Abstract
Background & Aims
Although nonalcoholic fatty liver disease (NAFLD) is increasingly common, only a minority of affected individuals develop fibrotic liver disease. Based on its role in liver growth and repair, we explored whether Kruppel-like factor 6 (KLF6) plays a role in NAFLD progression.
Methods
KLF6 expression in 31 fibrosis scored NAFLD liver biopsy specimens was assessed by real-time polymerase chain reaction. Transfected minigene constructs were used to study the effect of a polymorphism, KLF6-IVS1-27G>A, that promotes KLF6 alternative splicing in vitro. We genotyped KLF6-IVS1-27G>A in 3 groups of patients (UK group 1, n = 306; Italian group 2, n = 109; trio group 3, n = 61 children and parents).
Results
KLF6 expression was increased in association with increased steatosis, inflammation, and fibrosis in NAFLD livers. KLF6-IVS1-27G>A promoted alternative splicing of KLF6 and abrogated the up-regulation of both α-smooth muscle actin and collagen 1 in LX-2 cells. Group 1 genotyping identified KLF6-IVS1-27G>A in 44 of 306 (14.4%) patients. Notably, KLF6-IVS1-27G>A was associated significantly with milder NAFLD, with only 25% having more advanced fibrosis compared with 45% of wild-type (wt) individuals. This trend was confirmed in group 2. A linear regression analysis including all 415 patients, adjusted for age, sex, body mass index, and blood glucose level, confirmed that presence of the wt KLF6 allele was an independent predictor of fibrotic NAFLD. Furthermore, we have shown preferential transmission of the wt allele to children with fibrotic NAFLD.
Conclusions
We report a functional polymorphism in the KLF6 gene associated with advanced NAFLD and believe further study of KLF6 may enhance our understanding of this disease process.
Nonalcoholic fatty liver disease (NAFLD) is the liver manifestation of the metabolic syndrome, characterized by central obesity, atherogenic dyslipidemia, hypertension, and insulin resistance. NAFLD encompasses a spectrum of liver diseases from simple fatty liver to cirrhosis. The prevalence of NAFLD is growing rapidly and it is now the most common cause of chronic liver disease in Western countries.1 The cardiovascular health implications of the metabolic syndrome are significant, with the risk of progressive liver disease, cirrhosis, and liver cancer set to have a further major impact.2 Despite its high prevalence, however, less than a quarter of subjects with NAFLD ever progress beyond steatosis to significant fibrosis and liver cancer.3–5 The reasons for these differences in individual susceptibility to progressive disease are unclear, but family/ethnic studies suggest that genetic factors play a significant role.6,7 Although several candidate genes have been studied, as yet, no genetic associations with advanced NAFLD have been replicated in large studies.8
Kruppel-like factor 6 (KLF6) belongs to the Kruppel-like family of transcription factors known to play diverse roles in differentiation, development, cell growth, apoptosis, and angiogenesis.9 Similar to other members of the family, KLF6 has 3 contiguous C2H2 zinc fingers at the carboxyl-terminal domain and recognizes GC box motifs or CACCC motifs in responsive promoters.10 Although expression of some KLF transcription factors is tissue-specific, KLF6 is expressed ubiquitously.11 It was identified as an early gene expressed in activated hepatic stellate cells (HSCs) after liver injury,12,13 raising the possibility that it may be involved in the process of liver fibrogenesis. Furthermore, KLF6 transactivates several genes critical for the development of liver fibrosis, including collagen 1, transforming growth factor-β1, and types I and II transforming growth factor-β receptors in HSC.13,14 The recent demonstration of increased expression of KLF6 in response to oxidative stress in a methionine-choline– deficient diet model of nonalcoholic steatohepatitis, linked to increased transforming growth factor-β1 expression, provides support for a role of KLF6 in NAFLD.15 KLF6 also has been proposed as a tumor-suppressor gene, being located on chromosome 10p15 and frequently deleted in a number of cancers including prostate16 and hepatocellular cancer (HCC).17 Its expression is reduced in the majority of hepatitis B and C virus–associated HCCs.18 Moreover, retrovirally mediated over expression of KLF6 in HCC HepG2 cells is associated with reduced proliferation and increased differentiation,18 suggesting a key role in the regulation of hepatocyte growth.
A functional single nucleotide polymorphism (SNP) in the KLF6 gene has been identified recently.19 KLF6–IVS1-27G>A (rs3750861; NCBI Entrez SNP database, available at: http://www.ncbi.nlm.nih.gov/entrez) is a KLF6 SNP located within the first intron of the gene and is associated with an increased incidence of familial prostate cancer.19 Further characterization of the SNP showed that it created a novel binding site for the splicing factor, SRp40.20 Furthermore, the presence of the SNP in vitro in prostate cancer cells promoted alternative splicing of the KLF6 gene into dominant-negative alternative isoforms that fail to up-regulate p21, facilitating enhanced proliferation. Alternative splicing of KLF6 has now been accepted as an additional means of inactivation of its wild-type (wt) tumor-suppressor function in prostate20 and ovarian cancers,21 as well as in glioblastomas.22
Based on the suspected roles of KLF6 in hepatic fibrosis associated with oxidative stress, as well as in hepatocyte proliferation and differentiation, we have investigated and shown increased levels of KLF6 in liver biopsies of patients with advanced vs earlier stages of NAFLD. We also have shown that the KLF6-IVS1-27G>A SNP promotes splicing of KLF6 to its alternative forms in both HCC and HSC cell lines. In the latter, we have confirmed an increase in both α-smooth muscle actin (αSMA) and collagen I production stimulated by KLF6 wt, attenuated in the presence of SNP-associated alternative splice forms. We have identified wt KLF6 as an independent predictor of the presence of fibrosis in 2 independent cohorts of NAFLD patients, with those with the variant allele having less advanced fibrosis. Finally, we also have shown preferential transmission of the wt allele to children with fibrotic NAFLD using transmission disequilibrium testing in a family study. We believe KLF6 to be a strong candidate for further studies in this increasingly common disease.
Patients and Methods
Patients
We enrolled 415 patients with biopsy-proven NAFLD at different stages of disease from Newcastle and Turin hospitals according to the regulations and ethical requirements of the participating centers. All patients had clinical features and liver biopsies diagnostic of NAFLD. Women and men consuming greater than 14 or 21 units of alcohol per week, respectively, were excluded, as were any individuals with viral or autoimmune liver diseases. Clinical and laboratory data were collected on the date that a diagnostic liver biopsy was performed. Body mass index (BMI) was calculated using the formula: weight (kg)/height (m)2. The presence of diabetes mellitus (fasting glucose level ≥ 7.1 mmol/L or treatment with antidiabetic drugs) and hypertension (blood pressure ≥ 130/85 mm Hg or current treatment for hypertension) were recorded. Laboratory evaluation included liver biochemistry, blood count, total and high-density lipoprotein cholesterol and total triglyceride level, fasting glucose level, fasting insulin level, viral serology, and autoantibodies. The degree of insulin resistance was determined by the homeostatic model assessment.23 Patients included 306 northeast UK patients (group 1), and 109 Italian patients (group 2). The confirmation of results in 2 independent European NAFLD populations reduces the risk that any association observed in the first group was a chance finding subject to a type I error.
In addition, we assessed 71 Italian family trios with both parents alive and an index child with biopsy-proven NAFLD. Our particular interest was in those with fibrotic disease, of which there were 61 (stage 1, n = 59; stage 2, n = 2). We used transmission disequilibrium testing24 to look for preferential transmission of either KLF6 alleles to the affected children. This approach is not subject to the potential confounding effects inherent in case-control studies and is significantly more powerful at detecting true associations.
Liver Biopsy
Ultrasound-guided liver biopsy was performed and read by a single liver pathologist in each participating center. The severity of steatosis, necroinflammatory grade, and stage of fibrosis were scored according to modified Brunt et al25 criteria. Liver tissue from 31 patients undergoing liver biopsy for suspected NAFLD also had a 0.5- to 1-cm core of tissue stored in RNA later solution (Ambion, Warrington, UK).
RNA Extraction From Liver Tissues
Messenger RNA (mRNA) was isolated from liver tissue using TRIzol reagent (Invitrogen, Paisley, UK) according to the manufacturer’s instructions with a minor modification in the precipitation step: the aqueous phase with isopropanol, 5 mg/mL glycogen (Ambion), and a high-salt solution (1.2 mol/L NaCl, 0.8 mol/L Na-citrate) was incubated at −20°C for 1 hour before washing and resuspension in water. Complementary DNA (cDNA) (1 μg) was synthesized with Reverse Transcription System (Promega, Southampton, UK).
Real-Time Polymerase Chain Reaction Analyses
Semiquantitative reverse-transcription polymerase chain reaction (RT-PCR) assays were performed with either TaqMan (KLF6 quantification) or SYBR Green (KLF6, αSMA, collagen 1) technology on a 7900HT sequence detection system (Applied Biosystems, Warrington, UK). The SYBR Green primers used to compare the wt KLF6 mRNA (KLF6 wt) with all isoforms of KLF6 (KLF6 Total) were as previously described.19 TaqMan semiquantitative PCR was similarly performed using the following primer and probe sets: KLF6 Total forward – 5′ CGGACGCACACAGGAGAAAA 3′, reverse – 5′ GGTTAACTCATCACTTCTTGCAAA 3′, probe – 5′ AGGGTGTGAGTGGCGT 3′; KLF6 wt forward – 5′ AATTTGACAGCCAGGAAGATCTG 3′, reverse – 5′ CAG-TTCGGATTCCTCCTTTTTC 3′, probe – 5′ ACCAAAAT-CATTCTGGCTCGGG 3′; glyceraldehyde-3-phosphate dehydrogenase forward – 5′ GCACCGTCAAGGCTGAGAA 3′, reverse 5′ AGCATCGCCCCACTTGATT 3′, and probe 5′ CATCTTCCAGGAGCGAGAT 3′. The TaqMan master mix was used according to the manufacturer’s instructions (Applied Biosystems). cDNA (1 μL) was used per well in a total volume of 10 μL with standard cycling parameters (95°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C for 15 sec, 60°C for 1 min). All data were presented as relative quantity (RQ) values ± minimum/maximum for the relative amounts of target gene, normalized to glyceraldehyde-3-phosphate dehydrogenase, and relative to expression values in a chosen comparator sample. In the liver biopsy tissue analyses, the comparator sample served only as a reference for standardization of all other samples and was not included in any subsequent analyses. It was created from equal quantities of cDNA from 4 samples with previously determined low expression of KLF6, including one histologically normal sample and 3 samples with simple steatosis. The RQ of each target gene was determined from replicate samples analyzed on 3 separate occasions using the formula 2−ΔΔCT. Data generated were analyzed using SDS 2.2 (Applied Biosystems).
Cell Culture
HCC cell lines were obtained from the American Tissue Culture Collection, and the LX-2 cells were created and characterized as previously described.26 Cells were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum with 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 2 mmol/L L-glutamine (Sigma-Aldrich, Gillingham, Dorset, UK). Media was changed every 48 hours. Cells were passaged 1–2 times per week. Cells were seeded 24 hours before transient transfection experiments. LX-2 cells were maintained similarly, but in 1% serum.
Transient transfections of HCC cell lines were performed using 4 ng DNA and Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen). For analyses in LX-2 cells, which have a very low transient transfection efficiency, cell lines stably expressing minigene constructs were created and harvested at 1–2 weeks. LX-2 cells (2 × 105) were seeded into each well of a 6-well plate and grown overnight. Transfection was with Effectene reagent (Qiagen, Crawley, UK). A total of 0.5 μg of each minigene construct was combined with 0.5 μg of pCI-Neo and incubated in enhancer solution for 5 minutes at room temperature. The Effectene reagent was added for a further 10 minutes before adding to the cells in 1 mL of normal growth medium. The medium was replaced the following day and after 48 hours G418 was added at a concentration of 600 μg/mL to select for transfected cells.
Genotyping
Blood samples (5 mL) were collected from each subject, and DNA was extracted by Tissue DNeasyTM (Qiagen) according to the manufacturer’s instructions. The intron 1/exon 2 region of KLF6 was amplified as previously described.19 Successful DNA amplification was determined by the visualization of a 290-bp fragment on 1% agarose gel electrophoresis. The presence of the KLF6-IVS1-27G>A SNP (rs37508611) was determined by BsaAI (New England BioLabs Inc, Hitchin, Herts, UK) digestion, which cuts wt DNA at the recognition sequence CGCG at position 84 of the amplicon. The detection of 3 bands indicated heterozygosity. Positive and negative controls to confirm complete digestion were used on every gel. Every positive sample was confirmed by an independent PCR and restriction analysis. Each of these independent PCR reactions included random wt samples, similarly re-analyzed, to confirm a lack of cross-contamination.
Statistical Analysis
The data were analyzed using SPSS 14 (Statistical Package for Social Sciences, Chicago, IL) licensed to New-castle University. Comparisons of parametric data were analyzed using the Student t test or analysis of variance, whereas nonparametric data analyses were performed by the Mann–Whitney test for quantitative and the Pearson chi-square test for categoric variables. Comparisons of KLF6 expression associated with different stages of histologically scored disease were performed with the Kruskal–Wallis test. A P value of less than .05 was considered significant.
The main aim of this study was to evaluate the possible role of the polymorphism of KLF6 in predicting patients with progressive NAFLD. By using the web-based calculator available at www.tufts.edu/, we confirmed that the KLF6 polymorphism was in Hardy–Weinberg equilibrium in each of the British and Italian groups studied. The minimum sample size required to analyze wt KLF6 being a risk factor for fibrosis in NAFLD was calculated by QUANTO Software (http://hydra.usc.edu/gxe) as detailed in the supplementary methods (see supplementary material online at www.gastrojournal.org). We considered the presence of stage 2 fibrosis as indicative of progressive disease because stage 1 fibrosis in adults often does not, or can take many years to, progress. Logistic regression was used to determine the independent nature of a number of variables, including age, BMI, blood glucose level, platelet count, aspartate aminotransferase (AST)/alanine aminotransferase (ALT) ratio, and KLF6 genotype, on the presence of stage 2 or more of fibrosis. The Forward Stepwise Wald method was used. The goodness of fit of the model was assessed by the Hosmer–Lemeshow test for significance. Results are shown as odds ratios and 95% confidence intervals (Table 6).
Table 6.
KLF6 Genotype Is a Predictor of Fibrosis Stage in Patients With NAFLD: Binary Logistic Regression Analysis Presenting Factors Independently Associated With Advanced Fibrosis
| Groups 1 and 2 (n = 415) | Odds ratio | 95% Confidence interval | P |
|---|---|---|---|
| Age at biopsy, y | 1.03 | 1.00–1.05 | .042 |
| BMI, kg/m2 | 1.07 | 1.03–1.12 | .002 |
| Blood glucose level | 1.12 | 1.02–1.22 | .015 |
| Platelets, ×109 L | 0.99 | 0.99–1.00 | .000 |
| AST/ALT ratio | 2.88 | 1.12–7.40 | .028 |
| KLF wt | 2.77 | 1.30–5.91 | .009 |
The transmission disequilibrium test24 was used to look for intrafamilial allelic associations in the family study. The chi-square statistic (using R and UNPHASED, freeware software, http://www.mrc-bsu.cam.ac.uk/personal/frank/software/) was used to compare the observed number if parent-offspring transmission of the KLF6-IVSI-27G>A allele with the number of transmissions expected by chance. A P value of less than .05 was considered statistically significant.
Results
KLF6 Expression Is Increased in Association With More Advanced Stages of NAFLD
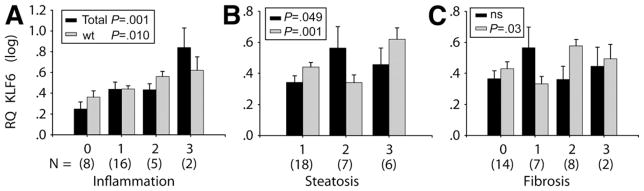
KLF6 is a ubiquitous transcription factor and immediate early gene induced in response to stressful stimuli. We have compared its expression at the mRNA level using 2 different sets of TaqMan real-time primers and probe, one of which quantifies all isoforms of KLF6 (KLF6 Total) and one of which quantifies the full-length, wild-type version only (KLF6 wt). Both are expressed as an RQ value, relative to the same control gene (glyceral-dehyde-3-phosphate dehydrogenase) and the same comparator sample. The difference between the 2 primer sets can be attributed to alternatively spliced KLF6 mRNA. Relative levels of Total KLF6, wt KLF6, and alternatively spliced KLF6 are summarized in Table 1. The fat, necroinflammatory, and fibrosis scores for each sample also are included. The levels of KLF6 expression subsequently were analyzed with respect to histologic scoring and there was clearly a significant increase in KLF6 expression in association with more advanced histologic disease, as shown in Figure 1. The expression of the full-length isoform, KLF6 wt, was increased significantly with more advanced stages of steatosis, inflammation, and fibrosis. The data for Total KLF6 isoforms were similar, but with some notable differences. Although a significant increase was seen in association with increasing inflammatory score, there were no significant differences in Total KLF6 isoforms relative to different stages of steatosis or fibrosis. These data suggest that although all isoforms of KLF6 are expressed in response to liver injury and ongoing inflammation, it is the predominance of the full-length wt isoform that is associated with more advanced fibrotic stages of NAFLD.
Table 1.
KLF6 in NAFLD Liver Biopsies
| Brunt et al25 score |
RQ total KLF6 | RQ wt KLF6 | |||
|---|---|---|---|---|---|
| Fat | N/I | Fib | |||
| 1 | 1 | 1 | 2 | 3.33 | 3.49 |
| 2 | 1 | 2 | 2 | 2.76 | 4.68 |
| 3 | 1 | 1 | 0 | 2.34 | 3.16 |
| 4 | 1 | 2 | 0 | 1.85 | 1.98 |
| 5 | 1 | 1 | 0 | 3.89 | 4.82 |
| 6 | 1 | 2 | 1 | 2.68 | 3.44 |
| 7 | 1 | 2 | 0 | 3.19 | 5.43 |
| 8a | 1 | 2 | 0 | 3.04 | 3.38 |
| 9 | 1 | 1 | 0 | 2.53 | 4.33 |
| 10 | 1 | 3 | 3 | 3.38 | 2.23 |
| 11 | 1 | 0 | 2 | 2.42 | 2.83 |
| 12 | 1 | 0 | 0 | 0.87 | 1.66 |
| 13 | 1 | 0 | 0 | 0.87 | 1.36 |
| 14 | 1 | 0 | 0 | 4.38 | 1.20 |
| 15a | 3 | 3 | 2 | 14.65 | 7.80 |
| 16a | 3 | 1 | 1 | 1.31 | 1.25 |
| 17 | 2 | 0 | 0 | 1.55 | 2.02 |
| 18 | 3 | 0 | 0 | 5.68 | 8.15 |
| 19 | 2 | 1 | 0 | 6.78 | 1.52 |
| 20 | 1 | 0 | 0 | 1.11 | 1.61 |
| 21 | 1 | 1 | 1 | 5.74 | 2.57 |
| 22 | 3 | 0 | 0 | 1.90 | 3.15 |
| 23 | 1 | 1 | 2 | 1.07 | 2.72 |
| 24 | 3 | 1 | 2 | 1.56 | 3.60 |
| 25 | 3 | 1 | 2 | 1.76 | 5.53 |
| 26 | 2 | 1 | 1 | 0.92 | 3.14 |
| 27 | 2 | 1 | 1 | 11.53 | 2.40 |
| 28 | 2 | 1 | 3 | 2.25 | 4.35 |
| 29 | 2 | 1 | 1 | 38.67 | 1.10 |
| 30 | 2 | 1 | 2 | 0.98 | 2.14 |
| 31 | 1 | 1 | 1 | 1.08 | 2.35 |
Fib, fibrosis; N/I, necroinflammation.
Denotes individuals heterozygous for KLF6 IVS1-27G>A.
Figure 1.
Increased wt KLF6 expression with advanced stages of NAFLD. Either Total KLF6 mRNA isoforms (represented by black bars) or the wt isoform only (grey bars) have been quantified by semiquantitative TaqMan RT-PCR. The individual Brunt et al25 histology scores and KLF6 RQ values are shown in Table 1, whereas the mean expression levels (RQ log) ± standard error within histologically defined grades of NAFLD are shown in this Figure (n = 31; P values are shown). (A) Stepwise increase in both Total KLF6 isoforms and wt KLF6 increased stepwise in association with increasing levels of inflammation (A). Although wt KLF6 also was increased significantly in association with advanced steatosis (B) and fibrosis (C), Total KLF6 isoforms were not. (A) ■, Total, P = .001;  , wt, P = .010; (B) ■, P = .049;
, wt, P = .010; (B) ■, P = .049;  , P = .001; (C) ■, NS;
, P = .001; (C) ■, NS;  , P = .03.
, P = .03.
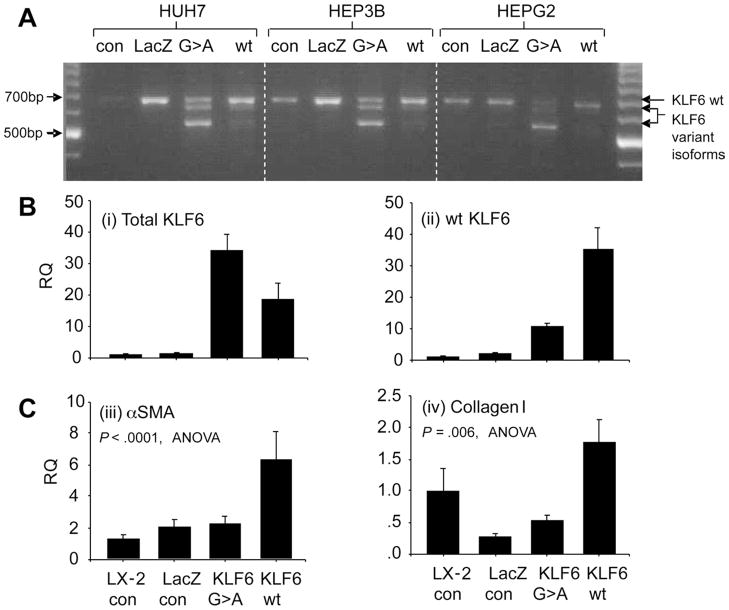
KLF6 IVS1-27G>A Enhances Splicing in Liver Cell Lines and Abrogates the KLF6 wt-Induced Transcriptional Increase in αSMA and Type 1 Collagen mRNA
By using transfected control LacZ-expressing and KLF6-expressing minigene constructs, the latter either with or without the KLF6 SNP, we showed that KLF6 alternative splicing to its smaller isoforms occurs readily in the presence of the SNP in a liver cell environment. Cell lines studied included the HepG2, Huh7, and Hep3B liver cancer cell lines, as shown in Figure 2A, and the human HSC cell line, LX-2, shown in Figure 2B. Both αSMA and type 1 collagen are markers of HSC activation, the latter being a known transcriptional target of wt KLF6. As shown in Figure 2C, both were increased at the mRNA level in LX-2 cells transformed with the KLF6 wt minigene. The increases in αSMA and collagen 1 expression were attenuated in the LX-2 cells expressing the KLF6 IVS1-27G>A minigene, producing increased levels of KLF6 splice forms.
Figure 2.
KLF6 IVS1-27G>A promotes alternative splicing of KLF6 and abrogates the up-regulation of αSMA and collagen 1. Cells were transfected with either transfection reagent alone (CON), a control minigene expressing LacZ (LacZ), a KLF6 minigene with the IVS1-27G>A SNP (G>A), or a KLF6 wt minigene construct (wt). (A) Agarose gel electrophoresis of cDNA products amplified using 5′ and 3′ flanking primers detecting all KLF6 isoforms identified is shown. KLF6 in the wt minigene transfected cells was predominantly the full-length wt isoform, whereas in the cells transfected with the KLF6 IVS1-27G>A minigene there were additional smaller bands representing alternative splice forms. Relative quantities of these isoforms were confirmed by semiquantitative RT-PCR (n = 3, data not shown). (B) Semiquantitative RT-PCR data from RNA extracted from stably transfected LX-2 cells harvested at 1 week. Total KLF6 was increased in cells transfected with either of the KLF6 minigene constructs, whereas only a relatively modest amount of this total KLF6 was the wt isoform in the G>A–transfected cells. (C) The marked increase in wt KLF6 in the LX-2 cells transfected with the wt KLF6 minigene was associated with significant increases in both αSMA and collagen I mRNA, which was not present in those cells transfected with the G>A SNP containing minigene-expressing dominant-negative KLF6 isoforms in addition to the wt isoform.
KLF6 IVS1-27G>A Is Associated With Reduced Fibrosis in NAFLD
To determine whether the KLF6 IVS1-27G>A SNP plays a role in susceptibility to NAFLD-related liver fibrosis, we examined whether the presence of the SNP was associated with histologic parameters in a large cohort of patients with NAFLD. The combined study populations comprised a total of 415 Caucasian European patients (group 1, 306 patients from Newcastle, UK; and group 2, 109 patients from Turin, Italy). Clinical details are reported in Table 2. Although there were some biologically relevant differences (eg, age, BMI, diabetes), and others that may reflect minor differences attributable to variation in laboratory, age, and sex normal ranges (alkaline phosphatase, bilirubin, and cholesterol), there was no difference in the prevalence of liver fibrosis, the homeostatic model assessment score reflecting insulin resistance, nor in the frequency of the KLF6 IVS1-27G>A SNP between the UK (14.4%) and Italian (13.8%) cohorts. The latter was similar to the KLF6 IVS1-27G>A SNP frequency in an independent group of Europeans (data available at NCBI Entrez SNP database: http://www.ncbi.nlm.nih.gov/entrez; rs3750861; heterozygous genotype incidence 15%, as reported by the International HAPMAP Project).
Table 2.
Clinical Features of Patients With NAFLD
| All | Group 1 | Group 2 | P | |
|---|---|---|---|---|
| Number | 415 | 306 | 109 | |
| Country/ethnicity | Caucasian | UK Caucasian | Italy Caucasian | |
| KLF6 IVS1-27G>A | 59 (14.2%) | 44 (14.4%) | 15 (13.8%) | .874 |
| Sex (male) | 290 (66.8%) | 193 (63%) | 87 (81%) | .001 |
| Age, y | 48.14 ± 12.66 | 49.56 ± 13.14 | 44.16 ± 10.27 | .000 |
| BMI, kg/m2 | 32.21 ± 6.25 | 34.10 ± 5.45 | 26.97 ± 5.29 | .000 |
| Hypertension | 165/379 (43%) | 134/272 (49%) | 31/107 (29%) | .000 |
| Diabetes | 124/406 (31%) | 116/299 (39%) | 8/107 (7.5%) | .000 |
| Platelets, ×109/L | 235 ± 72 | 239 ± 79 | 217 ± 63 | .010 |
| Total bilirubin level, μmol/L | 13.3 ± 10.1 | 12.44 ± 10.2 | 16.01 ± 8.90 | .000 |
| AST level, IU/L | 49 ± 33 | 51 ± 35 | 42 ± 26 | .001 |
| ALT level, IU/L | 78 ± 60 | 78 ± 63 | 77 ± 49 | .208 |
| AST/ALT ratio | 0.73 ± 0.36 | 0.78 ± 0.4 | 0.59 ± 0.19 | .000 |
| GGT, IU/L | 116 ± 167 | 125 ± 188 | 91 ± 88 | .016 |
| Alkaline phosphatase level, IU/L | 96 ± 44 | 99 ± 45 | 85 ± 41 | .000 |
| Fasting glucose level, mmol/L | 6.3 ± 2.9 | 6.67 ± 3.26 | 5.35 ± 1.19 | .000 |
| Fasting insulin level, mU/L | 22.8 ± 28.9 | 24.12 ± 20.89 | 20.67 ± 14.27 | .731 |
| HOMA score | 6.32 ± 2.92 | 6.51 ± 6.3 | 5.00 ± 3.99 | .259 |
| Total cholesterol, mmol/L | 5.51 ± 1.32 | 5.61 ± 1.37 | 5.24 ± 1.15 | .018 |
| HDL cholesterol, mmol/L | 1.24 ± 0.36 | 1.21 ± 0.35 | 1.32 ± 0.38 | .018 |
| Triglyceride level, mmol/L | 2.41 ± 1.61 | 2.71 ± 1.71 | 1.59 ± 0.91 | .000 |
| Fibrosis score | .263 | |||
| 0 | 156 (38%) | 119 (39%) | 37 (34%) | |
| 1 | 75 (18%) | 59 (19%) | 16 (15%) | |
| 2 | 71 (17%) | 49 (16%) | 22 (20%) | |
| 3 | 68 (16%) | 43 (14%) | 25 (23%) | |
| 4 | 45 (11%) | 36 (12%) | 9 (8%) |
The table shows the mean ± SD for continuous variables, number (%) for binary variables, and number (%) per group for categoric variables. GGT, γ-glutamyltransferase; HDL, high-density lipoprotein; HOMA, homeostatic model assessment.
Both groups of patient data subsequently were analyzed in relation to the presence or absence of KLF6 IVS1-27G>A. No significant differences were identified in a range of clinical and laboratory variables between those with and without the SNP. Data for the combined group are shown in Table 3. This was not the case, however, for the histologic fibrosis score, in which significant differences between the wt and heterozygous individuals were present. In group 1 (UK NAFLD patients), the KLF6 IVS1-27G>A heterozygotes had significantly less fibrosis (75% with fibrosis stage 0 or 1) compared with wt individuals (only 55% staged 0 or 1), as shown in Table 4. Those with fibrosis of stage 2 or more accounted for 45% of wt KLF6 individuals and only 25% of heterozygous patients. This difference was statistically significant (P = .015, Pearson chi-square test). The findings in group 2 (Italian NAFLD patients) were very similar, with 67% of heterozygous patients with stage 0 or 1 fibrosis compared with only 46% of wt individuals. This difference did not reach statistical significance (P = .134) owing to the smaller number of patients. In combination with group 1, however, the statistical significance for the protective effect of KLF6 IVS1-27G>A (73% with stage 0 or 1 fibrosis, compared with only 53% of wt KLF6 individuals) was enhanced (P = .004).
Table 3.
KLF6 Genotype Is Associated With the Fibrosis Score in NAFLD Patients: Clinical and Demographic Variables in the Combined Groups
| KLF6 IVS1-27G>A | KLF6 wt | P | |
|---|---|---|---|
| Number | 59 | 356 | |
| Sex (male) | 41 (69.5%) | 239 (67%) | .721 |
| Age, y | 47.76 ± 12.04 | 48.21 ± 12.78 | .729 |
| BMI, kg/m2 | 32.28 ± 5.45 | 32.20 ± 6.38 | .718 |
| Glucose level, mmol/L | 5.95 ± 2.16 | 6.38 ± 3.03 | .719 |
| Insulin level | 27.5 ± 21.2 | 22.0 ± 18.2 | .127 |
| Triglyceride level, mmol/L | 2.37 ± 1.47 | 2.42 ± 1.64 | .719 |
| Cholesterol level, mmol/L | 5.63 ± 1.54 | 5.49 ± 1.28 | .804 |
| HDL level, mmol/L | 1.21 ± 0.36 | 1.24 ± 0.36 | .702 |
| Total bilirubin level, μmol/L | 12.79 ± 8.07 | 13.36 ± 10.38 | .545 |
| ALT level | 77 ± 49 | 78 ± 61 | .520 |
| AST level | 53 ± 40 | 48 ± 32 | .656 |
| AST/ALT ratio | 0.71 ± 0.33 | 0.73 ± 0.37 | .624 |
| GGT level | 96 ± 94 | 120 ± 177 | .668 |
| Platelets, ×109 L | 239 ± 55 | 234 ± 74 | .680 |
| Hypertension | 27/53 (51%) | 138/326 (42%) | .241 |
| Diabetes | 19/58 (33%) | 105/348 (30%) | .693 |
| Fat score | .432 | ||
| 1 | 20 (34%) | 131 (38%) | |
| 2 | 30 (52%) | 119 (34%) | |
| 3 | 8 (14%) | 97 (28%) | |
| Inflammation | 38/58 (65%) | 232/348 (67%) | .864 |
| Fibrosis | .055 | ||
| 0 | 24 (41%) | 132 (37%) | |
| 1 | 19 (32%) | 56 (16%) | |
| 2 | 7 (12%) | 64 (18%) | |
| 3 | 5 (8%) | 63 (18%) | |
| 4 | 4 (7%) | 41 (11%) | |
| Fibrosis >2 | 16 (27%) | 168 (47%) | .004 |
| HCC | 1 (1.7%) | 3 (0.84%) | .535 |
GGT, γ-glutamyltransferase; HDL, high-density lipoprotein.
Table 4.
KLF6 Genotype Is Associated With the Fibrosis Score in NAFLD Patients: KLF6 Genotype Association in Individual and Combined Groups
| KLF6 IVS1-27 genotype | Group 1 (n = 306) |
Group 2 (n = 109) |
Combined (n = 415) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Fibrosis score |
P | Fibrosis score |
P | Fibrosis score |
P | ||||
| 0+1 | 2+3+4 | 0+1 | 2+3+4 | 0+1 | 2+3+4 | ||||
| GA or AA | 33 (75) | 11 (25) | .015 | 10 (67) | 5 (33) | .134 | 43 (73) | 16 (27) | .004 |
| GG (wt) | 145 (55) | 117 (45) | 43 (46) | 51 (54) | 188 (53) | 168 (47) | |||
Univariate analyses of differences between the defined control group (fibrosis 0 + 1) and the case group (fibrosis 2–4) are shown in Table 5. Variables analyzed included both reported risk factors for progressive disease (age, BMI, and serum glucose level) as well as 2 reported associations with progressive disease (platelet count and AST/ALT ratio).27 Logistic regression analysis was used on the combined data set to determine whether the association between KLF6 and advanced stage 2–4 fibrosis was independent of these factors. As presented in Table 6, wt KLF6 remained a highly significant and independent predictor of the presence of fibrosis, with an odds ratio of 2.76 (95% confidence interval, 1.295–5.908; P = .009).
Table 5.
KLF6 Genotype Is a Predictor of Fibrosis Stage in Patients With NAFLD: Univariate Analyses of Controls (Fibrosis 0 + 1) Versus Cases (Fibrosis 2 + 3 + 4)
| Fibrosis | 0–1 | 2–4 | P |
|---|---|---|---|
| Number | 231 | 184 | |
| Sex (male) | 1164 (71.0%) | 116 (63.0%) | .086 |
| Age, y | 45.39 ± 12.25 | 51.61 ± 12.35 | .000 |
| BMI, kg/m2 | 31.50 ± 6.46 | 33.15 ± 5.89 | .009 |
| Blood glucose level | 5.85 ± 2.39 | 6.93 ± 3.40 | .000 |
| Platelets, ×109 L | 253.45 ± 60.74 | 209 ± 77.95 | .000 |
| AST/ALT ratio | 0.654 ± 0.25 | 0.813 ± 0.45 | .000 |
| KLF6 IVS1-27G>A | 43 (18.60%) | 16 (8.69%) | .004 |
Wt KLF6 IVS1-27 Is Preferentially Transmitted to Children With Fibrotic NAFLD
We analyzed 71 families with 2 surviving parents and an index child with NAFLD and genotyped parents and children for the KLF6 IVS1-27G>A SNP. The majority of these family trios formed part of a previously reported and well-characterized series.28,29 Our particular interest was in those with fibrotic disease, of whom there were 61 (stage 1, n = 59; stage 2, n = 2). All of these children had nonalcoholic steatohepatitis, with a necro-inflammatory score of either stage 1 or 2. Forty were male (mean age, 11.47 ± 2.89 y; BMI, 25.46 ± 3.1) and 21 were female (mean age, 9.9 ± 2.31 y; BMI, 25.17 ± 4.75). Twenty-four families were informative in that 1 parent was heterozygous for the SNP. Transmission disequilibrium testing using these families revealed preferential transmission of the wt allele to the children (17 wt alleles transmitted vs 7 not transmitted, transmission disequilibrium testing, χ2 = 4.16667; P = .041; odds ratio of the wt allele being transmitted to affected children = 2.42).
Discussion
NAFLD is increasingly common in Western societies. Established predictors of those likely to develop progressive disease characterized by advanced fibrosis include age, increased BMI, and an increased blood glucose level.27 Why some individuals with these risk factors progress and others do not is unclear, but twin and ethnic studies suggest that genetic factors play a role.5,6 To date, SNPs in a number of genes have been associated with NAFLD severity, including microsomal triglyceride transfer protein, manganese superoxide dismutase,30 phosphatidylethanolamine N-methyltransferase,31 and tumor necrosis factor α.32 However, none of these studies were sufficiently large to exclude a type 1 error (number of patients, <110) and none have been replicated.
The identification of a functional alternative splicing promoting KLF6 SNP as a susceptibility factor for familial prostate cancer has lead to further studies establishing alternative splicing as a means of KLF6 deregulation in cancers.20,21,33 The suspected role of KLF6 in the regulation of both hepatocyte growth and HSC activation, as well as its induction in an established animal model of nonalcoholic steatohepatitis,13 provides justification for examining both the role of KLF6 in human NAFLD and the SNP as a susceptibility factor affecting disease stage. In this article, we have presented KLF6 mRNA expression data from liver biopsy samples of patients with NAFLD and showed increased total and wt isoforms in association with increased levels of inflammation. From these data alone we were unable to draw firm conclusions on the role of wt KLF6 or on the contribution of alternatively spliced KLF6 to the development of fibrotic disease. It was interesting, however, that wt KLF6 rather than the Total KLF6 isoforms was associated with the more advanced stages of fibrotic NAFLD. We hypothesized that, through promotion of fibrosis and inhibition of hepatocyte proliferation, wt KLF6 was involved in the pathogenesis of more advanced fibrotic liver disease. To test this hypothesis we investigated whether possession of the SNP promoting alternative splicing of KLF6 to its dominant-negative isoforms protected against the development of fibrotic disease in a large cohort of well-characterized patients with NAFLD.
Functionality of the KLF6 IVS1-27G>A SNP was confirmed by showing increased expression of alternatively spliced products in a liver cell environment and the subsequent attenuated up-regulation αSMA and collagen 1 in human HSCs. Subsequently, we performed both a classic allelic association study to determine if the frequency of the SNP differed according to the presence of more advanced fibrosis and an intrafamilial allelic association study to look for preferential transmission of the wt allele to children with fibrotic NAFLD. In our first group of 306 biopsy–proven UK NAFLD patients, we showed that the presence of the KLF6 IVS1-27G>A SNP was associated with less fibrosis. Not only was this trend confirmed in an independent group of Italian patients, analysis of the 2 groups combined identified the presence of wt KLF6 as a predictor of level 2 or more fibrosis independently of all other established risk factors of progressive disease. Clearly, using case-control methodology to study disease progression is subject to the potential bias that some controls (fibrosis stage, <2) could, with time, cross-over to become cases. Given the small age difference between the cases and controls (6 years) relative to the slow and infrequent progression of early NAFLD5 (5%–25% at 14 years depending on the presence of nonalcoholic steatohepatitis), and the fact that KLF6 remained significantly associated with more advanced fibrosis in the logistic regression analysis that included age, we believe this is unlikely to have affected our results significantly. With longer follow-up evaluations of our cohort, conducting a time-to-progression (survival)-type analysis should be possible in the future. In the family study, we have shown preferential transmission of the wt allele to affected children. The size of group 1 (n = 306), the replication in an independent cohort, the disproportionate transmission to affected children, and the confirmed functionality of the SNP make it extremely unlikely that the association is a result of type 1 error or that the SNP is a marker of some other disease-causing SNP in the same haplotype block. We therefore conclude that the wt KLF6 genotype is a significant susceptibility factor for fibrotic NAFLD, whereas KLF6 IVS1-27G>A protects against the development of fibrosis.
In the presence of inflammation and ongoing oxidative stress, we believe that increased wt KLF6 promotes HSC activation and fibrogenesis, as well as inhibits hepatocyte growth. However, in the presence of KLF6 IVS1-27G>A, the production of dominant-negative splice variants of KLF6 in addition to the wt KLF6 isoform will render wt KLF6 less active, delaying fibrogenesis, but also facilitating hepatocyte regeneration in the face of ongoing inflammation. It is interesting that an SNP in a tumor-suppressor gene associated with rapid tumor growth and an increase in the incidence of familial prostate cancers is so common in the populations studied. Our observation that this SNP has beneficial effects in promoting cell survival in a noncancer, chronic disease situation may offer an explanation. Whether individuals with KLF6 IVS1-27G>A who do get cirrhosis are at a greater risk of developing hepatocellular cancer remains to be determined. Further study of this interesting gene in other chronic hepatic and nonhepatic diseases and cancers will help to determine its increasingly recognized role in the regulation of these processes.
Supplementary Material
Acknowledgments
Supported by grants from GlaskoSmithKline (H.L.R.); Newcastle upon Tyne Hospitals Special Trustees (H.L.R.); European Association for Study of the Liver Sheila Sherlock fellowship (L.M.); European Research Advisory Board (C.P.D.; H.L.R.), and National Institutes of Health grant DK56621 (S.L.F.).
Abbreviations used in this paper
- αSMA
α-smooth muscle actin
- BMI
body mass index
- HSC
hepatic stellate cells
- KLF6
Kruppel-like factor 6
- NAFLD
nonalcoholic fatty liver disease
- PCR
polymerase chain reaction
- RQ
relative quantity
- SNP
single nucleotide polymorphism
Footnotes
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2008.04.004.
References
- 1.Neuschwander-Tetri BA, Caldwell SH. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003;37:1202–1219. doi: 10.1053/jhep.2003.50193. [DOI] [PubMed] [Google Scholar]
- 2.de Marco R, Locatelli F, Zoppini G, et al. Cause-specific mortality in type 2 diabetes. The Verona Diabetes Study Diabetes Care. 1999;22:756–761. doi: 10.2337/diacare.22.5.756. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Lymp JF, St Sauver J, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Bugianesi E, Leone N, Vanni E, et al. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123:134–140. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- 5.Ekstedt M, Franzen LE, Mathiesen UL, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44:865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 6.Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108:9–13. doi: 10.1016/s0002-9343(99)00315-0. [DOI] [PubMed] [Google Scholar]
- 7.Willner IR, Waters B, Patil SR, et al. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am J Gastroenterol. 2001;96:2957–2961. doi: 10.1111/j.1572-0241.2001.04667.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilfred de Alwis NM, Day CP. Genetics of alcoholic liver disease and nonalcoholic fatty liver disease. Semin Liver Dis. 2007;27:44–54. doi: 10.1055/s-2006-960170. [DOI] [PubMed] [Google Scholar]
- 9.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 10.Kojima S, Hayashi S, Shimokado K, et al. Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-beta1 in vascular endothelial cells. Blood. 2000;95:1309–1316. [PubMed] [Google Scholar]
- 11.Koritschoner NP, Bocco JL, Panzetta-Dutari GM, et al. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J Biol Chem. 1997;272:9573–9580. doi: 10.1074/jbc.272.14.9573. [DOI] [PubMed] [Google Scholar]
- 12.Lalazar A, Wong L, Yamasaki G, et al. Early genes induced in hepatic stellate cells during wound healing. Gene. 1997;195:235–243. doi: 10.1016/s0378-1119(97)00159-5. [DOI] [PubMed] [Google Scholar]
- 13.Ratziu V, Lalazar A, Wong L, et al. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, Ratziu V, Choi SG, et al. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem. 1998;273:33750–33758. doi: 10.1074/jbc.273.50.33750. [DOI] [PubMed] [Google Scholar]
- 15.Starkel P, Sempoux C, Leclercq I, et al. Oxidative stress, KLF6 and transforming growth factor-beta up-regulation differentiate non-alcoholic steatohepatitis progressing to fibrosis from uncomplicated steatosis in rats. J Hepatol. 2003;39:538–546. doi: 10.1016/s0168-8278(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 16.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 17.Tal-Kremer S, Reeves HL, Narla G, et al. KLF6 is a tumor suppressor gene that is lost and/or mutated in a majority of HCC. Hepatology. 2002;36:394A. [Google Scholar]
- 18.Kremer-Tal SNG, Chen Y, Hod E, et al. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645–654. doi: 10.1016/j.jhep.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narla G, Difeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 20.Narla G, DiFeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 21.DiFeo A, Narla G, Hirshfeld J, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–3739. doi: 10.1158/1078-0432.CCR-06-0054. [DOI] [PubMed] [Google Scholar]
- 22.Camacho-Vanegas O, Narla G, Teixeira MS, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–1395. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 24.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 25.Brunt EMJC, Di Bisceglie AM, Neuschwander-Tetri BA, et al. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Hui AY, Albanis E, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angulo P, Hui JM, Marchesini G, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 28.Nobili V, Marcellini M, Devito R, et al. NAFLD in children: a prospective clinical-pathological study and effect of lifestyle advice. Hepatology. 2006;44:458–465. doi: 10.1002/hep.21262. [DOI] [PubMed] [Google Scholar]
- 29.Nobili V, Marcellini M, Marchesini G, et al. Intrauterine growth retardation, insulin resistance, and nonalcoholic fatty liver disease in children. Diabetes Care. 2007;30:2638–2640. doi: 10.2337/dc07-0281. [DOI] [PubMed] [Google Scholar]
- 30.Namikawa C, Shu-Ping Z, Vyselaar JR, et al. Polymorphisms of microsomal triglyceride transfer protein gene and manganese superoxide dismutase gene in non-alcoholic steatohepatitis. J Hepatol. 2004;40:781–786. doi: 10.1016/j.jhep.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Wang J, Li C, et al. The phosphatidylethanolamine N-methyltransferase gene V175M single nucleotide polymorphism confers the susceptibility to NASH in Japanese population. J Hepatol. 2007;46:915–920. doi: 10.1016/j.jhep.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Tokushige K, Takakura M, Tsuchiya-Matsushita N, et al. Influence of TNF gene polymorphisms in Japanese patients with NASH and simple steatosis. J Hepatol. 2007;46:1104–1110. doi: 10.1016/j.jhep.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 33.Camacho-Vanegas O, Narla G, Teixeira MS, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–1395. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.