functionality by immunoblotting extracts from non-irradiated or UV-irradiated cells for UV-induced expression of p53 and its transactivation targets p21 and DDB2. As shown in Figure 4A, p53 functionality varied among the different cell lines, but exhibited normal responses in the normal melanocytes and in four of the melanoma lines (Sk-Mel-103, A375, Sk-Mel-5, Mel-537). With this information, we then re-examined the CPD repair data for the melanoma cell lines presented in Figure 3 by combining the repair data among the cell lines based on p53 status. Interestingly, we observed significantly more CPD repair in cells with wild-type, functional p53 compared to cells with mutant or inactive p53 (Figure 4B). Though less pronounced, [6-4] PP repair also correlated with p53 functionality (Supplementary Figure 4). To confirm the positive role for p53 in excision repair, we transfected p53-positive SK-Mel-103 and p53-inactive SK-Mel-187 cells with either a non-targeting control siRNA or a siRNA targeting p53 and then measured CPD repair. Consistent with the pooled cell line data shown in Figure 4B, knockdown of p53 in SK-Mel-103 cells, which did not have measurable effect on DDB2 level (Supplementary Figure 5), resulted in less CPD removal, near the level observed in p53-inactive SK-Mel-187 cells (Figure 4C). Based on these results, we conclude that p53 contributes to UV photoproduct removal in human melanoma cell lines.
Figure 4. Analysis of p53 and DDB2 functionality and in melanoma cells and melanocytes.
(A) Western blot analyses were performed with extracts from the indicated cell lines harvested at various times post UV-irradiation (12.5 J/m2) to monitor p53 functionality, as determined by induction of p21 and DDB2 protein expression. (B) Based on p53 functionality determined in (A), CPD repair measurements for the individual cell lines (Figure 3) were pooled and re-analyzed as a function of p53 status. Data show the average (and standard deviation) of CPD repair for the seven p53 mutant cell lines (RPMI was excluded) and four p53 wild-type cell lines. Asterisks indicate a statistically significant difference (p<0.01; two-tailed Student's t-test) in CPD repair between p53 mutant and wild-type cell lines. (C) Knockdown of p53 in SK-Mel-103 cells inhibits CPD repair. SK-Mel-103 (p53 wild-type) and SK-Mel-187 (p53 mutant) cells were transfected with non-targeting or p53 siRNAs, exposed to UV, and CPD repair measured at the indicated time points. (D) Electrophoretic mobility shift assay of UV-DDB binding to damaged DNA. Cell-free extract from SK-Mel-103 (p53 wild-type) and SK-Mel-187 (p53 mutant) cells were incubated with radiolabeled [6-4] PP substrate DNA and complexes separated on a 5% non-denaturing gel. Purified UV-DDB complex was used as a positive control and CHO-AA8 extract as a negative control. The cell-free extract protein concentrations ranged from 0.5, 1.0, 2.0 or 4.0 μgs per reaction. The experiment was repeated three times, and the average percent binding is indicated.
A role for p53 in excision repair has been reported to be due to transcriptional induction of the XPE gene encoding the DDB2 protein (41), which in the form of UV-DDB (DDB1-DDB2 heterodimer) directly binds to UV photoproducts. We therefore used an electrophoretic mobility shift assay (32, 33) to examine UV-DDB functionality in SK-Mel-103 and SK-Mel-187 cell extracts. As shown in Figure 4D, we observed similar levels of binding to an oligonucleotide containing a [6-4] PP in both extracts, but not in extract from CHO cells, in which the XPE (DDB2) gene is transcriptionally repressed (14). The levels of UV-DDB activities in the SK-Mel-103 and SK-Mel-187 were similar 6 hr after UV but slightly higher in SK-Mel-103 24 hr after UV (Supplementary Figure 6) by which time all of the CPDs were removed in the both cell lines. Thus, we conclude that though p53 contributes to the removal of UV lesions in DNA, UV-DDB does not appear to be a major contributor through which p53 promotes repair in melanoma cells.
Excision repair in highly metastatic melanoma cells
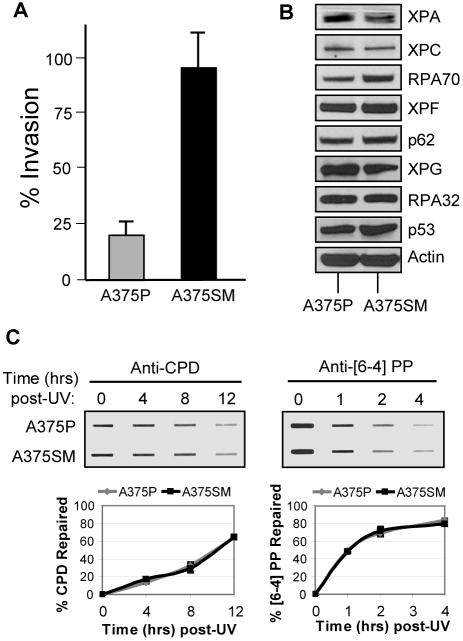
We next wished to address whether metastatic state in melanoma contributes to the efficiency of nucleotide excision repair, since a previous report indicated UV photoproduct removal occurred more rapidly in highly metastatic murine melanoma cells (24). However, since the repair assay used in that study did not directly measure repair of UV-damaged genomic DNA, we re-examined whether excision repair capacity is altered in melanoma cells of different metastatic states by comparing [6-4] and CPD removal in A375 cells and a derivative line with a higher metastatic potential (A375SM). As previously reported (42), we confirmed that A375SM cells are more invasive than its parental A375 line (A375P) (Figure 5A). We then examined the protein expression levels of excision repair factors and found no significant difference between the two lines (Figure 5B). Consistent with the similar expression levels, both A375P and A375SM cells repaired [6-4] PPs and CPDs at similar rates (Figure 5C). We conclude from these results that metastatic state does not necessarily alter nucleotide excision repair capacity in melanoma cell lines.
Figure 5. Excision repair in highly metastatic melanoma cells.
(A) Matrigel invasion assay of the highly metastatic cell line (A375SM) compared with its parent cell line (A375P). The results are the averages and standard deviations of three independent experiments. (B) Western blot analyses of the DNA excision repair protein levels of both A375P and A375SM cell lines. (C) Immunoslot blot analysis of CPD and [6-4] PP repair after UV irradiation (10 J/m2) of A375P and A375SM cells. The graphs show the average and standard deviation from two independent experiments.
Discussion
Melanoma is the most deadly form of skin cancer and in industrialized countries is rapidly growing in prevalence (43). Based on the high rates of melanoma in patients with xeroderma pigmentosum, most of whom lack one of the essential excision repair factors, the development or progression of melanoma may be associated with a reduced capacity for excision repair. However, here we show that excision repair capacity in normal human melanocytes is very similar to that in melanoma cells (Figure 2, 3), irrespective of B-Raf and N-Ras status, suggesting that altered repair capacity may not be a predominant cause of melanoma initiation or progression.
Tumors from metastatic melanoma patients are characterized by significant resistance to DNA damaging agents such as cisplatin, decarbazine, and melphalan (23), suggesting a broad underlying resistance to the effects of DNA damage. Though one report indicated that mouse melanoma cells with high metastatic potential exhibited elevated excision repair capacity (24), the study employed an indirect measure of DNA repair. By monitoring the direct removal of CPDs and [6-4] PPs from genomic DNA in human A375 cells and a super-metastatic derivative cell line (A375SM), here we observed no difference in nucleotide excision repair efficiency (Figure 5). We conclude that metastatic potential and excision repair capacity are not directly related to one another.
In contrast, our results do show that the functionality of the tumor suppressor p53 is an important determinant of UV photoproduct repair efficiency in melanoma cells (Figure 4, Table 1). These results are consistent with a variety of data from other cell types, including normal human fibroblasts (38, 39), though importantly our results provide the first evidence that p53 status affects excision repair specifically in melanoma cells. Even though only 1% of primary melanomas and 5% of metastatic melanomas show mutations in the p53 gene (15, 16, 44), the common loss of ARF function in metastatic melanomas with deletion of the CDKN2A locus (17-19) suggests that p53-dependent processes may contribute to melanoma development. Since p53 regulates many components of the cellular response to DNA damage induced by UV irradiation, including DNA repair, cell cycle checkpoint, and apoptosis, it is not clear how its many diverse functions ultimately control cell fate in melanoma. Similarly, though our data do not indicate that p53 regulation of DDB2 influences excision repair in melanoma cells, DDB2 may contribute to other aspects of the UV response in melanocytes or melanoma, such as cell survival and apoptosis (45, 46).
Table 1. Summary of N-Ras, B-Raf and p53 status and excision repair capacity in 12 melanoma cell lines and two normal human melanocyte lines used in our study. Gaddameedhi_Table 1.
| Cell line | Oncogene status | p53 status | Excision repair capacity |
|---|---|---|---|
| NHM16 | WT | Active | Normal |
| NHM21 | WT | Active | Normal |
| SK-Mel-187 | WT | Mutant/inactive | Normal |
| RPMI8332 | WT | Mutant/inactive | Low |
| SK-Mel-103 | N-Ras | Active | High |
| VMM39 | N-Ras | Mutant/inactive | Normal |
| A2058 | B-Raf | Mutant/inactive | Normal |
| A375 | B-Raf | Active | High |
| PMWK | WT | Mutant/inactive | Normal |
| Mel505 | WT | Mutant/inactive | High |
| Mel-537 | WT | Active | High |
| SK-Mel-78 | WT | Mutant/inactive | Normal |
| SK-Mel-88 | WT | Mutant/inactive | Normal |
| SK-Mel-5 | WT | Active | High |
Inefficient UV photoproduct repair has also been demonstrated in cells with mutations in the melanocortin 1 receptor (MC1R), which acts upstream in the microphthalmia-associated transcription factor (MITF) signaling pathway of eumelanin biosynthesis (47), and in cells with deletion of the CDKN2A locus that encodes the tumor suppressor genes p16 and ARF (48). Though the repair-deficient RPMI8332 cell line shows reduced expression of MITF and loss of p16, many of the other melanoma cell lines we examined also show reduced MITF levels (Mel505, SK-Mel-187, PMWK) or p16 loss (SK-Mel-103) (13, 26), indicating that other factors are responsible for the lack of excision repair in RPMI8332 cells.
In summary, 11 of 12 melanoma cell lines displayed normal rates of repair of UV-induced DNA photoproducts in comparison to normal melanocytes, indicating that functional inactivation of the excision repair pathway is uncommon in sporadic melanoma.
Supplementary Material
Acknowledgments
We thank Dr. Marila Cordeiro-Stone for her initial help on slotblot assay.
Supported by: National Institutes of Health grants ES014635 (SG, WKK, JS and AS), ES015856 (SS-R and WKK), ES10126 (WKK) and GM32833 (MGK, JTR and AS).
Footnotes
Note: Supplementary data for this work are available at Cancer Research Online (http://cancerres.aacrjournal.org/)
References
- 1.Markovic SN, Erickson LA, Rao RD, et al. Malignant melanoma in the 21st century, part 1: epidemiology, risk factors, screening, prevention, and diagnosis. Mayo Clin Proc. 2007;82:364–80. doi: 10.4065/82.3.364. [DOI] [PubMed] [Google Scholar]
- 2.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Ries LA, Wingo PA, Miller DS, et al. The annual report to the nation on the status of cancer, 1973-1997, with a special section on colorectal cancer. Cancer. 2000;88:2398–424. doi: 10.1002/(sici)1097-0142(20000515)88:10<2398::aid-cncr26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Geller AC, Miller DR, Annas GD, et al. Melanoma incidence and mortality among US whites, 1969-1999. Jama. 2002;288:1719–20. doi: 10.1001/jama.288.14.1719. [DOI] [PubMed] [Google Scholar]
- 5.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 6.Mu D, Park CH, Matsunaga T, et al. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–8. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 7.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16:6559–73. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–6. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–50. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 10.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 11.Daniotti M, Oggionni M, Ranzani T, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–77. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 12.Satyamoorthy K, Li G, Gerrero MR, et al. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res. 2003;63:756–9. [PubMed] [Google Scholar]
- 13.Shields JM, Thomas NE, Cregger M, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67:1502–12. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 14.Ford JM. Regulation of DNA damage recognition and nucleotide excision repair: another role for p53. Mutat Res. 2005;577:195–202. doi: 10.1016/j.mrfmmm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann A, Blaszyk H, Cunningham JS, et al. Overexpression and mutations of p53 in metastatic malignant melanomas. Int J Cancer. 1996;67:313–7. doi: 10.1002/(SICI)1097-0215(19960729)67:3<313::AID-IJC1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 16.Florenes VA, Oyjord T, Holm R, et al. TP53 allele loss, mutations and expression in malignant melanoma. Br J Cancer. 1994;69:253–9. doi: 10.1038/bjc.1994.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R, Sauroja I, Punnonen K, Jansen C, Hemminki K. Selective deletion of exon 1 beta of the p19ARF gene in metastatic melanoma cell lines. Genes Chromosomes Cancer. 1998;23:273–7. doi: 10.1002/(sici)1098-2264(199811)23:3<273::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 18.Tran TP, Titus-Ernstoff L, Perry AE, Ernstoff MS, Newsham IF. Alteration of chromosome 9p21 and/or p16 in benign and dysplastic nevi suggests a role in early melanoma progression (United States) Cancer Causes Control. 2002;13:675–82. doi: 10.1023/a:1019599629895. [DOI] [PubMed] [Google Scholar]
- 19.Grafstrom E, Egyhazi S, Ringborg U, Hansson J, Platz A. Biallelic deletions in INK4 in cutaneous melanoma are common and associated with decreased survival. Clin Cancer Res. 2005;11:2991–7. doi: 10.1158/1078-0432.CCR-04-1731. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Hu Z, Liu Z, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–32. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- 21.Kauffmann A, Rosselli F, Lazar V, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–73. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers AH, Lavin M, Atisoontornkul S, Mansbridge J, Kidson C. Resistance of human melanoma cells to ultraviolet radiation. Cancer Res. 1976;36:1930–4. [PubMed] [Google Scholar]
- 23.Hatton DH, Mitchell DL, Strickland PT, Johnson RT. Enhanced photoproduct repair: its role in the DNA damage-resistance phenotype of human malignant melanoma cells. Cancer Res. 1995;55:181–9. [PubMed] [Google Scholar]
- 24.Wei Q, Cheng L, Xie K, Bucana CD, Dong Z. Direct correlation between DNA repair capacity and metastatic potential of K-1735 murine melanoma cells. J Invest Dermatol. 1997;108:3–6. doi: 10.1111/1523-1747.ep12285608. [DOI] [PubMed] [Google Scholar]
- 25.Xu G, Snellman E, Bykov VJ, Jansen CT, Hemminki K. Cutaneous melanoma patients have normal repair kinetics of ultraviolet-induced DNA repair in skin in situ. J Invest Dermatol. 2000;114:628–31. doi: 10.1046/j.1523-1747.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann WK, Nevis KR, Qu P, et al. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–87. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2841–6. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsumi S, Kobayashi N, Imoto K, et al. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J Invest Dermatol. 2001;117:1156–61. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- 29.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 30.Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;35:e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulaksiz G, Reardon JT, Sancar A. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol Cell Biol. 2005;25:9784–92. doi: 10.1128/MCB.25.22.9784-9792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols AF, Itoh T, Graham JA, et al. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–8. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 33.Itoh T, Nichols A, Linn S. Abnormal regulation of DDB2 gene expression in xeroderma pigmentosum group E strains. Oncogene. 2001;20:7041–50. doi: 10.1038/sj.onc.1204909. [DOI] [PubMed] [Google Scholar]
- 34.Lancaster HO. Some geographical aspects of the mortality from melanoma in Europeans. Med J Aust. 1956;43:1082–7. [PubMed] [Google Scholar]
- 35.Lee JA, Merrill JM. Sunlight and melanoma. Lancet. 1971;2:550–1. doi: 10.1016/s0140-6736(71)90476-4. [DOI] [PubMed] [Google Scholar]
- 36.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–7. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riou L, Eveno E, van Hoffen A, et al. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–94. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 38.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–80. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 39.Ferguson BE, Oh DH. Proficient global nucleotide excision repair in human keratinocytes but not in fibroblasts deficient in p53. Cancer Res. 2005;65:8723–9. doi: 10.1158/0008-5472.CAN-05-1457. [DOI] [PubMed] [Google Scholar]
- 40.Haapajarvi T, Pitkanen K, Laiho M. Human melanoma cell line UV responses show independency of p53 function. Cell Growth Differ. 1999;10:163–71. [PubMed] [Google Scholar]
- 41.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96:424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 43.Ross PM, Carter DM. Actinic DNA damage and the pathogenesis of cutaneous malignant melanoma. J Invest Dermatol. 1989;92:293S–6S. doi: 10.1111/1523-1747.ep13076718. [DOI] [PubMed] [Google Scholar]
- 44.Akslen LA, Monstad SE, Larsen B, Straume O, Ogreid D. Frequent mutations of the p53 gene in cutaneous melanoma of the nodular type. Int J Cancer. 1998;79:91–5. doi: 10.1002/(sici)1097-0215(19980220)79:1<91::aid-ijc17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 45.Itoh T, Cado D, Kamide R, Linn S. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc Natl Acad Sci U S A. 2004;101:2052–7. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci U S A. 2009;106:10690–5. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hauser JE, Kadekaro AL, Kavanagh RJ, et al. Melanin content and MC1R function independently affect UVR-induced DNA damage in cultured human melanocytes. Pigment Cell Res. 2006;19:303–14. doi: 10.1111/j.1600-0749.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar-Agrawal P, Vergilis I, Sharpless NE, DePinho RA, Runger TM. Impaired processing of DNA photoproducts and ultraviolet hypermutability with loss of p16INK4a or p19ARF. J Natl Cancer Inst. 2004;96:1790–3. doi: 10.1093/jnci/djh307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.