Abstract
The protein-protein interaction of p53 and mdm2 is an important anticancer target. The interface, however, is very hydrophobic and naturally results in very hydrophobic antagonists. We used the Orru three component reaction (O-3CR) along with a rapid and efficient, recently discovered amidation reaction to dramatically improve the water solubility of our recently discovered low molecular weight p53/mdm2 antagonists. Arrays of amides were synthesized with improved hydrophilicity and retainment and/or improvement of p53/mdm2 inhibitory activity.
Protein-protein interfaces are often characterized by strongly hydrophobic interaction surfaces. Driven by these target requirements, antagonists of such sites are necessarily hydrophobic to accomplish high affinity. An example of medicinal relevance is the protein-protein interaction between the transcription factor p53 and its negative regulator mdm2.1 A wealth of data show that interruption of this protein-protein interaction drives cancer cells into apoptosis and cellular senescence, both in vitro and in vivo.2
Interestingly, no other protein-protein interaction has attracted so much attention: over 20 different low molecular weight backbones have been described as antagonists of the p53/mdm2 interaction in the past several years.3 The nature of the hydrophobic interface of this interaction, however, mostly leads to antagonists that lack sufficient water solubility and/or are promiscuous inhibitors.
We recently described a novel drug discovery approach geared towards the parallel discovery of several p53/mdm2 antagonizing low molecular weight scaffolds that employs a tight interplay of techniques, involving structure-based fragment generation, virtual chemistry, docking, efficient antagonist synthesis by multicomponent reactions (MCRs) and high content NMR-based screening (details of this method are described in an upcoming communication).4 We were able to discover 10 unprecedented scaffolds with low µM binding affinity to mdm2 by 2D NMR spectroscopy; these scaffolds are starting points for potential medicinal chemistry programs. In the following, we describe our optimization of several scaffolds towards potent p53/mdm2 antagonizing and cellular activity.
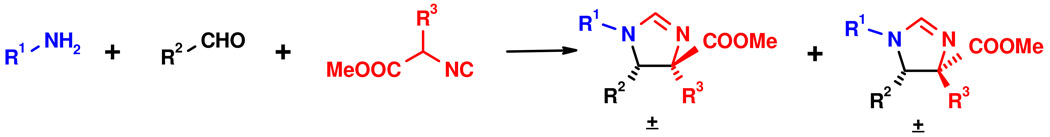
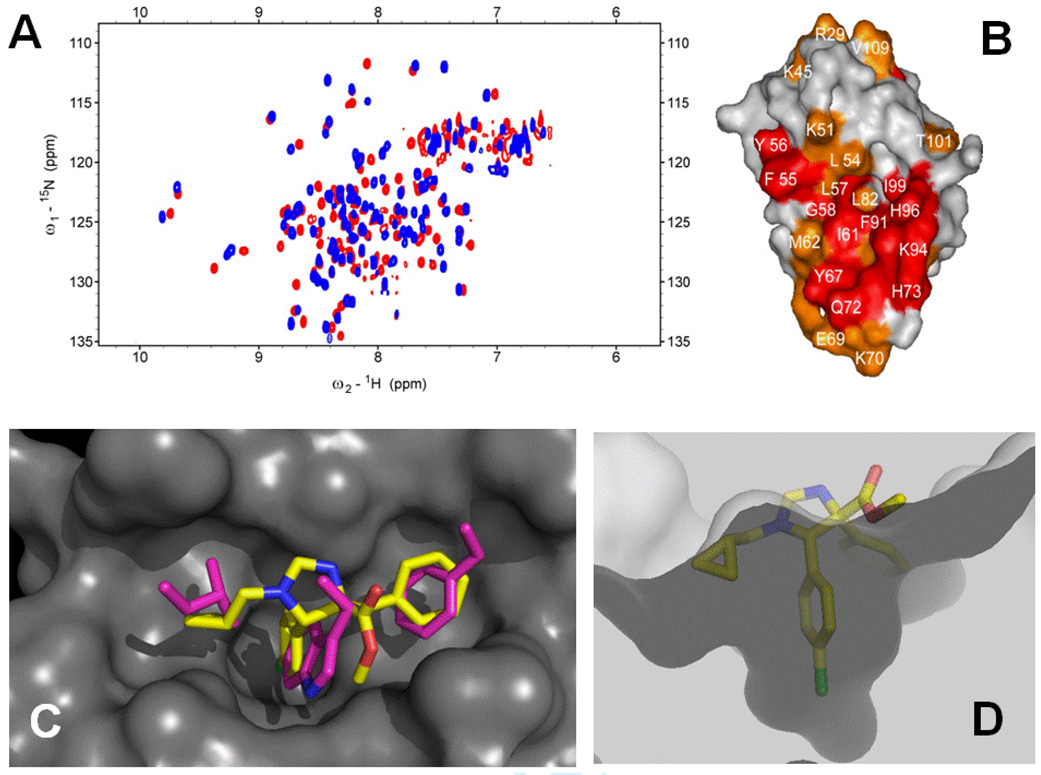
One of the scaffolds we predicted to bind to MDM2 is based on imidazoline, which can be conveniently accessed by the Orru 3-component reaction (O-3CR) of aldehydes, primary amines and amino acid derived isocyano esters (Scheme 1).5 Based on perturbation NMR data and the accompanying modeling of representative compounds, we were able to develop a binding model of the imidazoline scaffold in the p53 binding groove of mdm2 (Fig. 1).
Scheme 1.
Stereoselective synthesis of the imidazoline-4-carboxylmethylester by a 3-component reaction of primary amines, aldehydes and methyl isocyanoacetates (O-3CR). The reaction preferentially yields the cis-isomer.
Fig. 1.
Development of binding model of 1 in the p53 binding site of mdm2. A: 2D 15N-1H HSQC NMR of compound 1 (red without and blue with 1); B: Surface picture of mdm2 upon compound 1 binding based on the perturbation data of the 2D–HSQC (A, the more reddish the more signal shift in the NMR); this experiment suggests that compound 1 is binding in the p53 binding site as indicated by the mostly perturbed mdm2 amino acids L54, L57, L82, I99, H96, F91, I61, Y67 (red); from NMR titration experiments the affinity of 1 to mdm2 was determined Ki = 15 µM.; C: Docking pose of compound 1 (yellow stick) in the p53 binding site of mdm2 (grey surface) and overlapped with the p53 hot spot, the triad F19, W23 and L26 (pink sticks); the x-ray structure used for docking is based on the PDB identifier: 1YCR; D: cut-away view of compound 1 in mdm2 and showing the shape and depth of the binding site. Clearly the methyl ester moiety (yellow-red sticks) does not significantly contribute to the binding and points out of the binding site into the water space.6
According to our model, the cis-diastereomer should be more active than the trans-diastereomer. This has been confirmed by our NMR-based screening.4 The three residues of the imidazoline residues are designed to mimic the p53 amino acid “hot-spot” residues F19, W23 and L26 which are crucial for mdm2 binding: R1 relates to L26, R2 to W23 and R3 to F19, respectively (Scheme 1, Fig. 1 C). The carboxylicacid methylester derived from the isocyanide, however, is seemingly not involved in close contacts to the mdm2 surface (Fig. 1 C, D). Based on this design we synthesized several imidazoline derivatives and screened them in NMR-based 2D HSQC experiments to determine their affinity to mdm2. All the initially synthesized compounds showed activity in the single or double digit µM scale. Compounds derived from the amino acids leucine, phenylalanine, and phenylglycine (isocyanide input) together with p-chlorophenyl residue in 4-position (aldehyde input) and cyclopropylmethyl residue (amine input), however, showed improved low µM affinity as mixtures of the two enantiomers (Fig. 1; the detailed SAR and further biological studies will be communicated elsewhere).
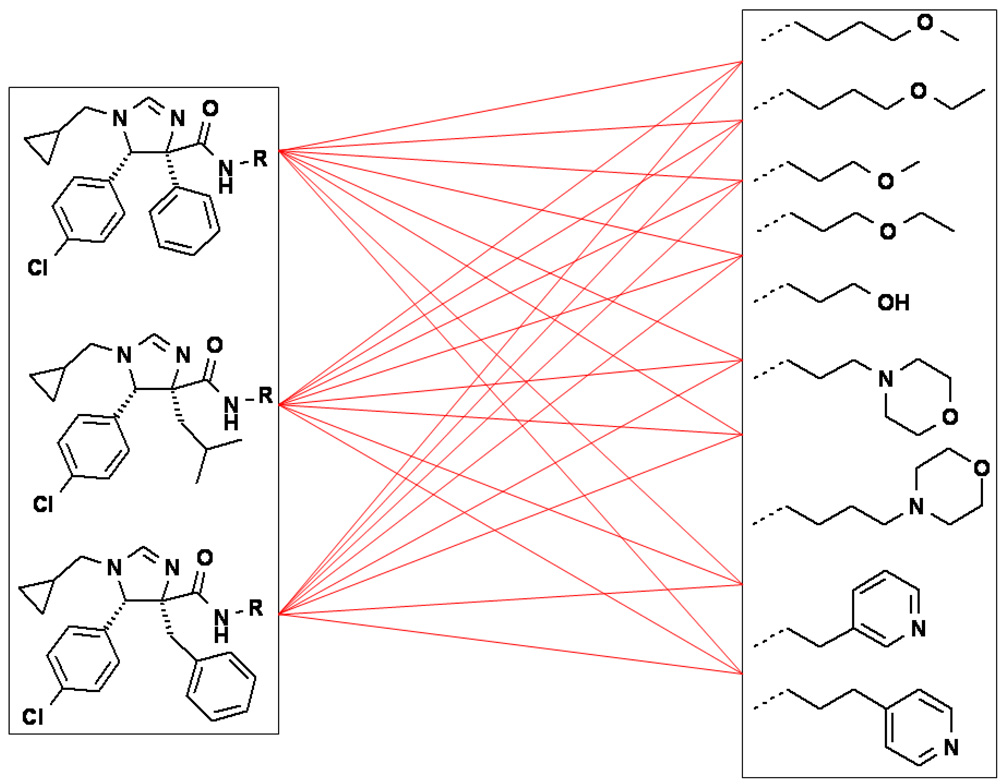
Our high content NMR-based screening provides a wealth of information, including affinity of the compounds to mdm2 and the approximate binding site (Fig. 1 A, B). In addition, we obtain information as to a compound’s solublility in the aqueous buffer, if the compound precipitates the protein, and whether the protein is undergoing major structural changes (denaturation). While optimizing, we realized that our compounds sometimes showed low solubility for NMR screening, which we could overcome by the addition of the solubilizer Tween20. We reasoned that compounds with good water solubility might be beneficial not only for the initial screening process, however, but also for later advantageous ADMET properties of lead compounds. Therefore, we investigated our model for the introduction of solubilizing substituents. According to our design, the pertinent carboxylic acid methyl ester should not contribute to the binding to mdm2 (Fig. 1 C, D). In order to prove the suitability of the methylester position for constructive derivatization, we synthesized a simple amide and, gratifyingly, found the same affinity of the compound to mdm2 as in the corresponding methylester (Fig. 2).
Fig. 2.
Array synthesis of imidazoline amide derivatives to improve affinity and water solubility.
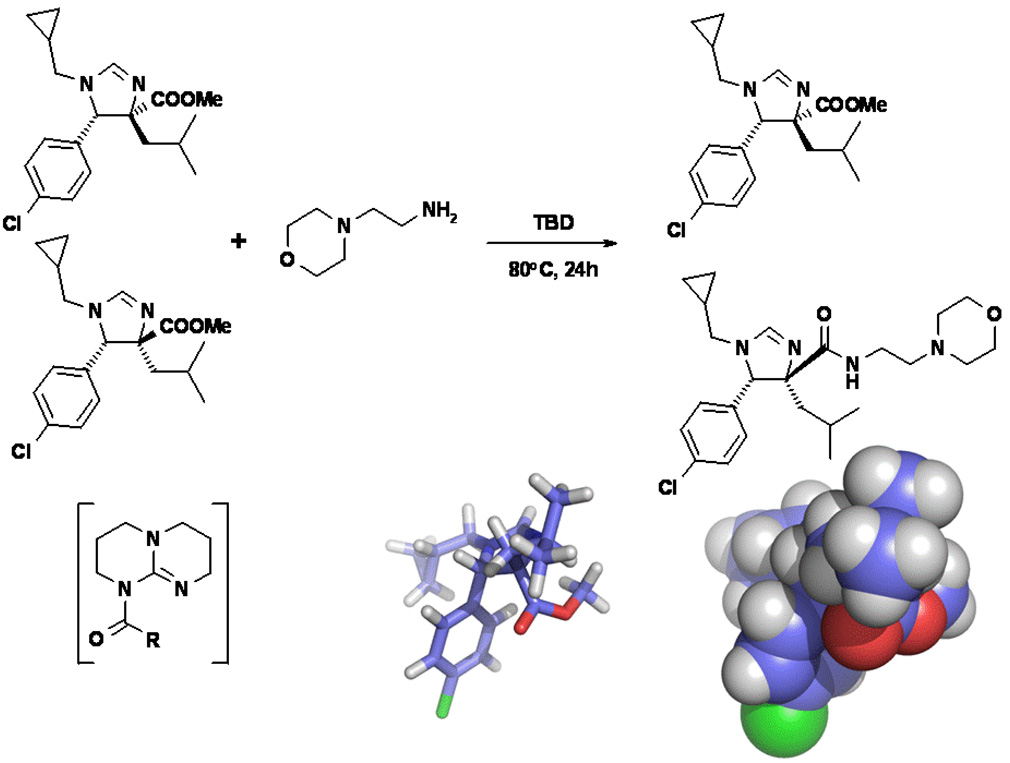
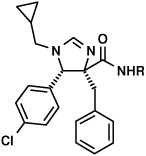
We thus prepared an array of amide derivatives aiming to improve solubility and affinity (Fig.2). The standard way to convert a carboxylic acid methyl ester into an amide is by saponification, activation and amide formation.7b Few protocols are described for one-pot transformation and these use harsh conditions not compatible with fragile molecules.7 In order to improve processes in terms of time and yields, we prefer to run one put-transformations.8 Based on our recently described and convenient one-pot transformation of isocyanomethylesters into amides, we reasoned that a similar process could potentially apply here, thus saving two steps over the traditional conversion.9 Additionally, Sabot et al. recently described an efficient way to convert carboxylic acid methyl esters in one step into their amides.10 Thus we transformed three different and potent mdm2-binding imidazoline methyl esters into their corresponding amides (Fig. 2). This reaction is catalyzed by 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD) under solvent-free conditions and yields the product amides in high yields. For reference purposes, we also tried to convert the trans imidazoline starting materials, but the reaction did not take place. This is presumably due to steric hindrance, (Fig 3) as indicated by the effective shielding of the ester group by the adjacent lateral p-chlorophenyl and frontal iso-valeric moiety. According to the proposed TBD catalysis, an intermediate covalent adduct between the ester and the catalyst must be formed,10 which is sterically rather impossible in the present conformation. This finding resulted in an efficient protocol where we used the mixture of the cis and trans-diasteroemers to convert stereoselectively only the less hindered cis-diastereomer. The cis-amide product could then be effectively separated from the unreacted trans-ester by chromatography.
Fig. 3.
Above: The cis-imidazolines give clean and high yielding conversions to the coresponding amides. The trans-imidazolines cannot be transformed into their amides under the TBD solvent free conditions due to steric hinderance. Below: A energy minimized model of the trans-diastereomer in stick and ball presentation clearly show the steric shielding of the ester bond, thus blocking access by the large bicyclic TBD catalyst (bracket).
Hydrophilicity and associated water solubility of compounds is clearly correlated with a good absorption,12 distribution, metabolism, excretion and toxicity profile of a drug;13 e.g., if the solubility of a compound remains poor in the gastrointestinal fluids, poor absorption and a low bioavailability may result. Sometimes the absorption issue can be overcome by formulation. If a drug has poor water solubility, however, it may not reach in sufficiently high concentrations the target tissue, or intracellular target compartment and thus prove to be non-efficacious. Lack of efficacy – also due to other reasons than poor pharmacokinetic-pharmacodynamic (PKPD) profile – is the major reason for current failure of drug candidates in clinical trials. Additionally, poor water solubility might result in a long half life time and an accumulation in fatty tissue and thus leading to toxicity.
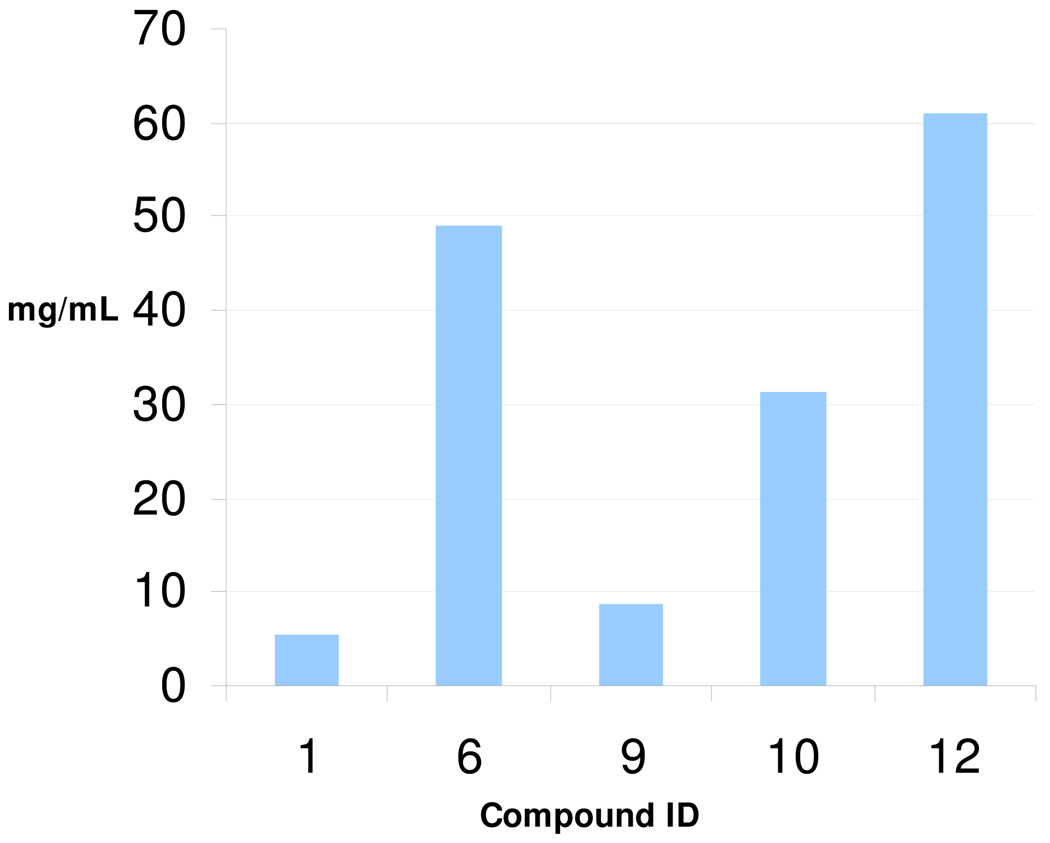
Apart from poor absorption-distribution-metabolism-excretion-toxicology (ADMET) properties, insufficiently water soluble compounds often lead to poor reproducibility and unreliable results or even false positive hits during in vitro screening; e.g., if a drug precipitates before reaching its cellular target, the target will be exposed to a concentration of drug lower than the nominal and could therefore yield a response that is diminished, undetectable, or independent of the input concentration.14 On the other hand, poorly soluble compounds often form hydrophobic aggregates potentially precipitating proteins and thus lead to false positive screening results.15 All the amides derivatives synthesized showed improved water solubility in our NMR-based affinity assay. In order to assess the exact values of solubility, we quantitatively measured six selected compounds (Fig. 4 and supplementary information). The cyclic amidine functionality of the imidazoline ring is basic and therefore can form salts. By forming the HCl salts, all compounds showed very high water solubility. Gratifyingly, the affinities of the amide derivatives retained the parental affinities or did even improve, indicating additional interactions of the substitutents with mdm2.
Fig. 4.
Water solubility of six selected compounds.
In summary, we described the synthesis of 25 novel imidazoline amides by a parallel synthesis approach. Many of the imidazolines have greatly improved water solubility over their parent methyl esters while retaining mdm2 affinity and are therefore more suitable as p53-mdm2 protein-protein interaction antagonists. The transformation was accomplished employing a convergent O-3CR followed by an efficient one-pot solvent-free amidation using TBD as a catalyst. The amidation reaction is stereoselective and the cis-isomer reacts much faster than the trans-isomer. This reaction has become instrumental for our drug discovery project to rapidly gain insight into SAR of this new class of p53-mdm2 antagonists via synthesis of hundreds of different derivatives. Additionally, the amidation reaction is general and has been employed in several other projects ongoing in our laboratory.11 We hope that the improved water solubility will also be reflected in the ADMET properties of improved lead candidates. In due course we will report about the biological properties of these compounds and in depth SAR studies.
Experimental Section
1. General experimental methods
Standard syringe techniques were applied for transfer of air sensitive reagents. Dry solvents and all purchased chemicals were purchased from Aldrich, Fisher Scientific, Acros Organics or Alfa Aesar and used as received. 1H- and 13C-NMR spectra were recorded on Ultrashield Plus 600, Bruker at 600MHz. Chemical shift values are in ppm relative to external TMS. Abbreviations used are s = singlet, brs = broad singlet, d = doublet, brd = broad doublet, m = multiplet; data in parenthesis are given in the following order: multiplicity, number of protons and coupling constants in Hz. Flash chromatography was performed with the indicated solvent mixture on Silica gel, MP Silitech 32–63 D, 60 Å, Bodman. Chromatotron chromatography was performed on Harrison Research Chromatotron, Ser. no. 65F with the indicated solvent mixture using Silica gel, Merck, TLC grade 7749, with gypsum binder and fluorescent indicator, Sigma Aldrich. Thin Layer Chromatography was performed using Whatmann flexible-backed TLC plates on aluminum with fluorescence indicator. Compounds on TLC were visualized by UV-detection. HPLC-MS measurements were done on a Shimadzu prominence UFLC (HPLC) and API 2000 LC/MS/MS System, Applied Biosystems MDS SCIEX, (MS) using a Dionex Acclaim 120 column (C18, 3µm, 120 Å, 2.1 × 150mm), mobile phase: water with 0.1% acetic acid and acetonitrile, gradient: 5–90% acetonitrile in 7 min, injection volume: 5 µl, detection wavelength 254 nm. HRMS measurements were performed at the Department of Chemistry, University of Pittsburgh with a Q-Tof spectrometer, ionization mode: ESI. Microwave reactions were performed on the Emrys Optimizer system from Personal Chemistry.
2. Representative Procedure for the Synthesis of the Imidazoline
To 30 mmol amine 30 mmol isocyanoacetic acid methyester were added and stirred over night at room temperature. If the product precipitated during the reaction, it was filtered off, washed 3 times with cold diethyl ether and dried under vacuum over night. If no precipitation was observable, cold diethyl ether was added to the reaction mixture and the product was allowed to crystallize in the freezer at −20°C. In the rare cases the product was formed as oil or no crystallization occurred, the product was purified by preparative chromatography with silica gel and ethyl acetate as eluent. Crystallization could also be enhanced in several cases by using ultrasound.
3. Representative Procedure for the Amidation Reaction
To 30 mmol amine and 30 mmol isocyanoacetic acid methyl ester, a catalytic amount of 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD) was added and stirred overnight at room temperature. If the product precipitated during the reaction, it was filtered off, washed three times with cold diethyl ether and dried under vacuum overnight. If no precipitation was observable, cold diethyl ether was added to the reaction mixture and the product was allowed to crystallize in the freezer at –20°C. In the rare cases, the product was formed as oil or no crystallization occurred. The product was then purified by preparative chromatography with silica gel and ethyl acetate as eluent. Crystallization could also be enhanced in several cases by using ultrasound.
4. Analytical data of described compounds
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid methyl ester (1)
81% yield; C21H21ClN2O2; MW 368.85 g/mol; HRMS calc. 369.1370; found 369.1365 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.05–0.07 (m, 2H), 0.44–0.50 (m, 1H), 0.56–0.62 (m, 1H), 0.85–0.91 (m, 1H), 2.55 (dd, J= 12 Hz & 6 Hz, 1H), 3.08 (dd, J= 12 Hz & 6 Hz, 1H), 3.77 (s, 3H), 5.62 (s, 1H), 6.88–7.04 (m, 9H), 7.44 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.38, 4.47, 9.05, 50.01, 52.72, 68.71, 84.11, 126.18, 126.82, 127.32, 127.52, 132.73, 134.05, 136.87, 156.29, 173.81.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (3-methoxypropyl)amide (4)
69% yield; C24H28ClN3O2; MW 425.96 g/mol; HRMS calc. 426.1948; found 426.1947 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.05 (m, 2H), 0.43–0.48 (m, 1H), 0.55–0.60 (m, 1H), 0.84–0.90 (m, 1H), 1.70–1.74 (m, 2H), 2.59 (dd, J= 14.22 Hz & 7.62 Hz, 1H), 3.07 (dd, J= 14.16 Hz & 6.18 Hz, 1H), 3.22 (s, 3H), 3.24–3.41 (m, 4H), 5.51 (s, 1H), 6.81–7.06 (m, 7H), 7.09–7.11 (m, 2H), 7.37 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.82, 4.77, 9.57, 29.16, 36.93, 50.35, 58.58, 68.70, 70.35, 83.87, 126.74, 126.89, 127.43, 127.60, 132.80, 135.09, 138.43, 155.31, 174.59.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (3-ethoxypropyl)amide (5)
84% yield; C25H30ClN3O2; MW 439.99 g/mol; HRMS calc. 440.2105; found 440.2086 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.03–0.07 (m, 2H), 0.43–0.49 (m, 1H), 0.55–0.60 (m, 1H), 0.84–0.91 (m, 1H), 1.13 (t, J= 6.96 Hz, 3H), 1.68–1.76 (m, 2H), 2.59 (dd, J= 14.16 Hz & 7.62 Hz, 1H), 3.07 (dd, J= 14.16 Hz & 6.18 Hz, 1H), 3.28–3.42 (m, 6H), 5.51 (s, 1H), 6.82–7.08 (m, 7H), 7.10–7.15 (m, 2H), 7.36 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.82, 4.77, 9.56, 15.12, 29.25, 37.18, 50.35, 66.20, 68.31, 68.72, 83.88, 126.74, 126.87, 127.41, 127.74, 132.78, 135.13, 138.46, 155.28, 174.57.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (2-methoxyethyl)amide (6)
66% yield; C23H26ClN3O2; MW 411.94 g/mol; HRMS calc. 412.1792; found 412.1767 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.07–0.06 (m, 2H), 0.40–0.49 (m, 1H), 0.53–0.61 (m, 1H), 0.82– 0.91 (m, 1H), 2.58 (dd, J= 14.10 Hz & 7.62 Hz, 1H), 3.08 (dd, J= 14.16 Hz & 6.12 Hz, 1H), 3.26 (s, 3H), 3.35–3.37 (m, 1H), 3.38–3.46 (m, 3H), 5.49 (s, 1H), 6.81–7.00 (m, 6H), 7.10–7.12 (m, 3H), 7.38 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.81, 4.79, 9.56, 39.26, 50.35, 58.76, 68.87, 70.85, 83.83, 126.76, 126.91, 127.43, 127.75, 132.81, 135.05, 138.27, 155.47, 174.79.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (2-ethoxyethyl)amide (7)
69% yield; C24H28ClN3O2; MW 425.96 g/mol; HRMS calc. 426.1949; found 426.1954 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ = 0.11-0.06 (m, 2H), 0.38–0.49 (m, 1H), 0.53–0.61 (m, 1H), 0.82–0.91 (m, 1H), 1.09 (t, J= 7.08 Hz, 3H), 2.59 (dd, J= 14.22 Hz & 7.62 Hz, 1H), 3.08 (dd, J= 14.16 Hz & 6.24 Hz, 1H), 3.30–3.52 (m, 6H), 5.50 (s, 1H), 6.81–7.01 (m, 6H), 7.02–7.12 (m, 3H), 7.37 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ = 2.81, 4.78, 9.57, 15.02, 39.43, 50.35, 66.37, 68.71, 83.87, 126.77, 126.89, 127.41, 127.75, 132.80, 135.09, 138.31, 155.43, 174.76.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (2-hydroxyethyl)amide (8)
71% yield; C22H24ClN3O2; MW 397.91 g/mol; HRMS calc. 398.1635; found 398.1600 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.05-0.04 (m, 2H), 0.42–0.50 (m, 1H), 0.54–0.60 (m, 1H), 0.83–0.90 (m, 1H), 2.58 (dd, J= 14.40 Hz & 7.8 Hz, 1H), 3.07 (dd, J= 13.8 Hz & 6 Hz, 1H), 3.32–3.44 (m, 2H), 3.59–3.67 (m, 2H), 5.52 (s, 1H), 6.77–7.07 (m, 9H), 7.17 (s, 1H), 7.37 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.83, 4.81, 9.52, 42.62, 50.38, 53.44, 61.87, 68.75, 83.76, 126.66, 127.12, 127.58, 127.81, 132.93, 134.71, 138.07, 155.60, 175.49.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid 3-(morpholin-4-yl)propylamide (9)
69% yield; C27H33ClN4O2; MW 481.04 g/mol; HRMS calc. 481.2370; found 481.2372 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.05 (m, 2H), 0.44–0.49 (m, 1H), 0.56–0.60 (m, 1H), 0.84–0.90 (m, 1H), 1.59–1.64 (m, 2H), 2.21–2.34 (m, 6H), 2.59 (dd, J= 14.22 Hz & 7.68 Hz, 1H), 3.07 (dd, J= 14.16 Hz & 6.18 Hz, 1H), 3.23–3.28 (m, 1H), 3.42–3.48 (m, 1H), 3.61–3.68 (m, 4H), 5.51 (s, 1H), 6.84–7.00 (m, 6H), 7.07–7.09 (m, 2H), 7.36 (s, 1H), 7.38–7.39 (m, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.82, 4.79, 9.53, 25.57, 38.51, 50.39, 53.65, 57.11, 66.83, 68.68, 83.98, 126.72, 126.90, 127.41, 127.73, 132.75, 135.12, 138.54, 155.25, 174.47.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid 2-(morpholin-4-yl)ethyl)amide (10)
68% yield; C26H31ClN4O2; MW 467.02 g/mol; HRMS calc. 467.2214; found 467.2198 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ 0.01–0.04 (m, 2H), 0.44–0.49 (m, 1 H), 0.56–0.61 (m, 1H), 0.85–0.91 (m, 1H), 2.27–2.46 (m, 6H), 2.60 (dd, J= 14.22 Hz & 7.68 Hz, 1H), 3.09 (dd, J= 14.22 Hz & 6.24 Hz, 1H), 3.30–3.35 (m, 1H), 3.39–3.44 (m, 1H), 3.53–3.61 (m, 4H), 5.54 (s, 1H), 6.98–7.07 (m, 8H), 7.10–7.11 (m, 2H), 7.38 (s, 1H); 13C–NMR (CDCl3, 150 MHz): δ 2.81, 4.77, 9.56, 36.20, 50.33, 53.26, 56.85, 66.84, 68.63, 83.96, 126.76, 126.92, 127.43, 127.78, 132.84, 135.04, 138.42, 155.35, 174.60.
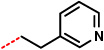
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid 2-(pyridin-4-yl)ethylamide (11)
67% yield; C27H27ClN4O; MW 459.00 g/mol; HRMS calc. 459.1952; found (s, 1H), 8.35 (d, J= 4.44 Hz, 2H); 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.06 (m, 2H), 0.44–0.50 (m, 1H), 0.56–0.61 (m, 1H), 0.82–0.89 (m, 1H), 2.58 (dd, J= 14.22 Hz & 7.68 Hz, 1H), 2.68–2.77 (m, 2H), 3.07 (dd, J= 14.22 Hz & 6.24 Hz, 1H), 3.36–3.42 (m, 1H), 3.61–3.67 (m, 1H), 5.50 (s, 1H), 6.86 (d, J= 4.62 Hz, 2H), 6.91–6.93 (m, 2H), 7.00–7.12 (m, 7H), 7.34 (s, 1H), 8.35 (d, J= 4.44 Hz, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.86, 4.77, 9.56, 34.90, 39.65, 50.34, 68.64, 83.61, 124.14, 126.68, 127.08, 127.56, 127.87, 133.02, 134.76, 138.11, 147.76, 149.71, 155.16, 174.63.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-phenyl-4,5-dihydro-1H-imidazole-4-carboxylic acid (pyridin-3-yl)methylamide (12)
64% yield; C26H25ClN4O; MW 444.97 g/mol; HRMS calc. 445.1795; found 445.1758 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.06 (m, 2H), 0.44–0.50 (m, 1H), 0.56–0.61 (m, 1H), 0.85–0.91 (m, 1H), 2.61 (dd, J= 14.16 Hz & 7.62 Hz, 1H), 3.08 (dd, J= 14.22 Hz & 6.3 Hz, 1H), 4.46 (dd, J= 6.18 Hz & 2.88 Hz, 2H), 5.54 (s, 1H), 6.89–7.03 (m, 6H), 7.09–7.12 (m, 2H), 7.15–7.17 (m, 1H), 7.28–7.32 (m, 1H), 7.36 (s, 1H), 7.43 (brd, J= 5.94 Hz, 1H), 8.44 (brs, 1H), 8.47 (brd, J= 4.8 Hz, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.82, 4.79, 9.60, 40.83, 50.33, 68.74, 83.85, 123.38, 126.71, 127.14, 127.57, 127.86, 133.00, 133.81, 134.78, 134.99, 137.99, 148.71, 148.92, 155.48, 174.89.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid methyl ester (2)
46% yield; C22H23ClN2O2; MW 382.8832; HRMS NA; 1H-NMR (CDCl3, 600 MHz): δ −0.03–0.05 (m, 2H), 0.43–0.49 (m, 1H), 0.52–0.58 (m, 1H), 0.83–0.90 (m, 1H), 2.36 (d, J= 12 Hz, 1H), 2.54 (d, J= 12 Hz, 1H), 2.63 (dd, J= 12 Hz & 6 Hz, 1H), 3.14 (dd, J= 12 Hz & 6 Hz, 1H), 3.59 (s, 3H), 5.04 (s, 1H), 6.97–7.01 (m, 2H), 7.11–7.17 (m, 3H), 7.31–7.37 (m, 5H); 13C–NMR (CDCl3, 150 MHz): δ 2.69, 4.80, 9.33, 14.19, 21.06, 21.61, 43.23, 50.45, 52.15, 60.39, 69.37, 81.36, 126.41, 127.80, 128.65, 129.23, 129.83, 131.04, 133.90, 134.04, 136.33, 155.71, 175.05.
4-Benzyl-5-(4-chloro-phenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-methoxypropylamide (13)
75% yield; C25H30ClN3O2; MW 439.99 g/mol; HRMS calc. 440.2106; found 440.1994 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.05 (m, 2H), 0.44–0.49 (m, 1H), 0.54–0.60 (m, 1H), 0.84–0.90 (m, 1H), 1.38–1.45 (m, 1H), 1.48–1.55 (m, 1H), 2.20 (d, J= 12 Hz, 1H), 2.38 (d, J= 12 Hz, 1H), 2.70 (dd, J= 12 Hz & 6 Hz, 1H), 2.98–3.07 (m, 2H), 3.10–3.19 (m, 2H), 3.19–3.25 (m, 4H), 5.04 (s, 1H), 6.64 (brt, 1H), 7.06–7.10 (brd, 2H), 7.13–7.21 (m, 3H), 7.23 (s, 1H), 7.28 (s, 1H), 7.39 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.70, 4.81, 9.44, 28.94, 36.25, 43.32, 50.40, 58.53, 69.18, 70.10, 80.82, 126.27, 127.64, 128.47, 130.27, 133.57, 134.68, 137.00, 155.41, 175.01.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-ethoxypropylamide (14)
62% yield; C26H32ClN3O2; MW 454.02 g/mol; HRMS calc. 454.2261; found 454.2273 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.05 (m, 2H), 0.42–0.48 (m, 1H), 0.53–0.58 (m, 1H), 0.83– 0.90 (m, 1H), 1.14 (t, J= 6 Hz, 3H), 1.38–1.45 (m, 1H), 1.47–1.54 (m, 1H), 2.19 (d, J= 12 Hz, 1H), 2.36 (d, J= 12 Hz, 1H), 2.68 (dd, J= 12 Hz & 6 Hz, 1H), 3.01–3.09 (m, 2H), 3.13–3.22 (m, 3H), 3.28–3.36 (m, 2H), 5.03 (s, 1H), 6.66 (brt, 1H), 7.06–7.07 (brd, 2H), 7.11–7.18 (m, 3H), 7.21 (s, 1H), 7.27 (s, 1H), 7.38 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.69, 4.81, 9.43, 15.14, 29.00, 36.47, 43.32, 50.40, 66.08, 68.05, 69.17, 80.83, 126.23, 127.60, 128.46, 130.26, 133.54, 134.71, 137.01, 155.33, 175.00.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-methoxyethylamide (15)
79% yield; C24H28ClN3O2; MW 425.96 g/mol; HRMS calc. 426.1948; found 426.1947 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.00–0.05 (m, 2H), 0.44–0.48 (m, 1H), 0.55–0.60 (m, 1H), 0.84–0.90 (m, 1H), 2.22 (d, J= 12 Hz, 1H), 2.37 (d, J= 12 Hz, 1H), 2.69 (dd, J= 12 Hz & 6 Hz, 1H), 2.86–2.91 (m, 1H), 3.09–3.19 (m, 5H), 3.23–3.28 (m, 1H), 3.32–3.37 (m, 1H), 5.05 (s, 1H), 6.75 (brt, 1H), 7.07–7.13 (brd, 2H), 7.13–7.21 (m, 3H), 7.23 (s, 1H), 7.28 (s, 1H), 7.39 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.69, 4.82, 9.43, 38.49, 43.40, 50.41, 58.58, 69.11, 70.77, 80.90, 126.24, 127.63, 128.74, 130.24, 133.58, 134.70, 137.02, 155.45, 175.06.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-ethoxyethylamide (16)
69% yield; C25H30ClN3O2; MW 439.99 g/mol; HRMS calc. 440.2105; found 440.2090 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.06 (m, 2H), 0.43–0.49 (m, 1H), 0.54–0.59 (m, 1H), 0.83–0.90 (m, 1H), 1.09 (t, J= 6 Hz, 3H), 2.22 (d, J= 12 Hz, 1H), 2.38 (d, J= 12 Hz, 1H), 2.69 (dd, J= 12 Hz & 6 Hz, 1H), 2.91 −2.96 (m, 1H), 3.10–3.19 (m, 2H), 3.24–3.37 (m, 4H), 5.05 (s, 1H), 6.76 (brt, 1H), 7.08–7.09 (brd, 2H), 7.12–7.21 (m, 3H), 7.23 (s, 1H), 7.28 (s, 1H), 7.39 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.70, 4.81, 9.43, 15.04, 38.69, 43.40, 50.40, 66.20, 68.70, 69.13, 80.91, 126.23, 127.60, 128.74, 130.23, 133.58, 134.71, 137.02, 155.44, 175.06.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-hydroxyethylamide (17)
71% yield; C23H26ClN3O2; MW 411.94 g/mol; HRMS calc. 412.1792; found 412.1758 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.00–0.07 (m, 2H), 0.45–0.50 (m, 1H), 0.56–0.61 (m, 1H), 0.85–0.90 (m, 1H), 2.26 (d, J= 12 Hz, 1H), 2.36 (d, J= 12 Hz, 1H), 2.70 (dd, J= 12 Hz & 6 Hz, 1H), 2.92–2.98 (m, 1H), 3.18 (dd, J= 12 Hz & 6 Hz, 1H), 3.31–3.42 (m, 3H), 5.06 (s, 1H), 6.85 (brt, 1H), 7.12–7.16 (brd, 2H), 7.18–7.26 (m, 4H), 7.28 (s, 1H), 7.40 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.74, 4.84, 9.42, 42.31, 43.41, 50.42, 61.92, 69.08, 80.68, 126.51, 127.74, 128.42, 130.40, 133.75, 134.41, 137.10, 155.50, 176.07.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-(morpholin-4-yl)propylamide (18)
66% yield; C28H35ClN4O2; MW 495.07 g/mol; HRMS calc. 495.2527; found 495.2479 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.05 (m, 2H), 0.43–0.49 (m, 1H), 0.54–0.59 (m, 1H), 0.84–0.91 (m, 1H), 1.26–1.34 (m, 1H), 1.38–1.44 (m, 1H), 2.03–2.08 (m, 1H), 2.12–2.29 (m, 6H), 2.37 (d, J= 12 Hz, 1H), 2.69 (dd, J= 12 Hz & 6 Hz, 1H), 2.97–3.02 (m, 1H), 3.16 (dd, J= 18 Hz & 6 Hz, 1H), 3.26–3.32 (m, 1H), 3.62–3.67 (m, 2H), 3.69–3.74 (m, 2H), 5.02 (s, 1H), 7.06 (d, J= 12 Hz, 2H), 7.11–7.18 (m, 4H), 7.20 (s, 1H), 7.28 (s, 1H), 7.39 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.71, 4.86, 9.44, 25.13, 38.16, 43.43, 50.45, 53.45, 57.18, 66.86, 69.28, 80.90, 126.16, 127.50, 128.36, 130.37, 133.50, 134.82, 137.21, 155.07, 174.97.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-(morpholin-4-yl)ethylamide (19)
75% yield; C27H33ClN4O2; MW 481.04 g/mol; HRMS calc. 481.2370; found 481.2400 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.06 (m, 2H), 0.44–0.48 (m, 1H), 0.54–0.60 (m, 1H), 0.83–0.90 (m, 1H), 1.99–2.05 (m, 1H), 2.13–2.18 (m, 2H), 2.19–2.31 (m, 4H), 2.35 (d, J= 12 Hz, 1H), 2.68 (dd, J= 12 Hz & 6 Hz, 1H), 3.09–3.20 (m, 3H), 3.55–3.62 (m, 4H), 5.06 (s, 1H), 6.76 (brt, 1H), 7.07–7.10 (brd, 2H), 7.11–7.13 (m, 1H), 7.16–7.18 (m, 2H), 7.23 (s, 1H), 7.29 (s, 1H), 7.39 (brs, 2H); 13C-NMR (CDCl3, 150 MHz): δ 6.29, 4.83, 9.44, 35.34, 43.54, 50.41, 53.28, 56.67, 66.84, 69.06, 80.93, 126.17, 127.59, 128.73, 130.28, 133.58, 134.70, 137.15, 155.29, 175.07.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-(pyridin-4-yl)ethylamide (20)
58% yield; C28H29ClN4O; MW 473.02 g/mol; HRMS calc. 473.2108; found 473.2088 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.01–0.05 (m, 2H), 0.44–0.48 (m, 1H), 0.54–0.59 (m, 1H), 0.81–0.87 (m, 1H), 2.22 (d, J= 12 Hz, 1H), 2.31–2.39 (m, 2H), 2.54–2.61 (m, 1H), 2.68 (dd, J= 12 Hz & 6 Hz, 1H), 3.13–3.22 (m, 2H), 3.37–3.43 (m, 1H), 5.01 (s, 1H), 6.60 (brt, 1H), 6.88 (d, J= 6 Hz, 2H), 7.11 (d, J= 6 Hz, 2H), 7.14–7.23 (4H, m, 4H), 7.28 (s, 1H), 7.40 (brs, 2H), 8.45 (d, J= 6 Hz, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.70, 4.81, 9.41, 34.86, 39.04, 43.30, 50.38, 69.18, 80.77, 123.96, 126.42, 127.70, 128.67, 130.39, 133.71, 134.45, 136.95, 147.89, 149.81, 155.49, 175.18.
4-Benzyl-5-(4-chlorophenyl)-1-cyclopropylmethyl-4,5-dihydro-1H–imidazole-4-carboxylic acid (pyridin-3-yl)methylamide (21)
71% yield; C27H27ClN4O; MW 459.00 g/mol; HRMS calc. 459.1952; found 459.1961 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ 0.02–0.07 (m, 2H), 0.45–0.50 (m, 1 H), 0.56–0.61 (m, 1H), 0.84–0.91 (m, 1H), 2.25 (d, J= 12 Hz, 1H), 2.40 (d, J= 12 Hz, 1H), 2.71 (dd, J= 12 Hz & 6 Hz, 1H), 3.19 (dd, J= 12 Hz & 6 Hz, 1H), 4.16 (dd, J= 12 Hz & 6 Hz, 1H), 4.33 (dd, J= 12 Hz & 6 Hz, 1H), 5.08 (s, 1H), 6.86 (brt, 1H), 7.04–7.09 (m, 3H), 7.10–7.19 (m, 4H), 7.23 (s, 1H), 7.28 (s, 1H), 7.41 (brs, 2H), 8.26 (brs, 1H), 8.46 (brd, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.67, 4.84, 9.45, 40.48, 43.23, 50.38, 69.29, 80.91, 123.38, 126.39, 127.80, 128.71, 130.26, 133.27, 133.77, 134.36, 135.42, 136.78, 148.65, 149.16, 155.69, 175.24.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid methyl ester (3)
70% yield; C19H25ClN2O2; MW 348.8670; HRMS NA; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.03 (m, 2H), 0.38–0.47 (m, 1H), 0.48–0.53 (m, 1H), 0.57 (d, J= 6 Hz, 3H), 0.73 (d, J= 6 Hz, 3H), 0.78–0.85 (m, 1H), 1.07 (dd, J= 12 Hz & 6 Hz, 1H), 1.18 (dd, J= 12 Hz & 6 Hz, 1H), 1.54–1.68 (m, 1H), 2.54 (dd, J= 12 Hz & 6 Hz, 1H), 3.10 (dd, J= 12 Hz & 6 Hz, 1H), 3.78 (m, 3H), 4.89 (s, 1H), 7.24–7.31 (m, 4H), 7.63 (d, J= 12 Hz, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.58, 3.36, 4.72, 9.18, 22.82, 24.60, 44.25, 44.87, 49.94, 50.36, 69.76, 79.67, 128.40, 128.46, 133.83, 155.85, 161.38, 171.99, 175.74.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-methoxypropylamide (22)
66% yield; C22H32ClN3O2; MW 405.97 g/mol; HRMS NA; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.03 (m, 2H), 0.43–0.48 (m, 1H), 0.53–0.58 (m, 1H), 0.69 (d, J= 6 Hz, 3H), 0.79–0.87 (m, 4H), 0.91 (dd, J= 12 Hz & 6 Hz, 1H), 1.14 (dd, J= 12 Hz & 6 Hz, 1H), 1.64–1.69 (m, 1H), 1.80–1.87 (m, 2H), 2.65 (dd, J= 12 Hz & 6 Hz, 1H), 3.10 (dd, J= 12 Hz & 6 Hz, 1H), 3.33–3.38 (m, 4H), 3.42–3.48 (m, 3H), 4.92 (s, 1H), 7.24 (s, 1H), 7.28–7.34 (m, 3H), 7.44–7.45 (m, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.69, 4.76, 9.38, 23.59, 24.37, 24.69, 29.30, 29.70, 36.92, 45.10, 50.33, 58.72, 70.09, 70.68, 79.73, 128.44, 133.47, 134.33, 155.55, 176.04.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-ethoxypropylamide (23)
58% yield; C23H34ClN3O2; MW 420.00 g/mol; HRMS calc. 420.2418; found 420.2429 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.03 (m, 2H), 0.42–0.46 (m, 1H), 0.52–0.56 (m, 1H), 0.69 (d, J= 6 Hz, 3H), 0.80–0.85 (m, 4H), 0.88 (dd, J= 12 Hz & 6 Hz, 1H), 1.11 (dd, J= 12 Hz & 6 Hz, 1H), 1.22 (t, J= 6 Hz, 3H), 1.63–1.70 (m, 1H), 1.79–1.86 (m, 2H), 2.63 (dd, J= 18 Hz & 6 Hz, 1H), 3.08 (dd, J= 12 Hz & 6 Hz, 1H), 3.33–3.38 (m, 1H), 3.41–3.52 (m, 5H), 4.87 (s, 1H), 7.16 (s, 1H), 7.31–7.32 (m, 4H); 13C-NMR (CDCl3, 150 MHz): δ 2.65, 4.76, 9.40, 15.21, 23.65, 24.42, 24.69, 29.44, 37.07, 45.17, 50.30, 66.36, 68.63, 70.02, 79.97, 128.40, 133.33, 134.61, 155.43, 176.24.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-methoxyethylamide (24)
76% yield; C21H30ClN3O2; MW 391.95 g/mol; HRMS calc. 392.2105; found 392.2099 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.04-0.03 (m, 2H), 0.41–0.46 (m, 1H), 0.50–0.56 (m, 1H), 0.69 (d, J= 6 Hz, 3H), 0.80–0.85 (m, 4H), 0.89 (dd, J= 12 Hz & 6 Hz, 1H), 1.10 (dd, J= 12 Hz & 6 Hz, 1H), 1.65–1.72 (m, 1H), 2.62 (dd, J= 12 Hz & 6 Hz, 1H), 3.09 (dd, J= 12 Hz & 6 Hz, 1H), 3.33–3.38 (m, 4H), 3.42–3.44 (m, 1H), 3.49–3.53 (m, 1H), 3.56–3.62 (m, 1H), 4.85 (s, 1H), 7.17 (s, 1H), 7.31–7.35 (m, 4H); 13C-NMR (CDCl3, 150 MHz): δ 2.62, 4.77, 9.37, 23.57, 24.41, 24.66, 38.96, 45.22, 50.31, 58.78, 70.01, 71.10, 79.96, 128.36, 133.32, 134.62, 155.58, 176.43.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-ethoxyethylamide (25)
60% yield; C22H32ClN3O2; MW 405.97 g/mol; HRMS calc. 406.2261; found 406.2225 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.04-0.03 (m, 2H), 0.41–0.46 (m, 1H), 0.50–0.56 (m, 1H), 0.69 (d, J= 6 Hz, 3H), 0.79–0.85 (m, 4H), 0.88 (dd, J= 12 Hz & 6 Hz, 1H), 1.10 (dd, J= 12 Hz & 6 Hz, 1H), 1.20 (t, J= 6 Hz, 3H), 1.66–1.72 (m, 1H), 2.62 (dd, J= 18 Hz & 6 Hz, 1H), 3.08 (dd, J= 18 Hz & 6 Hz, 1H), 3.36–3.41 (m, 1H), 3.47–3.56 (m, 4H), 3.57–3.62 (m, 1H), 4.85 (s, 1H), 7.16 (s, 1H), 7.28–7.39 (m, 4H); 13C-NMR (CDCl3, 150 MHz): δ 2.63, 2.76, 9.38, 15.12, 23.63, 24.43, 24.66, 39.12, 41.80, 45.24, 50.30, 66.44, 68.99, 70.00, 80.00, 128.35, 133.30, 134.66, 155.51, 176.41.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-hydroxyethylamide (26)
80% yield; C20H28ClN3O2; MW 377.92 g/mol; HRMS calc. 378.1948; found 378.1947 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.05 (m, 2H), 0.44–0.49 (m, 1H), 0.53–0.59 (m, 1H), 0.70 (d, J= 6 Hz, 3H), 0.77–0.88 (m, 4H), 0.94 (dd, J= 12 Hz & 6 Hz, 1H), 1.12 (dd, J= 12 Hz & 6 Hz, 1H), 1.64–1.71 (m, 1H), 2.64 (dd, J= 12 Hz & 6 Hz, 1H), 3.09 (dd, J= 18 Hz & 6 Hz, 1H), 3.40–3.47 (m, 1H), 3.48–3.55 (m, 1H), 3.73–3.80 (m, 2H), 4.88 (s, 1 H), 7.20 (s, 1H), 7.22–7.31 (m, 3H), 7.38–7.39 (m, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.69, 4.80, 9.34, 23.49, 24.36, 24.72, 42.79, 45.05, 50.36, 62.65, 70.18, 79.68, 128.49, 133.58, 134.13, 155.77, 177.71.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 3-(morpholin-4-yl)propylamide (27)
68% yield; C25H37ClN4O2; MW 461.05 g/mol; HRMS calc. 461.2683; found 461.2680 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.02 (m, 2H), 0.41–0.46 (m, 1H), 0.51–0.53 (m, 1H), 0.68 (d, J= 6 Hz, 3H), 0.79–0.84 (m, 4H), 0.87 (dd, J= 12 Hz & 6 Hz, 1H), 1.08 (dd, J= 12 Hz & 6 Hz, 1H), 1.63–1.69 (m, 1H), 1.70–1.76 (m, 2H), 2.40–2.48 (m, 6H), 2.62 (dd, J= 14 Hz & 8 Hz, 1H), 3.07 (dd, J= 12 Hz & 6 Hz, 1H), 3.26–3.31 (m, 1H), 3.40–3.48 (m, 1H), 3.70–3.79 (m, 4H), 4.84 (s, 1H), 7.14 (s, 1 H), 7.27–7.32 (m, 3H), 7.46–7.48 (m, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.66, 4.77, 9.38, 20.74, 23.73, 24.42, 24.93, 25.90, 37.85, 45.27, 50.31, 53.81, 57.06, 66.92, 70.01, 80.03, 128.36, 133.30, 134.66, 155.27, 176.32.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-(morpholin-4-yl)ethylamide (28)
74% yield; C24H35ClN4O2; MW 447.03 g/mol; HRMS calc. 447.2527; found 447.2506 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.07 (m, 2H), 0.40–0.45 (m, 1H), 0.50–0.55 (m, 1H), 0.68 (d, J= 6.6 Hz, 3H), 0.76–0.82 (m, 4H), 0.87 (dd, J= 12 Hz & 6 Hz, 1H), 1.09 (dd, J= 12 Hz & 6 Hz, 1H), 1.63–1.70 (m, 1H), 2.40–2.54 (m, 6H), 2.61 (dd, J= 14.4 Hz & 7.8 Hz, 1H), 3.08 (dd, J= 13.8 Hz & 6 Hz, 1H), 3.26–3.34 (m, 1H), 3.47–3.54 (m, 1H), 3.65–3.77 (m, 4H), 4.86 (s, 1H), 7.17 (s, 1H), 7.22–7.26 (m, 4H); 13C-NMR (CDCl3, 150 MHz): δ 2.64, 4.76, 9.39, 24.65, 35.74, 45.27, 50.27, 53.44, 57.24, 66.95, 69.98, 80.05, 128.39, 133.34, 134.59, 155.41, 176.38.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid 2-(pyridin-4-yl)ethylamide (29)
74% yield; C25H31ClN4O; MW 439.01 g/mol; HRMS calc. 439.2265; found 439.2237 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.02–0.03 (m, 2H), 0.43–0.48 (m, 1H), 0.53–0.58 (m, 1H), 0.66 (d, J= 6 Hz, 3H), 0.76–0.83 (m, 4H), 0.89 (dd, J= 12 Hz & 6 Hz, 1H), 1.10 (dd, J= 12 Hz & 6 Hz, 1H), 1.53–1.60 (m, 1H), 2.62 (dd, J= 12 Hz & 6 Hz, 1H), 2.84–2.96 (m, 2H), 3.09 (dd, J= 12 Hz & 6 Hz, 1H), 3.56–3.68 (m, 2H), 4.84 (s, 1H), 7.16–7.18 (m, 3H), 7.24–7.30 (m, 2H), 7.34 (brd, J= 12 Hz, 2H), 8.53 (brd, J= 6 Hz, 2H); 13C-NMR (CDCl3, 150 MHz): δ 2.69, 4.77, 9.33, 23.73, 24.40, 24.61, 29.70, 35.05, 39.26, 45.05, 50.32, 70.07, 79.84, 124.13, 128.45, 133.53, 134.20, 147.94, 149.86, 155.54, 176.39.
5-(4-Chlorophenyl)-1-cyclopropylmethyl-4-isobutyl-4,5-dihydro-1H–imidazole-4-carboxylic acid (pyridin-3-yl)methylamide (30)
54% yield; C24H29ClN4O; MW 424.98 g/mol; HRMS calc. 425.2108; found 425.2096 [M+H]+; 1H-NMR (CDCl3, 600 MHz): δ −0.03-0.02 (m, 2H), 0.42–0.47 (m, 1H), 0.53–0.57 (m, 1H), 0.63 (d, J= 6 Hz, 3H), 0.78 (d, J= 6 Hz, 3H), 0.79–0.85 (m, 1H), 0.91 (dd, J= 12 Hz & 6 Hz, 1H), 1.13 (dd, J= 12 Hz & 6 Hz, 1H), 1.60–1.65 (m, 1H), 2.63 (dd, J= 12 Hz & 6 Hz, 1H), 3.09 (dd, J= 12 Hz & 6 Hz, 1H), 4.49 (dd, J= 12 Hz & 6 Hz, 1H), 4.91 (s, 1H), 7.18–7.20 (brs, 1H), 7.24–7.26 (m, 3H), 7.33–7.34 (m, 3H), 7.66 (brd, J= 6Hz, 1H), 8.53 (d, J= 6 Hz, 1H), 8.57 (s, 1H); 13C-NMR (CDCl3, 150 MHz): δ 2.62, 4.77, 9.38, 23.63, 24.40, 24.62, 40.81, 45.04, 50.30, 70.07, 79.87, 123.45, 128.52, 133.61, 133.83, 135.49, 135.64, 148.91, 149.33, 155.74, 176.41.
Supplementary Material
Table 1.
Structures and yields of synthesized imidazolinemethylesters by the O-3CR and subsequent amidation.
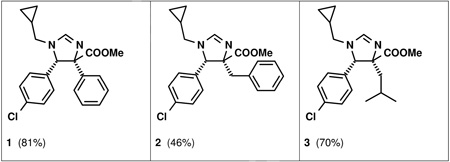 |
Table 2.
Structures and yields of synthesized amide derivatives.
| Amine input | 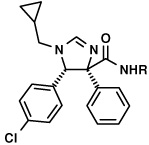 |
 |
 |
|---|---|---|---|
| 4 (69%) | 13 (75%) | 22 (66%) | |
| 5 (84%) | 14 (62%) | 23 (58%) | |
| 6 (66%) | 15 (79%) | 24 (76%) | |
| 7 (69%) | 16 (69%) | 25 (60%) | |
| 8 (71%) | 17 (71%) | 26 (80%) | |
| 9 (69%) | 18 (66%) | 27 (68%) | |
| 10 (68%) | 19 (75%) | 28 (74%) | |
| 11 (67%) | 20 (58%) | 29 (74%) | |
 |
12 (64%) | 21 (71%) | 30 (54%) |
Acknowledgements
AD is grateful to the Drug Discovery Institute of The University of Pittsburgh for providing an excellent working environment and to the National Cancer Institute for their interest in my p53/mdm2 project, allowing me to participate in a Rapid Access to NCI Discovery Resources (R-A-N-D) program. Part of this work was funded by CMCR grant U19AI068021. This work has also been supported by the Deutsche Krebshilfe (German Cancer Aid), Grant 108354.
Footnotes
Dedicated to Sir Jack Baldwin on the occasion of his 81st birthday
Supporting Information: Protocol for water solubility measurements and 1H and 13C NNMR spectra of all new compounds.
REFERENCES
- 1.Vassilev LT. Trends. Mol. Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Canc. Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Dömling A. Curr. Op. Chem. Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]; b Holak T. 2009 in press. [Google Scholar]
- 4.Czarna A, et al. 2009 unpublished. [Google Scholar]
- 5.a Bon RS, Hong C, Bouma MJ, Schmitz RF, De Kanter FJJ, Lutz M, Spek AL, Orru RVA. Org. Lett. 2003;5:3759–3762. doi: 10.1021/ol035521g. [DOI] [PubMed] [Google Scholar]; b Bon RS, Van Vliet B, Sprenkels NE, Schmitz RF, De Kanter FJJ, Stevens CV, Swart M, Bickelhaupt FM, Groen MB, Orru RVA. J. Org. Chem. 2005;70:3542–3553. doi: 10.1021/jo050132g. [DOI] [PubMed] [Google Scholar]; c Elders N, Schmitz RF, De Kanter FJJ, Ruijter E, Groen MB, Orru RVA. J. Org. Chem. 2007;72:6135–6142. doi: 10.1021/jo070840x. [DOI] [PubMed] [Google Scholar]; d Bon RS, Sprenkels NE, Koningstein MM, Schmitz RF, de Kanter FJJ, Dömling A, Groen MB, OrruRomano VA. Org. Biomol. Chem. 2008;6:130–137. doi: 10.1039/b713065a. [DOI] [PubMed] [Google Scholar]
- 6.The modeling pictures were rendered using PYMOL
- 7.a Barrow RA, Hemscheidt T, Liang J, Paik S, Moore RE, Tius ME. J. Am. Chem. Soc. 1995;117:2479–2490. [Google Scholar]; b Smith MB, March J. Advanced Organic Reactions: Reactions, Mechanisms and Structure. 5th ed. New York: J. Wiley & Sons; 2001. p. 510. [Google Scholar]; c Perreux L, Loupy A, Delmotte M. Tetrahedron. 2003;59:2185–2189. and references cited wherein. [Google Scholar]; d Riviere-Baudet M, Morere A, Dias M. Tetrahedron Lett. 1992;33:6453–6456. [Google Scholar]; e Weinreb SM, Anderson GT, Nylund CSIn. In: Encyclopedia of Reagents for Organic Synthesis. Paquette LA, editor. Vol. 3. Wiley: Chichester; 1995. p. 1997. [Google Scholar]; f Houghton RP, Williams CS. Tetrahedron Lett. 1967;8:3929–3931. [Google Scholar]
- 8.Dömling A. Chem. Rev. 2006;106:17–89. doi: 10.1021/cr0505728. [DOI] [PubMed] [Google Scholar]
- 9.Dömling A, Beck B, Fuchs T, Yazbak A. J. Comb. Chem. 2006;8:872–880. doi: 10.1021/cc060068w. [DOI] [PubMed] [Google Scholar]
- 10.Sabot C, Kumar KA, Meunier S, Mioskowski C. Tetrahedron Lett. 2007;48:3863–3866. [Google Scholar]
- 11.Wei W, Dömling A. J. Comb. Chem. 2009 [Google Scholar]
- 12.Bergstrom CAS, Strafford M, Lazorova L, Avdeef A, Luthman K, Artursson P. J. Med. Chem. 2003;46:558–570. doi: 10.1021/jm020986i. [DOI] [PubMed] [Google Scholar]
- 13.van der Waterbeemd H, Smith DA, Beaumont K, Walker DK. J. Med. Chem. 2001;44:1312–1331. doi: 10.1021/jm000407e. [DOI] [PubMed] [Google Scholar]
- 14.Bhattachar SN, Deschenes L, Wesley JA. Drug Discovery Today. 2006;11:1012–1018. doi: 10.1016/j.drudis.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 15.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.