Abstract
Embryonic stem cells (ESCs) maintain self-renewal while ensuring a rapid response to differentiation signals, but the exact mechanism of this process remains unknown. PD2 is the human homolog of the RNA polymerase II-associated factor 1 (Paf1). The Paf1/PD2 is a member of the human PAF complex that consists of four other subunits, hCdc73, hLeo1, hCtr9, and hSki8, and is involved in the regulation of transcriptional elongation and further downstream events. Here, we show that Paf1/PD2 is overexpressed in mouse ESCs and is involved in the maintenance of mouse ESCs. The Paf1/PD2 knockdown and knockout ESCs grown under self-renewal conditions express substantially reduced levels of self-renewal regulators, including Oct3/4, SOX2, Nanog, and Shh. We observed that the level of Paf1/PD2 expression is much higher in self-renewing mouse embryonic carcinoma cells than in the differentiating cells. Knockout of Paf1/PD2 altered ESC phenotype by increasing apoptosis and decreasing the percentage of cells in S-phase of the cell cycle. Interestingly, we found that the key genes that regulate endodermal differentiation (Gata4, Gata6, and Fgf8) are induced in the Paf1/PD2 heterozygous knockout ESCs. This suggests that Paf1/PD2 plays a specific role in regulating early commitment of ESCs to endodermal differentiation. Furthermore, for the first time, we showed that Paf1/PD2 protein interacts with Oct3/4 and RNA polymerase II, and through this interaction Paf1/PD2 may regulate Oct3/4-mediated gene expression. Thus, the Paf1/PD2 protein is a newly discovered element of the interconnected regulatory network that maintains the self-renewal of mouse ESCs.
Keywords: Paf1/PD2, PAF complex, Oct3/4, Self-renewal, Embryonic stem cells
Introduction
The development and function of multicellular organisms is based on the activity of stem cells. During early embryonic development, pluripotent stem cells give rise to all cell types in the body, including the germline [1]. Since this property can be exploited for genetic engineering and holds great promise for applications in regenerative medicine, an important goal is to understand the molecular pathways unique to pluripotent and self-renewal cells. Although the precise differentiation potential varies between different stem cell types, all stem cells share the fundamental property of self-renewal, that is, the maintenance of their population and existing in an undifferentiated state [2, 3]. Embryonic stem cells (ESCs), derived from the inner cell mass of the blastocyst, are the most commonly used cell types in studies of early embryonic development and the pluripotent state [4–6], largely because of their ability to self-renew in tissue culture for extended periods.
The pluripotency of ESCs is regulated by a unique network of ESC-specific transcription factors, including Oct3/4, Nanog, SOX2, and their binding partners [7–9]. Recent reports show a defined combination of Oct3/4, SOX2, and other transcription factors is sufficient to induce mouse fibroblast cells to take on an ESC-like phenotype, with the resulting cells labeled as induced pluripotent stem cells (iPSs) [10]. The ESC lines are capable of unlimited growth in culture while retaining their undifferentiated state and full developmental potential. Murine ESCs are typically cultured on fibroblast feeder cells with serum and the addition of leukemia inhibitory factor (LIF), a cytokine that activates the gp130/Stat3 signaling pathway essential for self-renewal [11]. In addition, there is a distinct genetic pathway regulating ESC self-renewal, which involves the transcription factors Tbx3 and Esrrb and the signaling cofactor Tcl1 [12].
The 19q13 amplicon in pancreatic cancer cells contains a novel pancreatic differentiation 2 (PD2) gene, which was identified by differential screening analysis [13]. PD2 is the human homolog of the yeast RNA polymerase II-associated factor 1 (yPaf1) and is part of the human (h) PAF complex. hPAF is composed of five subunits that include PD2/hPaf1, parafibromin/Cdc73, hLeo1, hCtr9, and hSki8 [13, 14]. This multifaceted complex was first identified in yeast (y) PAF and subsequently in Drosophila and humans. Recent advances in the study of the hPAF complex have revealed various functions of this complex in humans are similar to those of the yPAF complex, including efficient transcription elongation, mRNA quality control, and cell cycle regulation. We have previously shown that PD2/hPaf1 is overexpressed in poorly differentiated pancreatic cancer cells compared with well-differentiated cells. In addition, a recent study showed that one of the PAF complex subunits, parafibromin, plays a role in mammalian development and survival in adults [15].
In the present study, we elucidate the role of Paf1/PD2 in the maintenance of ESCs. We show that the knockout of Paf1/PD2 in ESCs loses its self-renewal function and leads to the induction of key endoderm genes, indicating a specific role of Paf1/PD2 in the early commitment to an endodermal lineage. Our result demonstrates, for the first time, the association of Paf1/PD2 with Oct3/4 for self-renewal maintenance.
Materials and Methods
ESC Culture
E-14 ESCs, PD2 heterozygous knockout (PD2+/−) ESCs, J1 ESCs, and F9 embryonic carcinoma (EC) cells were cultured in gelatinized tissue culture dishes in ESC medium containing Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY, http://www.invitrogen.com) supplemented with 15% ESC-specific fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, http://www.invitrogen.com), L-glutamine (Gibco), 100 nM nonessential amino acids (Gibco), 1000 U/ml LIF (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com), and 100 μM β-mercaptoethanol (Sigma-Aldrich).
Generation of Paf1/PD2 Heterozygous Knockout ESCs by Gene Trap Method
The Paf1/PD2 knockout mouse ESCs were established using the ESC line RRO233 from the BayGenomics Gene trap resource (Los Angeles, http://baygenomics.ucsf.edu) [16]. The linearized Gene trap vector was inserted in intron 1 of the Paf1/PD2 gene, resulting in a Paf1/PD2-β-galactosidase fusion protein that lacked 512 amino acids from the C-terminal of the full-length protein (535 amino acids). These RRO233 ESCs were used for further experiments to elucidate the role of Paf1/PD2 in ESCs.
RNA Isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction
Total cellular RNA was extracted from ESCs using the RNAeasy kit (Qiagen, Hilden, Germany, http://www1.qiagen.com) and processed for reverse transcription. The initial polymerase chain reaction (PCR) activation step was at 94°C for 4 minutes, followed by the denaturation step at 94°C for 1 minute, primer-annealing step at 58°C for 30 seconds, extension step at 72°C for 1 minute, and the final extension step at 72°C for 10 minutes. PCR reaction products were then separated by electrophoresis using a 2% agarose gel. Gels were stained using 0.5 μg/ml of ethidium bromide, illuminated with UV light. Total cell RNA was reverse-transcribed and assayed by quantitative real-time PCR using SYBR Green incorporation. The expression of all genes was normalized to that of internal control β-actin and expressed relative to the indicated reference sample (average ± SD of triplicate reactions). The expressions of lineage-specific genes were compared between the ESCs and PD2+/− ESCs by the two-tailed Student's t test. A p value of <.05 was considered statistically significant.
RNA Interference
The region of mouse Paf1/PD2 was targeted with specific siRNA (sequence 5′-CGAGTCAAGTACTGCAATA-3′). Synthetic sense and antisense oligonucleotides (Dharmacon, Lafayette, CO, http://www.dharmacon.com) were annealed in 100 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), and 2 mM magnesium acetate for 1 minute at 90°C and 1 hour at 37°C, and frozen. Oligonucleotides were transfected into cells with TransIT-TKO (Mirus Bio LLC, Madison, WI, http://www.mirusbio.com) in accordance with the supplier's recommendations.
Immunoblot Assay
ESC and F9 EC cell lines were processed for protein extraction and Western blotting using standard procedures. Briefly, the cells were washed twice in phosphate-buffered saline (PBS) and lysed in RIPA buffer (100 mM Tris, 5 mM EDTA, 5% Nonidet P40; pH 8.0) containing protease inhibitors (1 mM phenyl-methyl sulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin) and kept at 4°C and supernatants were collected. Resolved proteins were transferred onto the polyvinylidene fluoride (PVDF) membrane. After quick washing in phosphate-buffered saline and 0.1% Tween 20 (PBST), the membranes were blocked in 5% nonfat dry milk in PBS for at least 2 hours and then incubated with primary antibodies (anti-Paf1/PD2, anti-Leo1, anti-Cdc73, anti-Ski8, anti-Oct3/4, anti-SOX2, anti-β-actin) (diluted in 3% BSA in PBS) overnight at 4°C. Then the membrane was washed (3 × 10 minutes) in PBST at room temperature and probed with 1:2,000 diluted horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies for 1 hour at room temperature and washed 5 × 10 minutes with PBST. The signal was detected with an enhanced chemiluminescence (ECL) chemiluminescence kit (Amersham Biosciences, Buckinghamshire, U.K., http://www. amersham.com).
Alkaline Phosphatase Staining
Alkaline phosphatase was visualized using stemTAGTM alkaline phosphatase staining kit (Cell Biolabs, Inc., San Diego, http://www.cellbiolabs.com). The kit was used according to the manufacturer's specifications with the following modification: cell layers were fixed in 4.5 mM citric acid, 2.25 mM sodium citrate, 3 mM sodium chloride, 65% methanol, and 4% paraformaldehyde prior to washing and staining.
Confocal Microscopy
Cells were plated onto sterile round coverslips (CIR 18-1 Fisher brand 12-545-10; Fisher Scientific International, Hampton, NH, http://www.fisherscientific.com) and grown in 12-well plates for 24 hours. Cells were fixed in acetone/methanol (1:1; prechilled to −20°C) and permeabilized with 0.1% Triton X-100 in PBS. Then the cells were washed in PBS and incubated with primary (for 2 hours) and fluorescent-tagged secondary (for 30 minutes) antibodies at room temperature. Antibodies were diluted in 5% goat serum. Finally, coverslips were mounted with vectashell containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com).
Immunoprecipitation Analysis
Equal amounts of protein (20 μg) were incubated overnight with anti-PD2 (rabbit polyclonal), anti-Oct3/4 (mouse monoclonal), and anti-RNA polymerase II (Pol II; mouse monoclonal) antibodies in a 500-μl total volume. Protein A+ G-Sepharose beads (Sigma-Aldrich Corp. St. Louis, MO, USA, http://www.sigmaaldrich. com) were added to the lysate-antibody mix and incubated on a rotating platform for 3 hours at 4°C and then washed four times with lysis buffer. The immunoprecipitates or total cell lysates were electrophoretically resolved on SDS-polyacrylamide gel electrophoresis (10%). Resolved proteins were transferred onto the PVDF membrane. After quick washing in PBST, the membranes were blocked in 5% nonfat dry milk in PBS for at least 2 hours and then incubated with primary antibodies (anti-Paf1/PD2, Oct3/4, and RNA Pol II). The immunoblots were washed five times (5 × 10 minutes), incubated for 1 hour with horseradish peroxidase-conjugated secondary antibodies, washed five times (5× 10 minutes), reacted with enhanced chemiluminescence ECL reagent (Amersham Biosciences), and exposed to x-ray film to detect the signal.
Colony-Forming Analysis
PD2 heterozygous knockout and wild-type (WT) ESCs were seeded in triplicate at 200, 400, 800, and 2,000 cells per 10-cm dishes in complete ESC medium. After 2 weeks of growth, the cells were fixed and stained with crystal violet stain (0.1%, wt/vol) in 20 nm 4-morpholinepropanesulfonic acid (Sigma-Aldrich), and the grossly visible colonies were counted; all experiments were repeated at least three times. Plating efficiency was determined as the number of colonies formed, divided by the total number of cells plated.
Apoptosis Assay
Apoptosis was measured using the annexin V-fluorescein isothiocyanate apoptosis detection kit (Roche Diagnostics, Indianapolis, IN, http://www.roche-diagnostics.us). Apoptosis was detected by staining the cells with annexin V and propidium iodide solution followed by flow cytometry.
Cell Cycle Analysis
Confluent cells were detached from the flask with trypsin, washed in PBS, and fixed in 2 mM glycine (pH 2)/70% ethanol. Fixed cells were then washed and resuspended in 1 ml of Telford reagent (90 mM EDTA, 2.5 mU of RNase A/ml, 50 μg of propidium iodide/ml, and 0.1% Triton X-100 in PBS). The total DNA content was analyzed in a FACSCalibur analyzer using ModFit LT software (Becton, Dickinson and Company, Franklin Lakes, NJ, http://www.bd.com).
Embryoid Body Formation
Embryoid bodies (EBs) were made as described elsewhere [17], with minor modifications. Cells were plated in a V-bottom 96-well plate at a final concentration of 5,000 cells per well in EB medium (DMEM; Gibco) supplemented with 15% ESC-specific FBS (Invitrogen), L-glutamine (Gibco), 100 nM nonessential amino acids (Gibco), and 100 μM β-mercaptoethanol (Sigma-Aldrich). After 2 days, EBs were transferred with DMEM medium to an ultra-low-binding 6-well plate. Then EBs were photographed using a phase-contrast microscope and collected for RNA extraction to analyze the lineage-specific genes.
Teratoma Formation
To induce teratoma formation, 1 × 106 cells from each cell population were injected into the quadriceps muscle of the hind legs of immunocompromised mice. Teratomas were dissected after 16 days. A small piece of each tumor was processed in PCR analyses to confirm genotypes. Half of the remaining tissue was fixed in 0.4% paraformaldehyde and embedded in optimal cutting temperature medium (Sakura Finetek Inc., Zoeterwoude, The Netherlands, http://www.sakuraeu.com) after incubating overnight in 30% sucrose; the remaining half was fixed in 10% formalin overnight, transferred to 70% ethanol, and paraffin embedded. Paraffin-embedded tissue sections were analyzed by hematoxylin and eosin staining.
Histological Analysis
Isolated teratomas were fixed in 4% paraformaldehyde in phosphate-buffered saline overnight and subsequently embedded in paraffin wax. Sections were cut at a thickness of 2 μm and stained with hematoxylin and eosin. The different structures were photographed and analyzed for structural variation.
ChIP-Re-ChIP Assay
ChIP-Re-ChIP experiment indicates sequential chromatin immunoprecipitations with two antibodies to study the simultaneous presence of two proteins in the genome sequence of interest [18]. ESCs were fixed with 1% formaldehyde, washed, harvested, and resuspended in 200 μl SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1 mM PMSF, and 1 μg/ml aprotinin). Samples were sonicated and diluted in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, 1 mM PMSF, and 1 μg/ml aprotinin). For input control, 14 μl of sonicated samples was separated. Immunoprecipitation was performed with ChIP-specific Oct3/4 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com) as previously described. Chromatin extracts were pulled down with a protein G bead. The samples were washed extensively with wash buffers (low salt, high salt, LiCl, and Tris/EDTA buffers) and incubated at 65°C for 4 hours in 0.3 M NaCl. The eluted Oct3/4 pull-down chromatin extract was used for Re-ChIP assay. For the Re-ChIP assay, PD2 ChIP-specific antibody (Abcam, Cambridge, U.K., http://www.abcam.com) was used for immunoprecipitation and was washed using the above-mentioned method. MUC4 antibody was used as a control for the ChIP-Re-ChIP assay. The eluted Re-ChIP DNA and input samples were amplified by SYBR Green quantitative PCR (QPCR) with specific promoter primers of FGF-4, Nanog, and Lefty-1. All the nonspecific primers were chosen from the 3′ region of these genes. All primers used in this study are available in supporting information Table 1.
RESULTS
Paf1/PD2 Expression in Mouse Embryonic Stem Cells
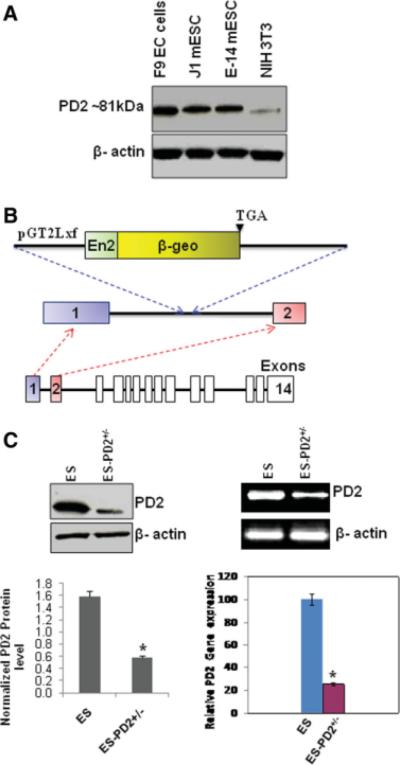
Paf1/PD2 is a member of the hPAF complex that comprises four other subunits, Cdc73/parafibromin, hLeo1, hCtr9, and hSki8. This complex is involved in the regulation of RNA elongation and the events downstream of transcription [13]. We began by assessing the levels of Paf1/PD2 in J1 and E-14 mouse ESCs and F9 EC cells under self-renewal conditions. Western blot analysis showed that the ESCs and EC cells had higher levels of Paf1/PD2 expression compared with the differentiated cells (Fig. 1A). Furthermore, we sought to determine the function of Paf1/PD2 in ESC and EC cell maintenance. For these studies we have obtained Paf1/PD2 heterozygous knockout (PD2+/− ) mouse ESC line (RRO233) from BayGenomics. The PD2+/− ESC was developed by gene trap mutagenesis method, wherein gene trap vector (pGT2Lxf) was inserted in-frame into the Paf1/PD2 gene immediately after exon 1, leading to the formation of a truncated protein having 23 amino acids from the N-terminal (Fig. 1B). The WT ESCs and PD2+/− ESCs were grown with LIF in ESC-specific culture condition. We performed Western blot and its densitometry (*, p = .01) (Fig. 1C), reverse-transcription (RT)-PCR (Fig. 1C), and quantitative RT-PCR (*, p .004) (Fig. 1C) to examine the Paf1/PD2 expression in both WT ESCs and PD2+/− ESCs. The PD2+/− ESC line had a substantially lower level of Paf1/PD2 transcript and protein than the control mouse ESCs.
Figure 1.
Expression of Paf1/PD2 in ESCs and heterozygous knockout ESCs. (A): Western blot analysis showed that Paf1/PD2 was overexpressed in F9 embryonic carcinoma cells, and J1 and E-14 mouse embryonic stem cells compared with differentiated NIH3T3 cells. (B): Generation of Paf1−/− mice. Schematic diagrams of the Gene trap vector pGT2Lxf insertion between exons 1 and 2. pGT2Lxf contains En2, the splice acceptor/Engrailed-2 exon, and β-geo. β-geo was inserted downstream of exon 1 of the Paf1 gene, producing a chimerical mRNA of Paf1 and β-geo mRNAs. Paf1 mRNA encodes a fusion protein containing the N-terminal 23 a residues and β-gal. (C): Expression of Paf1/PD2 (~81 kDa) in heterozygous knockout (PD2+/−) ESCs and wild-type ESCs by Western blot method with densitometry (left lower panel: *, p = .01) and mRNA expression by reverse-transcription polymerase chain reaction (RT-PCR) and quantitative RT-PCR analysis (right lower panel: *, p = .004). Abbreviations: β-geo, β-gal gene; EC, embryonic carcinoma; mESC, mouse ESC; PD2+/−, PD2 heterozygous knockout.
Unique Expression of the Paf1/PD2 Subunit in Mouse ESCs
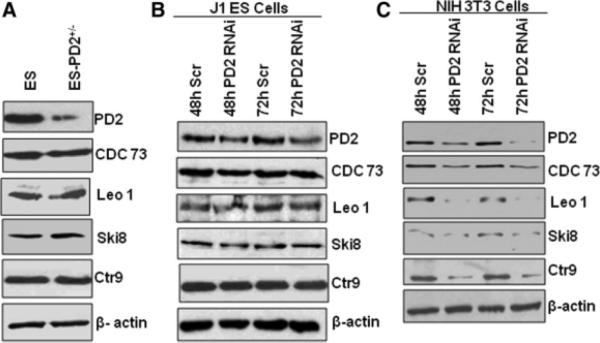
The hPaf1 is part of the PAF complex composed of four other subunits: Cdc73, Leo1, Ctr9, and Ski8. Previously established functions of the PAF complex revealed a coordinated role of all the members of the complex. The PAF complex coordinates events during transcription and RNA quality control [13]. In light of the previous study, our aim was to analyze the expression of other PAF complex subunits (Cdc73, Leo1, Ski8, and Ctr9) in Paf1/PD2 heterozygous knockout ESCs and PD2 RNAi (short interfering RNA [siRNA] against PD2 mRNA)-treated J1 ESCs. Interestingly, in contrast to the previous report [13, 19], we did not observe any coordinated expression of the other PAF complex subunits in the mouse ESCs. Western blot analysis of Cdc73 (parafibromin), Leo1, Ski8, and Ctr9 proteins showed no variation in their expression levels in the PD2+/− ESCs compared with the control ESCs (Fig. 2A). In addition, the transient knockdown of Paf1/PD2 in the J1 mouse ESCs also showed no decrease in the expression of other PAF complex members: Cdc73, Leo1, Ski8, and Ctr9 (Fig. 2B). Interestingly, the knockdown of Paf1/PD2 in the NIH3T3 mouse fibroblast cell line (a differentiated cell line) showed a decrease in the expression of other PAF complex members such as Cdc73, Leo1, Ski8, and Ctr9 (Fig. 2C). Thus, our results suggest that the expression of the PAF complex subunits is coordinated in differentiated mouse fibroblast cells but not in the undifferentiated ESCs. These findings suggest that Paf1/PD2 has a unique role in embryonic stem cell maintenance.
Figure 2.
Expression of PAF complex molecules in Paf1/PD2 knockout and RNAi-treated ESCs. (A): Western blot analysis showed an expression of other PAF complex molecules (Cdc73, Leo1, sKi8, and Ctr9) in wild-type ESCs and Paf1/PD2 heterozygous knockout ESCs. (B): Transient knockdown of Paf1/PD2 protein using specific RNAi at 48 and 72 hours in J1 ESCs. Expression of Paf1/PD2 protein and other PAF complex proteins (Cdc73, Leo1, sKi8, and Ctr9) in Paf1/PD2 RNAi-treated J1 ESCs by Western blot. β-actin served as a loading control. (C): Transient knockdown of Paf1/PD2 protein (NIH3T3 cells) using specific RNAi for both 48 and 72 hours. Expression of Paf1/PD2 protein and other PAF complex proteins (Cdc73, Leo1, sKi8, and Ctr9) in Paf1/PD2 RNAi-treated NIH3T3 cells by Western blot. β-actin served as a loading control. Abbreviations: h, hour; PD2+/−, PD2 heterozygous knockout, Scr, Scrambled RNAi.
Paf1/PD2 Regulates Self-Renewal in Mouse ESCs
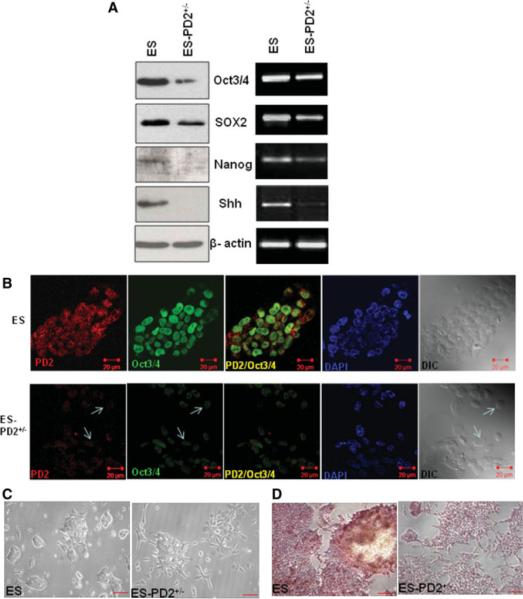
ESCs are pluripotent cells that have the potential for both indefinite self-renewal and differentiation into the three germ layers of the body [20]. Oct3/4, SOX2, and Nanog are considered a part of the core set of factors associated with the maintenance of pluripotency and self-renewal character in ESCs [21–23]. In our study, we have analyzed the level of the self-renewal markers Oct3/4, SOX2, and Nanog in PD2+/− ESCs and compared it with their expression in WT ESCs. Western blot and RT-PCR results for all these self-renewal proteins and mRNA showed a decreased level of expression in the PD2+/− ESCs compared with WT ESCs (Fig. 3A). The expression of sonic hedgehog (Shh), a marker of self-renewal property, was almost completely lost in the PD2+/− ESCs (Fig. 3A). Confocal analysis also showed a decreased expression of PD2 and Oct3/4 in the PD2+/− ESCs compared with the WT ESCs (Fig. 3B). To determine any changes in the cell morphology due to reduced Paf1/PD2 expression, cells were seeded at low density and observed under an optical microscope (×100 magnification). A vast majority of colonies in the PD2+/− ESCs appeared differentiated compared with the WT ESCs (Fig. 3C). To confirm this result, we performed alkaline phosphatase staining [24], which is indicative of the self-renewal propensity of ESCs. As expected, in the PD2+/− ESCs, almost 50% of the colonies did not stain with alkaline phosphatase, whereas all the WT ESC colonies were completely stained with alkaline phosphatase (×100 magnification) (Fig. 3D).
Figure 3.
Expression of self-renewal markers and morphological variation of Paf1/PD2 knockout and wild type ES cells. (A): Expression of Oct3/4, SOX2, Nanog, and Shh proteins and mRNA in Paf1/PD2 knockout ESCs and wild-type (WT) ESCs by Western blot and reverse-transcription polymerase chain reaction analysis. (B): Confocal analysis to show Paf1/PD2 and Oct3/4 to localization in WT ESCs and Paf1/PD2 knockout ESCs. The arrows indicate the decreased expression of PD2 and Oct3/4 in Paf1/PD2 knockout ESCs. (C): Phase-contrast images of control and Paf1/PD2 knockout ESCs (scale bar = 0.8 mm). (D): Alkaline phosphatase analysis of WT ESCs and Paf1/PD2 knockout ESCs (scale bar = 0.8 mm). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; PD2+/−, PD2 heterozygous knockout.
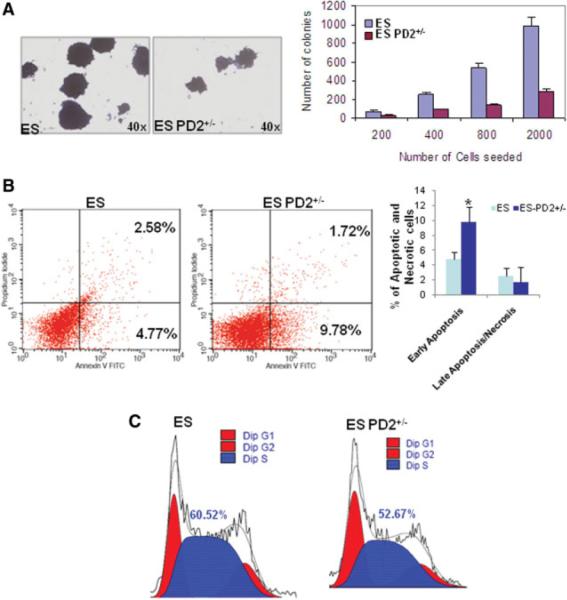
We have also analyzed the functional consequences of Paf1/PD2 heterozygous knockout in ESCs by assessing its effect on circular colony formation, cell cycle, and apoptosis. Colony-forming assay was performed to investigate the ESC potency of PD2+/−. The PD2+/− ESCs showed a substantial reduction in short-term colony formation compared with WT ESCs with a differential concentration of cells (×40 magnification) (Fig. 4A). Analysis of propidium iodide and annexin V-positive cells by flow cytometry indicated a significant (*, p = .005) increase in early apoptosis in PD2+/− ESCs compared with WT ESCs (Fig. 4B). Late apoptosis or necrosis did not show any significant variation in either cell line (Fig. 4B). A comparison of cell cycle distribution in PD2+/− and WT ESCs revealed a decreased S-phase population in the PD2 knockout ESCs (52.67%) compared with WT ESCs (60.52%) (Fig. 4C). These results indicate that Paf1/PD2 knockout causes a growth pattern of ESCs.
Figure 4.
Loss of Paf1/PD2 abolishes ESC functions. (A): Qualitative and quantitative analysis for short-term colony formation in WT and Paf1/PD2 knockout ESCs. (B): Annexin V and propidium iodide staining analysis by flow cytometry to identify apoptosis in WT ESCs and Paf1/PD2 knockout ESCs. The statistical analysis showed significant (*, p = .05) variation in early apoptosis but not in late apoptosis or necrosis in Paf1/PD2 knockout ESCs. (C): Cell cycle analysis of WT ESCs and Paf1/PD2 knockout ESCs by flow cytometry using propidium iodide. Abbreviations: FITC, fluorescein isothiocyanate; PD2+/−, PD2 heterozygous knockout.
To confirm the aforementioned results, we transiently knocked down Paf1/PD2 in J1 mouse embryonic stem cells (supporting information Fig. 1A). Similar to PD2+/− ESCs, the expression of self-renewal factors Oct3/4, SOX2, and Nanog was reduced in the Paf1/PD2 knockdown cells (supporting information Fig. 1A). An examination of the morphology of the Paf1/PD2 RNAi-treated cells showed features of differentiation compared with scrambled RNAi-treated cells, which retained their undifferentiated character (original magnification ×200) (supporting information Fig. 1B). These results confirm that Paf1/PD2 is essential for self-renewal of embryonic stem cells.
Paf1/PD2 Expression Decreases During the Differentiation of F9 Cells Along with Self-Renewal Markers
The F9 cells were treated with 9-cis retinoic acid (1 μM) for 5 days to investigate the effect of differentiation on the level of Paf1/PD2 expression. Retinoic acid (RA) treatment in vitro induces F9 EC cells to differentiate into the primitive endoderm layer [25]. The F9 EC cells and ESCs are developmentally pluripotent cells derived from murine embryos. In RA-treated F9 cells, a gradual change in cell morphology was observed, characterized by less tightly packed cells within the colonies and the generation of long filamentous structures compared with the untreated cells (original magnification ×100) (supporting information Fig. 2). Similarly, the J1 mESCs also showed the same morphological variation in RA-treated cells (supporting information Fig. 3). F9 cell differentiation by retinoic acid treatment led to decreased levels of Oct4 and Oct5 proteins [26]. In our study, we analyzed the expression of Paf1/PD2, the other PAF complex molecules, and self-renewal markers in both retinoic acid-treated and untreated F9 cells. As expected, expression of the Paf1/PD2 protein progressively decreased along with that of the self-renewal proteins Oct3/4, SOX2, and Nanog in the differentiating F9 cells following RA treatment (Fig. 5A). However, other PAF complex molecules, Cdc73, Leo1, Ski8, and Ctr9 proteins, did not show any change in either the differentiated or the undifferentiated F9 cells (Fig. 5A, 5B). Furthermore, J1 mESCs also confirmed the same observation (supporting information Fig. 4). The differentiation of EC cells and ESCs leads to the loss of Paf1/PD2 expression similarly like self-renewal markers. Furthermore, it appears that there is no coordinated expression of the PAF complex molecules during the differentiation of the primitive endoderm layer from EC cells. The retinoic acid-treated cells were also analyzed by confocal microscopy to look at the pattern of Paf1/PD2 and Oct3/4 expression. Paf1/PD2 (red) and Oct3/4 (green) expression gradually decreased in differentiating F9 EC cells (original magnification ×630) (supporting information Fig. 4).
Figure 5.
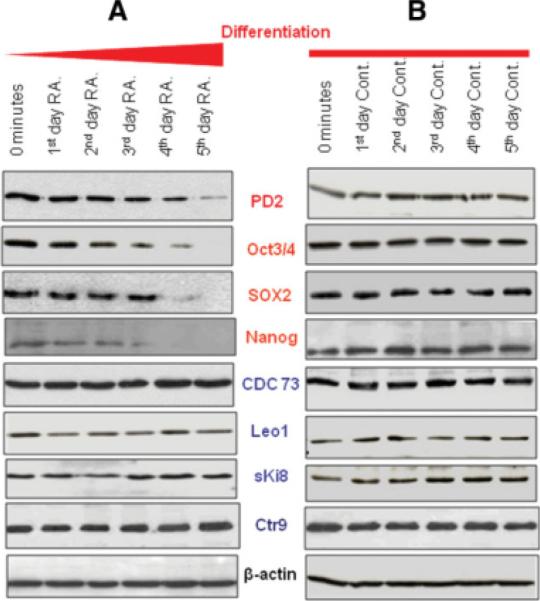
Expression of PAF complex and self-renewal proteins in retinoic acid-treated F9 embryonic carcinoma (EC) cells. (A): Expression of Paf1/PD2, Cdc73, Leo1, sKi8, and Ctr9, and self-renewal proteins Oct3/4, SOX2, and Nanog in differentiated F9 EC cells by Western blot analysis. (B): Western blot analysis showed the expression of Paf1/PD2, Cdc73, Leo1, sKi8, and Ctr9 and self-renewal proteins Oct3/4, SOX2, and Nanog in undifferentiated F9 EC cells. Abbreviations: Cont, control; RA, retinoic acid.
Paf1/PD2 Deficiency Results in Increased Expression of Lineage-Specific Markers in ESCs
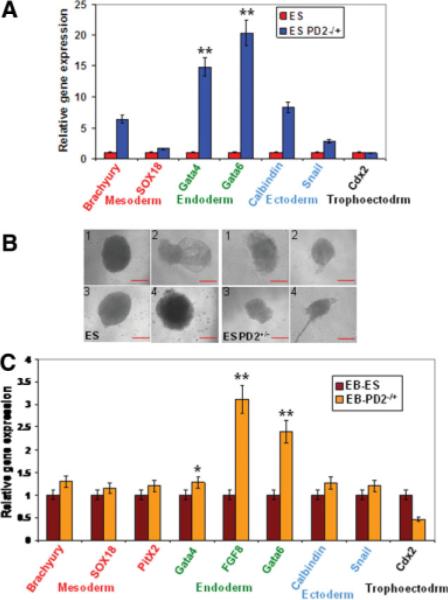
Cell fate decision of pluripotent embryonic stem cells is dictated by the activation and repression of specific sets of lineage-specific genes. To determine the lineages into which PD2+/− ESCs differentiate upon their exit from Paf1/PD2-mediated self-renewal, we performed quantitative RT-PCR (QRT-PCR) assays for lineage-specific genes in PD2+/− ESCs and WT ESCs growing under self-renewal conditions (in the presence of LIF). PD2+/− ESCs showed a significantly increased expression of endoderm (Gata4 [**, p < .0001] and Gata6 [**, p < .0001])-specific markers and little to no variation in mesoderm (Brachyury and Sox18)-, ectoderm (Calbindin and Snail)-, and trophoectoderm (Cdx2)-specific markers in comparison to WT ESCs (Fig. 6A). Expression of all of these differentiation markers was either absent or detectable at very low levels in the self-renewing WT ESCs (Fig. 6A). We carried out EB formation analysis to identify the Paf1/PD2 and its role in ESC potency. Embryoid bodies are aggregates of cells derived from ESCs and EB culture is used to examine the differentiation potential of the ESC line. PD2+/− ESCs and WT ESCs grew normally in short-term differentiation cultures without LIF. However, deficient EB formation was observed in the PD2+/− ESCs compared with WT ESCs (Fig. 6B). The differentiation of PD2+/− ESCs into multiple lineages while growing under self-renewal conditions prompted us to determine the differentiation characteristics of EB cells derived from wild-type ESCs. QRT-PCR of PD2+/− ESC line-derived EBs showed a significantly increased expression of endoderm (Gata4 [*, p = .01], Fgf8 [**, p < .0001], and Gata6 [**, p < .0001])-specific markers and little variation in mesoderm (Brachyury, PitX2, and Sox18)-, ectoderm (Calbindin and Snail)-, and trophoectoderm (Cdx2)-specific markers compared with WT ESC-derived EBs (Fig. 6C). Some of these markers were either absent or markedly reduced in WT ESC-derived EBs (Fig. 6C). Therefore, although EBs derived from both wild-type and PD2+/− ESCs started expressing multiple differentiation markers, the PD2+/− ESC-derived EB cells expressed a greater number of endoderm-specific lineage markers than the EB cells derived from WT mouse ESCs. This suggests that PD2+/− predisposes mouse ESCs to endodermal differentiation. Furthermore, we induced teratoma formation to analyze the differentiation potential of PD2+/− ESCs. This teratoma formation was to test whether the embryonic stem cells are pluripotent by allowing the cells to differentiate spontaneously in cell culture, manipulating the cells so they will differentiate to form ESCs characteristic of the three germ layers. Both ESCs and PD2+/− ESCs (1 × 106) were injected into the immunocompromised mice and teratomas were dissected after 16 days. The hematoxylin and eosin staining of WT ESC-derived teratoma sections showed mixture of many differentiated or partly differentiated cell types, an indication that the WT ESCs are capable of differentiating into multiple cell types (supporting information Fig. 5). In contrast PD2+/− ESC-derived teratoma showed defective germ layer formation (neural epithelium, skeletal muscle cells, and gut epithelium) compared with WT ESCs (supporting information Fig. 5).
Figure 6.
Paf1/PD2 affects lineage-specific genes in ESCs. (A): Analysis of lineage-specific markers for ectoderm (Calbindin and Snail), endoderm (Gata4 [**, p < .0001] and Gata6 [**, p < .0001]), mesoderm (Brachyury and Sox18), and trophoectoderm (Cdx2) layers in WT ESCs and PD2+/− ESCs by QRT-PCR. (B): Induced embryoid body (EB) formation in both WT ESCs and PD2+/− ESCs (scale bar = 0.8 mm). Defective and smaller embryoid body formation in Paf1/PD2 knockout ESCs compared with ESCs. (C): QRT-PCR analysis showed the expression of lineage-specific markers for ectoderm (Calbindin, Snail, and PitX2), endoderm (Gata4 [*, p = .01], Fgf8 [**, p < .0001], and Gata6 [**, p < .0001]), mesoderm (Brachyury and Sox18), and trophoectoderm (Cdx2) layers in embryoid body of WT ESCs and PD2+/− ESCs. Abbreviations: EB, embryoid body; PD2+/−, PD2 heterozygous knockout.
Paf1/PD2 Interacts with Oct3/4 and RNA Polymerase II in ESCs
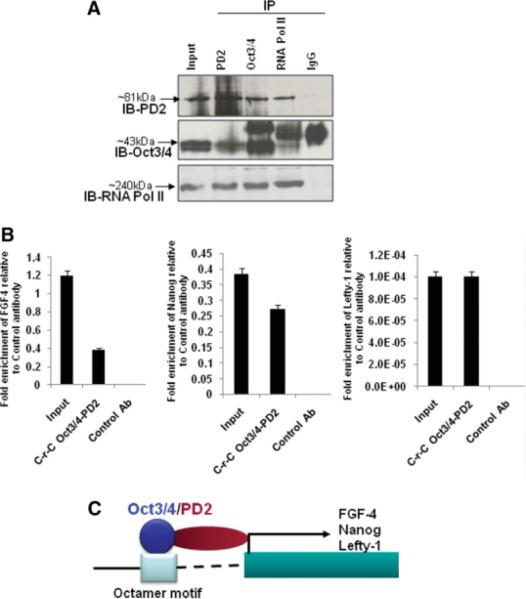
To investigate the molecular mechanism underlying the role of Paf1/PD2 in the self-renewal of embryonic stem cells, we looked for any protein-protein interaction between Paf1/PD2 and Oct3/4 in ESCs. Protein-protein interaction is of fundamental importance for transcriptional regulation [27–29]. We performed reciprocal immunoprecipitation studies, which revealed a clear interaction between Paf1/PD2, Oct3/4, and RNA Pol II in mouse ESCs (Fig. 7A). Our previous study showed that Paf1/PD2 is part of the transcription machinery, acting as a docking protein in between the complexes and is directly involved in transcription initiation and elongation [14]. Our current results show that Paf1/PD2 interacts directly with RNA Pol II as well as Oct3/4 in ESCs. These results suggest that Paf1/PD2 may be an interacting partner for Oct3/4 to regulate the transcription machinery of Oct3/4 in ESC maintenance.
Figure 7.
Mechanism of Paf1/PD2 in the maintenance of ESCs. (A): Reciprocal immunoprecipitation of Paf1/PD2, Oct3/4, and RNA Pol II in mouse ESCs. (B): The ChIP-Re-ChIP analysis for Oct3/4 and PD2 in embryonic stem cells. ChIP-Re-ChIP DNA was used to amplify FGF-4, Nanog, and Lefty-1 promoter fragment by SYBR Green QPCR analysis. The nonspecific primers were not amplified in any of the three genes. MUC4 antibody was used as a control antibody. The fold enrichment of FGF-4, Nanog, and Lefty-1 is related to MUC4 control antibody. (C): Mechanisms through which Oct3/4-PD2 interaction can regulate Oct3/4-mediated gene expression. A protein-protein interaction between Oct3/4 and Paf1/PD2 may have different possible implications through which Oct3/4-mediated gene expression can be regulated. An interaction between Oct3/4 with Paf1/PD2 may help to regulate expression of a subset Oct3/4 target genes such as FGF-4, Nanog, and Lefty-1 for the maintenance of ESCs. Abbreviations: Ab, antibody; C-r-C, sequential chromatin immunoprecipitations with two antibodies to study the simultaneous presence of two proteins in the genome sequence of interest; IB, immunoblot; IP, immunoprecipitation; Pol II, polymerase II.
Paf1/PD2 and Oct3/4 Regulate FGF-4, Nanog, and Lefty-1 in ESCs
ChIP-re-ChIP assay represents a direct strategy to determine the in vivo colocalization of proteins interacting with or in close contact to a chromatinized template on the basis of double and independent rounds of immunoprecipitations with high-quality ChIP-grade antibodies [18]. We have performed the ChIP-Re-ChIP assay to determine the role of Paf1/PD2 in ESC maintenance. Oct3/4 and PD2 ChIP-specific antibodies were used to pull down chromatin extracts of ESCs, and their binding to the promoter regions of FGF-4, Nanog, and Lefty-1 genes was examined by QPCR using promoter-specific primers. QPCR analysis showed a significant enrichment of FGF-4, Nanog, and Lefty-1 promoters in the Oct3/4-PD2 pull-down extracts relative to those pulled down with the control antibody (Fig. 7B). The nonspecific primers for FGF-4, Nanog, and Lefty-1 were not amplified in ChIP-Re-ChIP DNA. This result suggests that the complex of Oct3/4 and PD2 together regulates self-renewal genes such as FGF-4, Nanog, and Lefty-1. It is possible that Oct3/4 may interact indirectly with RNA pol II via Paf1/PD2, and thereby regulate the expression of a subset of self-renewal and pluripotency genes.
Discussion
A detailed molecular understanding of how specific factors influence the balance between pluripotency and differentiation of mammalian cells is crucial to develop ESCs for their potential therapeutic use. The importance of this fact is becoming increasingly evident as it is now possible to generate ESC-like cells from somatic cells via direct reprogramming [30–35], avoiding many ethical issues. Induced nuclear reprogramming through iPS cell technology is a pioneering achievement in this direction [33, 34]. The repercussions of iPS technology are vast: it provides a way to create patient-specific stem cells that bypasses ethical and technical issues surrounding human ESC derivation and somatic cell nuclear transfer [36, 37], a state-of-the-art model for studying genetic diseases in vitro [38, 39].
In this study, we have investigated the biological role of Paf1/PD2 in mouse ESCs. The human homolog of the yeast transcription elongation factor Paf1 was recently identified by our laboratory and others [13, 14, 19]. The hPaf1/PD2 is part of the human PAF complex along with hCdc73 (parafibromin), hCtr9, hLeo1, and Ski8 [13]. The human PAF complex lacks the Rtf1 protein, a component of the yeast core complex [13], and mouse Paf1 has 98% homology to the human equivalent. In our study, we found that Paf1/PD2 heterozygous knockout in ESCs affects the maintenance of embryonic stem cells, by downregulating factors such as Oct3/4 and SOX2. Oct3/4 and SOX2 are highly expressed in the early embryonic developmental stage for maintaining the pluripotency and self-renewal in embryonic stem cells [7–9]. Ronin, Zfx, REST, Sall4, and Klf5 are also involved in the maintenance of embryonic stem cells by different regulating mechanisms [40–44]. A recent study has shown that PAF complex proteins coordinate the maintenance of ESCs [45]. Our findings show, for the first time, that PAF complex proteins work individually (and not as a complex) for the maintenance of mouse embryonic stem cells.
Our previous report showed that Paf1/PD2 is overexpressed in poorly differentiated rather than in well-differentiated cells [14]. Similarly, Paf1/PD2 is overexpressed in undifferentiated mouse ESCs and EC cells compared with the well-differentiated cells. It is known that the hPAF complex coordinates events during transcription (initiation, promoter clearance, and elongation) and RNA quality control in HEK293F cells [13]. In contrast to the previous reports, we observed that the PAF complex molecules Cdc73, Leo1, and sKi8 did not show any coordinated expression in the Paf1/PD2 heterozygous knockout and knockdown ESCs. A recent study shows that knockdown of all PAF complex molecules is required for maintenance of ESC identity [45] but did not clarify whether these molecules work as a complex or independently. Interestingly, in our study we found that knockout of Paf1/PD2 does not affect the expression of other PAF complex molecules. This uncoordinated effect of PAF complex was observed only in ESCs and not in mouse fibroblast NIH3T3 cells [14]. These results suggest that coordinated expression of PAF complex molecules may play a role only in differentiated cells and not in the undifferentiated ESCs.
The PD2+/− ESCs and Paf1/PD2 siRNA-treated J1 ESCs showed a decreased expression of the self-renewal markers Oct3/4, SOX2, and Nanog compared with the WT ESCs. As expected, a reduced expression of Paf1/PD2 resulted in ESC differentiation as shown by morphological examination and alkaline phosphatase staining. These results strongly suggest that Paf1/PD2 plays an important role in the self-renewal of mouse ESCs. Other studies have reported that Ronin, Zfx, REST, Sall4, and Klf5 are also involved in the maintenance of embryonic stem cells by different regulating mechanisms [40–44]. We also found that Paf1/PD2 downregulation in ESCs drastically reduced the number of ESC colonies compared with WT ESCs. Similar phenotypes have also been observed after the inducible deletion of Sall4, Ronin, and Klf5 in ESCs [40, 41, 44]. furthermore, Paf1/PD2 knockout ESCs showed an increase in apoptosis, suggesting a defect in long-term self-renewal due to impaired survival. A previous report showed that deficiency of Zfx, an ESC maintaining gene, resulted in increased apoptosis in ESCs [42]. Paf1/PD2 deficiency also leads to a reduction in the percentage of ESCs in the S-phase, consistent with an exit of these cells from self renewal. This suggests a role for Paf1/PD2 in cell cycle regulation in ESCs. Consistent with this idea, a previous report showed that forced G1-to-S-phase cell cycle progression by Akt1 kinase overexpression induces a LIF-independent self-renewal in ESCs, suggesting a possible link between cell cycle progression and the maintenance of pluripotency [41].
An interesting observation in our study was the progressive loss of Paf1/PD2, but not other PAF complex molecules, upon differentiation of F9 EC cells (induced by RA). This pattern of decrease in Paf1/PD2 expression is similar to that observed for the self-renewal markers Oct3/4 and SOX2. It has been reported that RA treatment induces F9 cells to differentiate into the primitive endoderm layer [25]. Our result suggests that Paf1/PD2 alone may be involved in the differentiation of ESCs into an endodermal lineage. Interestingly, the endoderm differentiation markers showed increased expression in the embryoid body derived from PD2+/− ESCs. Based on these findings, we speculate that the absence of Paf1/PD2 leads to the induction of key endoderm genes, indicating a specific role of Paf1/PD2 in early commitment to an endodermal lineage. The role of other PAF complex molecules in ESC maintenance remains to be explored. furthermore, the development of teratomas with defective germ layers in PD2+/− ESCs suggests that deficiency of Paf1/PD2 leads to a loss of ESC potency.
Although transcriptional regulation of Oct3/4 gene expression by PAF complex proteins has been delineated [45], the other possible mechanisms through which Paf1/PD2 itself can regulate Oct3/4 protein function remained unexplored. Depending on the cellular context, Oct3/4 can either activate or repress target gene expression. To mediate its function, in addition to its binding to DNA, Oct3/4 interacts with other proteins to form homo- or heterodimers [20, 23, 26, 46–48]. Experimental evidence has shown that due to presence of two distinct DNA binding domains, Oct3/4 has the flexibility to interact with other transcription factors [49]. In the present study, through reciprocal coimmunoprecipitation, we were the first to show a protein-protein interaction between Oct3/4-PD2 and Oct3/4-RNA pol II. Oct3/4 may interact with Paf1/PD2 and RNA pol II individually or through a ternary complex, whereas Paf1/PD2 can act as a cross-linker between Oct3/4 and RNA pol II. Previous studies have shown that the PAF complex interacts with RNA pol II for a transcriptional process [13, 14]. Furthermore, ChIP-Re-ChIP result showed that Oct3/4 and PD2 interact together to regulate several key self-renewal genes including FGF-4, Nanog, and Lefty-1. Therefore, it is possible that Oct3/4 may interact indirectly with RNA pol II via Paf1/PD2, and regulate the expression of a subset of self-renewal and pluripotency genes. As mentioned previously, it is also equally possible that an independent interaction between Oct3/4 and Paf1/PD2 (without RNA pol II) may regulate expression of some genes, whereas Paf1/ PD2 will act as a coactivator/corepressor. The coactivator/corepressor activity of Paf1/PD2 may be due to its possible role in chromatin modification and/or the inhibition of interaction between Oct3/4 and other proteins. The specific domains involved in the interaction of Paf1/PD2 with Oct3/4 need to be explored in ESCs.
Conclusion
We have found that Paf1/PD2 can regulate the function of Oct3/4 partly by regulating its interaction with the Oct3/4 protein. Overall, our finding identifies Paf1/PD2 as a novel self-renewal protein that interacts with Oct3/4 and thereby helps in the maintenance of ESCs by regulating self-renewal genes such as FGF-4, Nanog, and Lefty-1 (Fig. 7C). Furthermore, a practical application of our findings could be the development of lineage-specific cell types from ESCs by knockdown of Paf1/PD2. This can be combined with other methods to induce ESC differentiation and thus improve the efficiency of generating specific cell types for experimental and therapeutic stem cell transplantation.
Supplementary Material
Acknowledgments
The authors in this research article were supported by grants from the National Institutes of Health (CA78590, CA131944, 133774) and the Department of Defense (BC061220). We thank Janice A. Taylor and James R. Talaska of the confocal laser scanning microscope core facility at the University of Nebraska Medical Center, Nebraska Research and the Eppley Cancer Center for their support of the core facility.
Footnotes
Disclosure of Potential Conflicts of Interest The authors indicate no potential conflicts of interest.
References
- 1.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 2.Mimeault M, Batra SK. Concise review: recent advances on the significance of stem cells in tissue regeneration and cancer therapies. Stem Cells. 2006;24:2319–2345. doi: 10.1634/stemcells.2006-0066. [DOI] [PubMed] [Google Scholar]
- 3.Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- 4.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 5.Moorthy PP, Kumar AA, Devaraj H. Expression of the Gas7 gene and Oct4 in embryonic stem cells of mice. Stem Cells Dev. 2005;14:664–670. doi: 10.1089/scd.2005.14.664. [DOI] [PubMed] [Google Scholar]
- 6.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 7.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;20(23):7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova N, Dobrin R, Lu R, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Mandal SS, Pham AD, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moniaux N, Nemos C, Schmied BM, et al. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25:3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Bowl MR, Bender S, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stryke D, Kawamoto M, Huang CC, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burridge PW, Anderson D, Priddle H, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 18.Furlan-Magaril M, Rincon-Arano H, Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–266. doi: 10.1007/978-1-60327-015-1_17. [DOI] [PubMed] [Google Scholar]
- 19.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 21.Avilion AA, Nicolis SK, Pevny LH, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 23.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 24.Pritsker M, Ford NR, Jenq HT, et al. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strickland S, Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15:393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- 26.Schöler HR, Balling R, Hatzopoulos AK, et al. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 1989;8:2551–2557. doi: 10.1002/j.1460-2075.1989.tb08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990;61:1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne M, Gann AA. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 29.Schöler HR, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 30.Esteban MA, Xu J, Yang J, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JB, Sebastiano V, Wu G, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Loh YH, Agarwal S, Park IH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei D. The magic continues for the iPS strategy. Cell Res. 2008;18:221–223. doi: 10.1038/cr.2008.21. [DOI] [PubMed] [Google Scholar]
- 34.Tweedell KS. New paths to pluripotent stem cells. Curr Stem Cell Res Ther. 2008;3:151–162. doi: 10.2174/157488808785740361. [DOI] [PubMed] [Google Scholar]
- 35.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson JA, Kalishman J, Golos TG, et al. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilmut I, Schnieke AE, McWhir J, et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 38.Ebert AD, Yu J, Rose FF, Jr., et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dejosez M, Krumenacker JS, Zitur LJ, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ema M, Mori D, Niwa H, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Galan-Caridad JM, Harel S, Arenzana TL, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;20(129):345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh SK, Kagalwala MN, Parker-Thornburg J, et al. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Tam WL, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 45.Ding L, Paszkowski-Rogacz M, Nitzsche A, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 47.Saxe JP, Tomilin A, Scholer HR, et al. Post-translational regulation of Oct4 transcriptional activity. Plos One. 2009;4:4467. doi: 10.1371/journal.pone.0004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolkunova E, Malashicheva A, Parfenov VN, et al. PIAS proteins as repressors of Oct4 function. J Mol Biol. 2007;374:1200–1212. doi: 10.1016/j.jmb.2007.09.081. [DOI] [PubMed] [Google Scholar]
- 49.Reményi A, Lins K, Nissen LJ, et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.