Abstract
Autophagy is activated in response to cellular stressors and mediates lysosomal degradation and recycling of cytoplasmic material and organelles as a temporary cell survival mechanism. Defective autophagy is implicated in human pathology, as disruption of protein and organelle homeostasis enables disease-promoting mechanisms such as toxic protein aggregation, oxidative stress, genomic damage and inflammation. We previously showed that autophagy-defective immortalized mouse mammary epithelial cells (iMMECs) are susceptible to metabolic stress, DNA damage and genomic instability. We now report that autophagy deficiency was associated with ER and oxidative stress, and deregulation of p62-mediated keratin homeostasis in mammary cells and allograft tumors and in mammary tissues from genetically engineered mice. In human breast tumors, high phospho(Ser73)-K8 levels inversely correlated with Beclin 1 expression. Thus, autophagy preserves cellular fitness by limiting ER and oxidative stress, a function potentially important in autophagy-mediated suppression of mammary tumorigenesis. Furthermore, autophagy regulates keratin homeostasis in the mammary gland via a p62-dependent mechanism. High phospho(Ser73)-K8 expression may be a marker of autophagy functional status in breast tumors and, as such, could have therapeutic implications for breast cancer patients.
Keywords: autophagy, keratin homeostasis, breast cancer, ER stress, p62
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is a cellular self-consumption process whereby cytoplasm, long-lived proteins and organelles are engulfed in double-membrane vesicles and delivered to lysosomes for degradation. Basal autophagy maintains cellular homeostasis by eliminating damaged proteins and organelles. Autophagy defects have been implicated in the pathogenesis of diseases, such as myopathy, neuronal degeneration, microbial infection, inflammatory bowel disease, aging and cancer. In addition to its basal function, autophagy is induced by nutrient deprivation, metabolic stress, ER stress, radiation and chemotherapy, mostly functioning as a temporary survival mechanism (1).
Apoptosis inactivation occurs frequently in tumors, indicating that aberrant cell survival contributes to cancer progression. Loss of a survival pathway, such as autophagy, might have been expected to undermine tumorigenesis; however, the essential autophagy regulator beclin 1 is a haploinsufficient tumor suppressor (2, 3) arguing against this simplistic scenario. Recent studies described non-cell-autonomous and cell-autonomous mechanisms for autophagy-mediated tumor suppression (4, 5). Autophagy may suppress tumorigenesis by limiting necrosis-associated inflammation (6), and by preserving genome integrity and cellular fitness, as autophagy defects result in accumulation of ubiquitin-positive protein aggregates in neurons and liver (7); deformed mitochondria (8) and peroxisomes (9); DNA damage and genomic instability in tumor cells (4, 10); accumulation of oxidative and ER stress-sensing proteins and reactive oxygen species (ROS) in kidney epithelial cells (5).
Since autophagy mediates protein degradation, we used a proteomic approach to investigate how beclin 1+/+ and beclin 1+/− iMMECs respond to metabolic stress. We found that ER chaperones, oxidative stress-mitigating mitochondrial proteins, enzymes involved in glucose metabolism, and cytoskeletal proteins were upregulated in mammary cells under stress, preferentially in beclin 1+/− iMMECs. Defective autophagy was also associated with accumulation of p62, a scaffolding protein involved in cell signaling, receptor internalization and protein turnover (11) during metabolic stress and recovery. Furthermore, autophagy defects deregulated keratin homeostasis in mammary cells and phospho(Ser73)-K8, which is involved in stress-induced keratin remodeling, accumulated in metabolically stressed iMMECs in a p62-dependent manner upon autophagy inhibition. Higher levels of p62, keratins and ER chaperones were also observed in autophagy-deficient mammary tissues and beclin 1+/− iMMEC-generated tumors. Evaluation of a human breast cancer tissue microarray (TMA) revealed that ER chaperone, p62 and keratin upregulation reliably discriminated tumors from normal adjacent tissue, but only high phospho-K8 levels inversely correlated with Beclin 1 expression. Thus, elevated phospho-K8 may be an epithelial cell marker of autophagy deficiency in breast tumors.
MATERIALS AND METHODS
Stable cell line generation and culture conditions
Primary mouse mammary epithelial cells from beclin 1+/+ and beclin 1+/− mice were immortalized to generate iMMEC cell lines, which were engineered to stably express Bcl-2 as previously described (10, 12). Metabolic stress was induced as previously described (12). NAC (Sigma-Aldrich) was used at 1 nM concentration.
Western blotting, IF and IHC
Antibodies against the following antigens were used: GRp78 (Stressgen); ATF6α, Beclin 1 (BECN1 H-300), calnexin, SOD2 (Santa Cruz); aconitase (Atlas); K19 (GeneTex); PDI (Sigma); p62 (Biomol); K8/18, K17, peroxiredoxin 3, PGAM1, phospho(Ser73)-K8 (Abcam); actin (Oncogene); β-catenin (Zymed); K8 (University of Iowa Developmental Studies Hybridoma Bank); LC3B (Cell Signaling).
Three-dimensional (3D) morphogenesis
3D-culture of iMMECs and IF on mammary acini generated by Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs were performed as previously described (10, 12).
Generation of mammary gland-specific atg7-deficient mice
atg7Flox/Flox (atg7F/F) mice were obtained from Dr. Komatsu and crossed to WAP-cre mice obtained from the MMHCC Repository. atg7F/+;WAP-cre progeny were crossed to atg7F/+ mice, and atg7+/+;WAP-cre, atg7F/+;WAP-cre and atg7F/F;WAP-cre mice were identified by genotyping. Female mice of the above genotypes were bred with C57BL/6 males and Cre recombinase was activated under the WAP promoter after two rounds of pregnancy and lactation.
IHC staining quantification and statistical analysis
Beclin 1, GRp78, GRp170, p62, K8, K17, and phospho(Ser73)-K8 levels were evaluated by IHC in a human breast cancer TMA. Protein expression, i.e. staining intensity, in epithelial cells was manually quantified by two study investigators, Drs. Barnard and Karantza. The following scale was used: 0–1+, 2+ and 3+ staining intensity corresponded to low, moderate, and high protein levels, respectively. A two-sided exact Wilcoxon test was used to compare Beclin 1 levels in human breast tumors to NAT. A two-sided test was used to compare high levels (3+ staining) of p62, GRp170, GRp78 or phospho(Ser73)-K8 in breast tumors to NAT. Correlation between absolute Beclin 1 levels and high expression (3+ staining) of p62, GRp170, GRp78 or phospho(Ser73)-K8 in breast tumors was examined using logistic regression.
RESULTS
Elevated ER stress in autophagy-deficient mammary cells under metabolic stress and in recovery
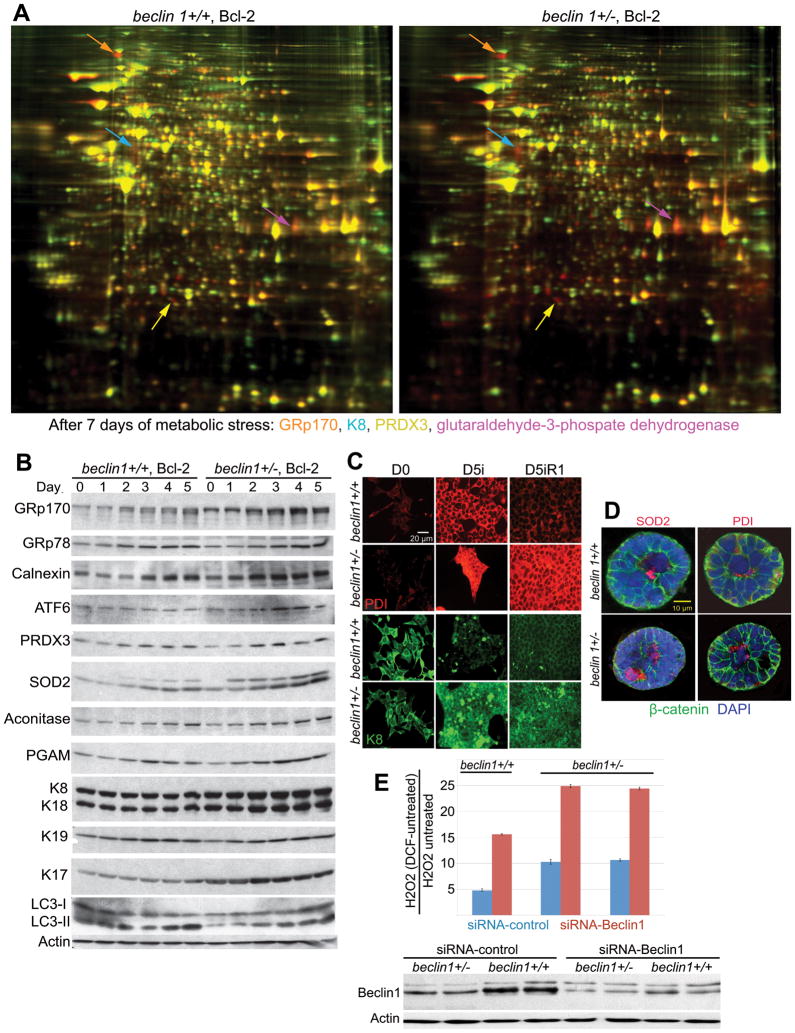
beclin 1 monoallelic loss renders iMMECs susceptible to metabolic stress, DNA damage and genomic instability, and enhances mammary tumorigenesis (10). Since autophagy is a lysosomal protein degradation pathway, we used proteomics to investigate the role of autophagy in mammary cell response to metabolic stress and assess possible mechanisms of stress management failure. iMMECs were engineered to express Bcl-2, as autophagy assessment is facilitated in an apoptosis-defective background and Bcl-2 expression is functionally equivalent to Bax and Bak deficiency regarding autophagy modulation and tumorigenesis (5, 6, 10, 13). Apoptosis-defective autophagy-competent beclin 1+/+ and autophagy-defective beclin 1+/− iMMECs (10) were exposed to metabolic stress (glucose and oxygen limitation), and differential protein expression after 4 and 7 days of stress was determined by two dimensional fluorescence difference gel electrophoresis (2D-DIGE). 106 differentially regulated proteins were identified by mass spectroscopy. Representative images of differential protein expression (red, upregulated; green, downregulated) for beclin 1+/+ and beclin 1+/− iMMECs after 7 days of stress are presented in Fig. 1A and quantified in Supplementary Table 1.
Figure 1. Metabolic stress causes preferential upregulation of ER chaperones, mitochondrial enzymes, metabolism-related proteins and keratins in autophagy-deficient mammary cells.
A, 2-DIGE gels showing differential regulation of ER chaperones (GRp170, orange arrows), mitochondrial enzymes (PRDX3, yellow arrows), metabolism-related proteins (GPDH, magenta arrows) and keratins (K8, blue arrows) in Bcl-2-expressing beclin 1+/+ (left panel) and beclin 1+/− (right panel) iMMECs in response to metabolic stress (7 days). Total protein from unstressed or metabolically stressed iMMECs were labeled with Cy3 (unstressed) or Cy5 (stressed) and analyzed by 2-DIGE. Images show 2-DIGE gels with proteins that are induced (red), repressed (green) or unchanged (yellow) under stress. 106 differentially expressed protein spots were identified by mass spectroscopy.
B, Western blots showing levels of ER chaperones (GRp170, GRp78, calnexin), ATF6, mitochondrial enzymes (PRDX3, SOD2, aconitase), glycolytic enzyme (PGAM) and keratins (K8/18, K17 and K19) in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs under metabolic stress for 0–5 days.
C, PDI (red) and K8 (green) IF in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs following 5 days metabolic stress and 1 day of recovery.
D, SOD2 and PDI IF (red) in mammary acini generated by Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs. β-catenin (green) was used as an epithelial cell marker and DAPI (blue) for counterstaining nuclei.
E, ROS (H2O2) measurements in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs using beclin 1 versus scramble (control) siRNA.
Proteins upregulated under stress included ER chaperones [Glucose Regulated protein (GRp) 170, GRp94, GRp78, Protein Disulfide Isomerase (PDI)], mitochondrial proteins with antioxidant activity [peroxiredoxin (PRDX) 3, superoxide dismutase (SOD) 2], enzymes involved in glucose metabolism (pyruvate kinase, enolase, phosphoglycerate mutase, triose phosphate isomerase), and cytoskeletal proteins, including keratins and annexins. Upregulation was higher in beclin 1+/− iMMECs for all above protein families (Supplementary Table 1), indicating that defective autophagy is associated with increased ER and oxidative stress, elevated metabolic demands and cytoskeletal alterations in mammary cells under stress. Similarly to iMMECs, autophagy-deficient immortalized baby mouse kidney (iBMK) cells showed preferential induction of ER chaperones under metabolic stress (5) and the microsomal fractions of atg7-deficient livers had high PDI, GRp78 and GRp94 levels (14), indicating that defective autophagy is associated with elevated ER stress, independently of tissue type and mode of autophagy defect.
Since protein level estimation by spot volume ratios is complicated by representation of individual proteins by multiple spots in 2D-DIGE, results were validated by Western blotting. ER chaperones (GRp170, calnexin), the unfolded protein response (UPR) mediator (ATF6), mitochondrial enzymes (PRDX3, SOD2, aconitase), glycolytic enzymes (PGAM) and keratins (K8/18, K19, K17) were preferentially upregulated in beclin 1+/− iMMECs under stress (Fig. 1B), whereas stress-induced processing of LC3-I to LC3-II was attenuated in these cells, consistently with their previously reported decreased autophagy potential (10). Other markers of UPR were also examined (Suppl. Fig. 1A) and elevated ER stress in autophagy-deficient iMMECs was confirmed by accumulation of stress-induced phospho-eIF2α and by increased XBP-1 splicing under normal growth conditions and upon metabolic stress induction (Suppl. Fig. 1B).
PDI and K8 expression in beclin 1+/+ and beclin 1+/− iMMECs under stress and recovery was also examined by immunofluorescence (IF). PDI expression dramatically increased in both autophagy-competent and -defective iMMECs under stress (Fig. 1C). Upon recovery (in normoxia and regular growth medium), PDI expression decreased to near-baseline levels in beclin 1+/+ iMMECs, but remained high in beclin 1+/− iMMECs, indicating that autophagy-defective mammary cells have persistent need for PDI function during recovery due to remaining unfolded protein load, and thus revealing that autophagy mediates misfolded protein clearance during recovery from metabolic stress. Expression of keratins 18 and 19 increased upon stress in beclin 1+/+ iMMECs and to a higher degree in beclin 1+/− iMMECs (Supplementary Table 1, Fig. 1B), whereas K17 was significantly upregulated only in stressed autophagy-defective iMMECs (Supplementary Table 1). In autophagy-competent iMMECs, K8 levels remained stable (Fig. 1B) or decreased (Fig. 1C) with stress and slowly recovered after 1 day of normal growth conditions, whereas in autophagy-defective iMMECs, K8 levels greatly increased under stress and remained elevated in recovery (Fig. 1C).
The response of beclin 1+/+ and beclin 1+/− iMMECs to stress was also examined by three dimensional (3D)-morphogenesis. SOD2 and PDI levels were upregulated in the acinar center, particularly in beclin 1+/− iMMECs (Fig. 1D), indicating that central acinar cells, which are under increased metabolic stress (10), are also under oxidative stress. Higher oxidative stress levels in autophagy-defective iMMECs were confirmed by ROS measurements in beclin 1+/+ and beclin 1+/− iMMECs and upon beclin1 knockdown (Fig. 1E).
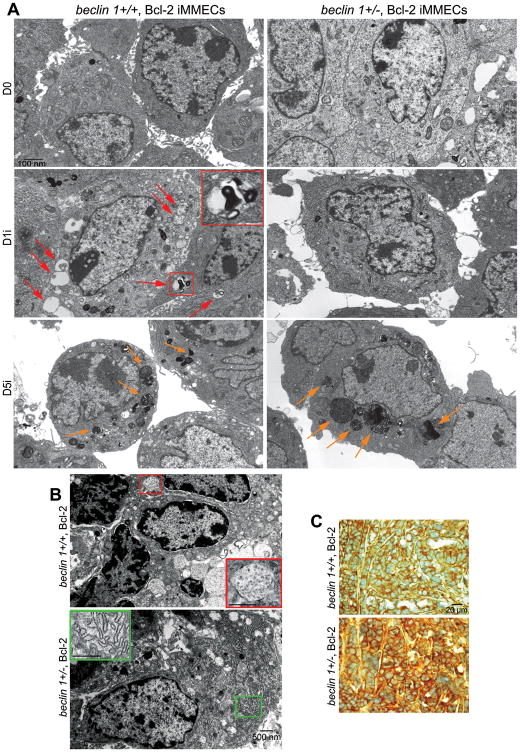
Metabolic stress-induced morphological changes in beclin 1+/+ and beclin 1+/− iMMECs were examined by transmission electron microscopy (TEM) (Fig. 2A). Autophagosomes were apparent in beclin 1+/+ iMMECs after 1 day of metabolic stress, whereas beclin 1+/− iMMECs were defective in autophagosome formation and progressively filled with electron-dense material suggestive of “cellular garbage”, due to decreased clearance and/or increased protein aggregate formation, similarly to autophagy-deficient iBMK cells (5).
Figure 2. Metabolic stress causes autophagy induction in beclin 1+/+ iMMECs, preferential accumulation of cytoplasmic “garbage” in beclin 1+/− iMMECs and elevated ER stress in autophagy-deficient mammary tumors.
A, Representative electron micrographs of Bcl-2-expressing beclin 1+/+ (left column) and beclin 1+/− (right column) iMMECs following 5 days of metabolic stress. Arrows point to autophagosomes (red) in beclin 1+/+ iMMECs and to electron-dense cytoplasmic material (yellow) preferentially accumulating in beclin 1+/− iMMECs.
B, Representative electron micrographs of apoptosis-defective beclin 1+/+ (top panel) and beclin 1+/− (bottom panel) iMMEC-generated mammary tumors. An autophagosome is magnified in the red insert (top panel), whereas prominent ER expansion is presented in the green insert (bottom panel).
C, GRp170 IHC in mammary tumors generated by beclin 1+/+ (top panel) and beclin 1+/− (bottom panel) iMMECs.
Elevated ER stress in autophagy-deficient mammary tumors
Mammary tumors generated by orthotopic implantation of Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs in nude mice were also examined by TEM. Autophagosomes were present only in mammary tumors generated by apoptosis-defective beclin 1+/+ iMMECs, whereas beclin 1+/− iMMEC-generated tumors displayed prominent ER expansion indicative of ER stress (Fig. 2B). Thus, autophagy induction is likely a frequent occurrence in breast cancer, whereas ER proliferation and chaperone upregulation are characteristics of autophagy-defective mammary tumors.
iMMEC-generated tumors were also examined for ER stress by immunohistochemistry (IHC). GRp170 levels were higher in mammary tumors generated by beclin 1+/− iMMECs (Fig. 2C), indicating that defective autophagy was associated with increased ER stress in mammary tumors in vivo. Similarly, tumor allografts generated by autophagy-deficient iBMK cells displayed ER chaperone upregulation, and spontaneous lung and liver tumors from beclin 1+/− mice showed high GRp170 expression (5), indicating that autophagy defects lead to ER stress in epithelial tumors, independently of tissue type.
Autophagy-defective mammary tumor cells are sensitized to proteasome inhibitors and ER stress-inducers
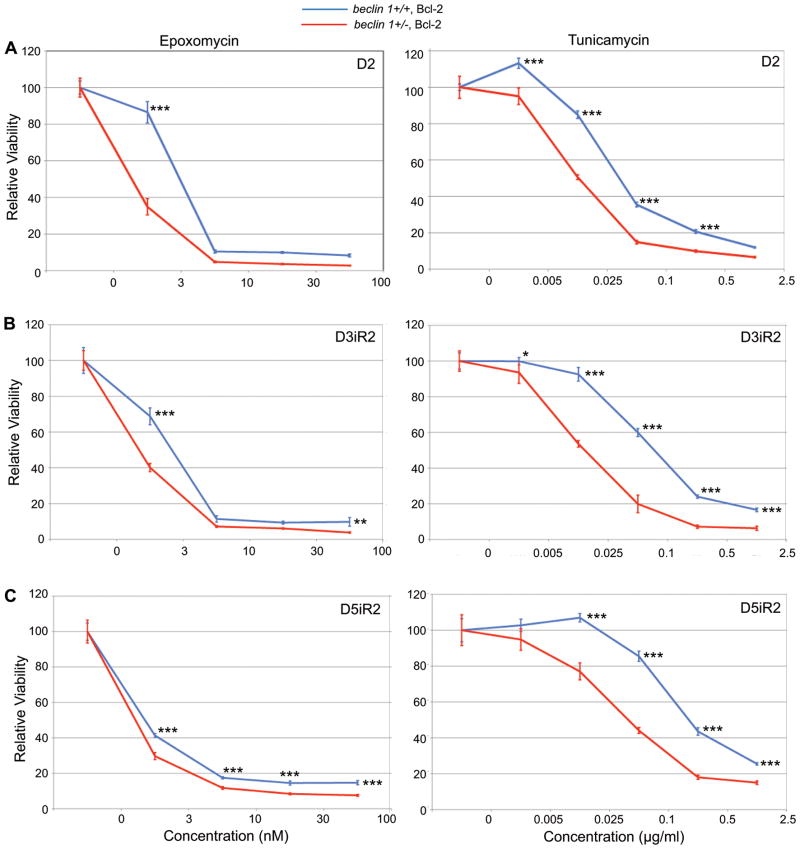
A prediction based on the induction of ER stress in beclin 1+/− iMMECs under metabolic stress is that autophagy-defective mammary cells may be sensitive to ER stress-inducing agents and to proteasome inhibitors. To test this hypothesis, beclin 1+/+ and beclin 1+/− iMMECs were treated with the ER stress inducer tunicamycin and the proteasome inhibitor epoxomycin under normal growth conditions and upon recovery from metabolic stress, when the difference in ER stress levels between autophagy-competent and -defective mammary cells is maximal (Fig. 1C). Similarly to autophagy-defective iBMK cells (5), beclin 1+/− iMMECs were more sensitive than beclin 1+/+ iMMECs to both agents (Fig. 3), suggesting that induction of ER stress and/or proteasome inhibition may be useful in autophagy-defective tumor treatment.
Figure 3. Defective autophagy sensitizes mammary tumor cells to ER stressors and proteasome inhibitors.
MTT assays showing sensitivity of Bcl-2-expressing beclin 1+/+ (blue) and beclin 1+/− (red) iMMECs to increasing concentrations of epoxomycin (left column) and tunicamycin (right column) after 2 days of metabolic stress (A), and after a 2-day recovery following metabolic stress for 3 days (B) and 5 days (C). Data is presented as mean ± SD. p-values were calculated by two way ANOVA.
***, <0.001; **, <0.01; *, <0.05.
Autophagy regulates keratin homeostasis in mammary epithelial cells in a p62-dependent manner
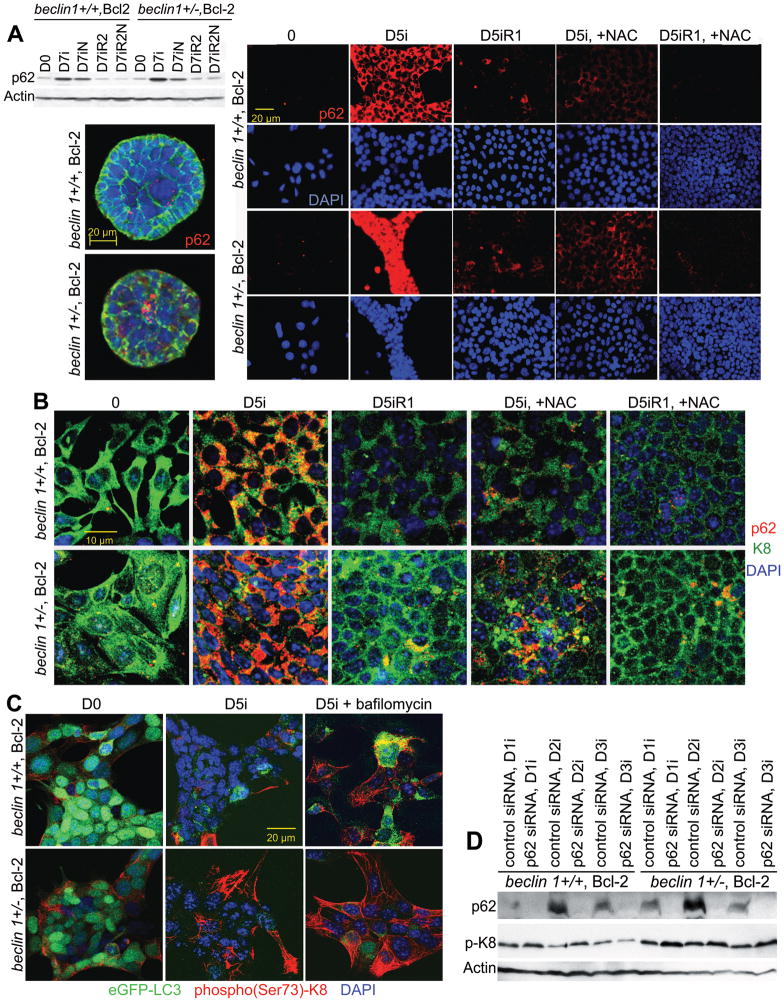
p62 (SQSTM1) is an oligomerizing signaling adaptor protein (11, 15) that binds to ubiquitin (16) and LC3 (17), shuttles ubiquitinated proteins to proteasome (16) and autophagosomes for degradation (18), and is induced by oxidative stress (19). beclin 1+/+ and beclin 1+/− iMMECs were examined for p62 expression under metabolic stress and recovery (Fig. 4A). Similarly to iBMK cells (5), p62 levels increased in stressed iMMECs, more so in autophagy-defective cells, whereas they decreased during recovery, especially in beclin 1+/+ iMMECs, indicating that oxygen and nutrient limitation is a potent stimulus for p62 expression. The ROS-scavenger N-acetylcysteine suppressed stress-induced p62 upregulation, particularly in autophagy-competent iMMECs. Higher p62 levels under metabolic stress and recovery in beclin 1+/− iMMECs, both in the absence and presence of NAC at a concentration adequate to reduce p62 accumulation in beclin 1+/+ iMMECs, indicated that autophagy-deficient mammary cells exhibit increased oxidative stress upon hypoxia and glucose deprivation, as well as during recovery. These results complement recently reported higher ROS levels in autophagy-defective iBMK cells (5). p62 accumulation in beclin 1+/− iMMECs was also observed in metabolically stressed central acinar cells in 3D-morphogenesis (Fig. 4A).
Figure 4. p62 and keratin accumulation in autophagy-deficient iMMECs under metabolic stress and recovery.
A (left top), Western blot showing p62 levels in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs following 7 days of metabolic stress (D7i) and 2 days of recovery (R2) in the absence and presence of the ROS-scavenger N-acetylcysteine (NAC, N); (left bottom) p62 IF (red) in mammary acini generated by Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs. β-catenin (green) was used as an epithelial cell marker and DAPI (blue) for counterstaining nuclei; (right) p62 IF (red) in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs following 5 days of metabolic stress (D5i) and 1 day of recovery (R1) in the absence and presence of NAC.
B, p62 (red) and K8 (green) IF in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs following 5 days of metabolic stress (D5i) and 1 day of recovery (R1) in the absence and presence of NAC.
C, phospho(Ser73)-K8 (red) IF in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs stably expressing eGFP-LC3 (green) following 5 days of metabolic stress (D5i) in the absence and presence of bafilomycin.
D, Western blots showing levels of p62, phospho(Ser73)-K8, and actin in Bcl-2-expressing beclin 1+/+ and beclin 1+/− iMMECs following metabolic stress for 1–3 days after transfection with control or p62 siRNA.
In beclin 1+/+ iMMECs under stress, p62 co-localized with K8 in perinuclear puncta reminiscent of autophagosomes, whereas p62 masked keratin epitopes in beclin 1+/− iMMECs, possibly due to higher expression levels and oligomerization around aggregated keratin structures (Fig. 4B). K8 accumulation in autophagy-defective cells became apparent in recovery when p62 levels decreased.
Intermediate filament reorganization under stress is known to involve keratin phosphorylation (20), particularly K8 phosphorylation at Ser73 (21). Phospho(Ser73)-K8 basal levels were similarly low in beclin 1+/+ and beclin 1+/− iMMECs (Fig. 4C, left column), but increased in autophagy-defective iMMECs under stress (Fig. 4C, middle column) and even further upon pharmacologic inhibition of autophagy with bafilomycin, more so in beclin 1+/− iMMECs (Fig. 4C, right column), suggesting that K8 phosphorylation is an intermediate step in autophagy-mediated keratin remodeling in mammary epithelial cells under stress. After 5 days of metabolic stress, autophagosomes could not be visualized as they were degraded in lysosomes (Fig. 4C, middle column), unless lysosomal function was inhibited with bafilomycin (Fig. 4C, right column). p62 knockdown by siRNA (Dharmakon) also increased phospho(Ser73)-K8 levels in iMMECs under stress (Fig. 4D), particularly in beclin 1+/− cells, indicating that p62 is involved in this process. Thus, both p62-deficiency and abnormal p62 accumulation due to defective autophagy result in phospho(Ser73)-K8 accumulation, a finding that at a first glance appears paradoxical. However, in both cases autophagy-mediated keratin homeostasis is impaired, either due to failure of keratin delivery to autophagosomes in the absence of its carrier (p62) or due to a defect in the autophagic process, which also leads to abnormally increased p62 levels.
Defective autophagy leads to ER chaperone, p62, oxidative stress marker and keratin accumulation in vivo
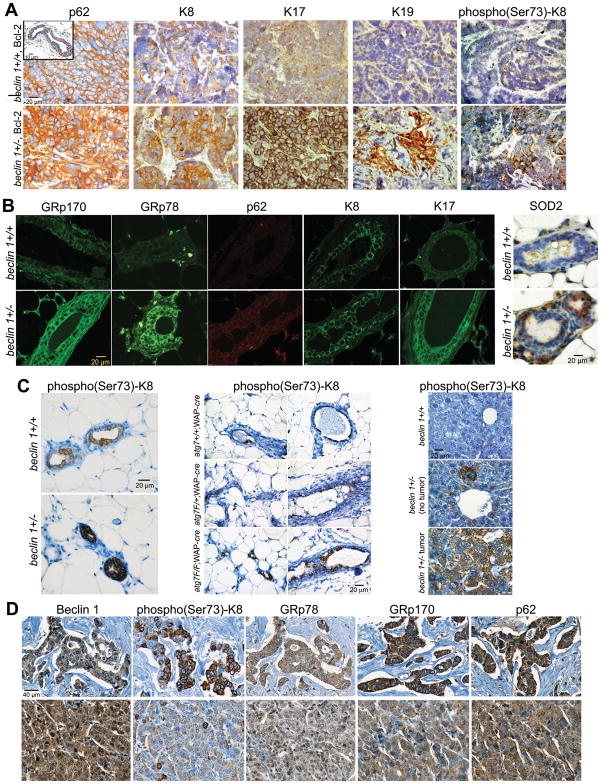
Higher p62 levels were also observed in mammary tumors generated by orthotopic implantation of beclin 1+/− iMMECs (Fig. 5A), similarly to autophagy-deficient iBMK cell allograft tumors, and lung and liver tumors arising spontaneously in beclin 1+/− mice (5), indicating that p62 accumulates in autophagy-deficient epithelial tumors, independently of tissue type and nature of autophagy defect. Mammary tumors generated by autophagy-defective iMMECs also showed higher K8, K17, K19 and phospho-K8 levels (Fig. 5A), similarly to in vitro results (Fig. 1B-C; 4B-D). Mammary tissues from beclin 1+/+ and beclin 1+/− 9-month old retired breeders also exhibited increased expression of GRp78, GRp170, p62, SOD2, K8, K17 and phospho-K8 (Fig. 5B-C), suggesting that concurrent upregulation of these proteins may constitute an autophagy deficiency signature in vivo. Elevated ER chaperone and oxidative stress marker (p62, SOD2) levels were also observed in atg7-null mammary tissues (Suppl. Fig. 2). Furthermore, phospho-K8 levels were higher in atg7-deficient mammary glands (Fig. 5C), indicating that autophagy-defective mammary epithelial cells accumulate keratin independently of autophagy defect type. Metabolically stressed liver tissue (near portal vein) from beclin 1+/− mice also demonstrated elevated phospho(Ser73)-K8 expression, which further increased in tumors (Fig. 5C), suggesting that phospho(Ser73)-K8 accumulation is not mammary gland-specific, but may be a general characteristic of autophagy-defective epithelial cells and tumors.
Figure 5. ER chaperone, p62 and keratin accumulation in beclin 1+/− iMMEC-generated tumors, in autophagy-deficient mammary tissues and in human breast cancers with low Beclin 1 levels.
A, p62 and keratin IHC on mammary tumors generated by Bcl-2-expressing beclin 1+/+ (top row) and beclin 1+/− (bottom row) iMMECs. Rectangular insert in top left panel, p62 IHC in normal mammary duct.
B, ER chaperone (GRp170, GRp78), p62, and keratin IF, and SOD2 IHC on mammary tissues from 9-month old beclin 1+/+ (top row) and beclin 1+/− (bottom row) female mice.
C, phospho(Ser73)-K8 IHC on (left) mammary tissues from 9-month old beclin 1+/+ and beclin 1+/− mice; (middle) mammary tissues from atg7+/+;WAP-cre, atg7F/+;WAP-cre and atg7F/F;WAP-cre mice three weeks after completion of second pregnancy and lactation; (right) liver tissues from 2-year old beclin 1+/+ and beclin 1+/− mice.
D, Beclin 1, phospho(Ser73)-K8, GRp78, GRp170, and p62 IHC on human breast tumors. Top row, breast tumor with low (1+) Beclin 1 expression; bottom row, breast tumor with high (3+) Beclin 1 expression.
p62, ER chaperone and keratin accumulation in human breast tumors
To examine whether concurrent ER chaperone, p62 and keratin accumulation constitutes an autophagy-deficiency signature in human breast cancer, a breast cancer tissue microarray (TMA BR804, US Biomax), consisting of 40 pairs of matched tumor (T) and normal adjacent tissue (NAT), was examined for Beclin 1, GRp170, GRp78, p62, K8, K17, and phospho(Ser73)-K8 expression (Fig. 5D and Table 1). Beclin 1 levels were higher in T than in NAT in 13/25 evaluable T-NAT pairs, indicating that induction of autophagy regulators, and thus autophagy, is likely a frequent occurrence in breast tumors. 7/25 evaluable pairs had comparable and 5/25 evaluable pairs had lower Beclin 1 expression in T compared to NAT. Given that Beclin 1 was upregulated in 52% of breast tumors examined, lower and equal Beclin 1 levels may together constitute relative Beclin 1 deficiency in tumors compared to NAT, similarly to an earlier study that reported relative Beclin 1 loss in 40% of breast tumors using a different Beclin 1 antibody (22). K8 and K17 levels varied in tumors and NAT in a non-specific pattern, whereas ER chaperone and p62 expression was consistently higher in T than NAT (Table 1), but did not correlate with Beclin 1 levels. High (3+ staining) phospho(Ser73)-K8 levels were also more common in tumors than NAT and inversely correlated with Beclin 1 absolute levels, indicating that phospho(Ser73)-K8 accumulation may be a marker of Beclin 1 -and thus autophagy- deficiency in breast tumors. Elevated ER chaperone and p62 expression, even though not significantly correlating with Beclin 1 expression in this breast cancer TMA, may still be part of an autophagy deficiency signature in breast cancer, as the functional level of autophagy in tumors is likely determined by additional parameters beyond Beclin 1 levels, such as PI3K/AKT/mTOR pathway activation (23).
Table 1.
ER chaperone, p62 and keratin expression in human breast cancer
| A. High phospho(Ser73)-K8, GRp78, GRp170 and p62 levels and Beclin 1 expression in breast tumors | ||||
|---|---|---|---|---|
| Beclin 1 | Phospho-K8 | GRp78 | GRp170 | p62 |
| 0–1+ (6) | 4#/6 | 1#/5 | 3#/5 | 2#/6 |
| 2+ (10) | 3#/10 | 4#/9 | 4#/7 | 4#/9 |
| 3+ (16) | 1#/16 | 6#/15 | 11#/13 | 7#/16 |
| 8#/32 | 11#/29 | 18#/25 | 13#/31 | |
| B. High phospho(Ser73)-K8, GRp78, GRp170 and p62 levels and Beclin 1 expression in normal breast tissue | ||||
|---|---|---|---|---|
| Beclin 1 | Phospho-K8 | GRp78 | GRp170 | p62 |
| 0–1+ (4) | 0#/4 | 0#/3 | 1#/3 | 0#/2 |
| 2+ (18) | 0#/16 | 0#/15 | 1#/7 | 0#/13 |
| 3+ (3) | 0#/2 | 0#/3 | 0#/3 | 0#/2 |
| 0#/22 | 0#/21 | 2#/13 | 0#/17 | |
Specimens with 3+ staining
( ) Number of specimens
1) Beclin 1 levels in breast tumors versus normal adjacent tissue were compared using a two-sided exact Wilcoxon test of the equality of distributions of Beclin 1 staining levels. A p-value of 0.0349 was obtained, indicating a statistically significant difference between Beclin 1 levels in breast tumors versus normal adjacent tissue.
2) Regarding high levels of phospho(Ser73)-K8, GRp78, GRp170, and p62, any of these discriminates between breast tumor and normal breast tissue (two-sided test of the equality of two proportions; p-value < 0.003 in all cases).
3) Regarding trend tests of the breast tumors with high expression (3+ staining) of phospho(Ser73)-K8, GRp78, GRp170 or p62 in relationship with absolute Beclin-1 levels, the following p-values were obtained using logistic regression: phospho(Ser73)-K8 0.0105*; GRp78 0.543; GRp170 0.216; p62 0.706.
statistically significant
DISCUSSION
Defective autophagy, ER stress and mammary tumorigenesis
ER is responsible for folding of secreted proteins and those destined to cell surface and intracellular organelles. Nutrient deprivation, hypoxia, deregulation of calcium homeostasis and toxic chemicals disrupt ER protein folding and cause unfolded protein accumulation (ER stress) resulting in activation of the unfolded protein response (UPR), a tightly regulated cellular process that adjusts the cell’s folding capacity, protein synthesis and degradation. Recent studies showed that autophagy is induced by ER stress and is involved in polyubiquitinated protein aggregate removal (24–27). We provide evidence for another aspect in the relationship between autophagy and ER stress: not only does ER stress induce autophagy, but also defective autophagy induces ER stress in mammary tumor cells in vitro and allograft mammary tumors in vivo. Prolonged ER stress and UPR activation have been linked to cancer progression, as downstream effects include NF-κB activation, G1 arrest and p38 activation (28). A connection between elevated ER stress and mammary tumorigenesis has also been established, as the ER chaperone GRp78/BiP is overexpressed in aggressive breast tumors (29), whereas GRp78 heterozygocity prolongs mammary tumor latency and impedes tumor growth (30). Thus, ER stress may be a mechanism contributing to defective autophagy-associated mammary tumorigenesis in parallel with DNA damage and genomic instability (10). Indeed, ER stress and the resultant oxidative stress (31) may be major etiologies of genotoxic damage associated with autophagy defects (10, 13). In this case, antioxidants may rescue defective autophagy-induced genotoxic effects and tumorigenicity. Furthermore, chronic ER stress suppression by pharmacologic upregulation of autophagy may represent a cancer preventative strategy for preneoplastic lesions with autophagy defects. The association between autophagy defects and elevated ER stress in tumors has treatment implications, as autophagy-defective mammary tumor cells are sensitized to ER stress-inducing agents and proteasome inhibitors. This finding indicates that the functional status of autophagy in tumors may determine rational cancer treatment design. GRp170 upregulation in autophagy-defective mammary tumors may also be exploited for therapeutic benefit, as vaccination with tumor-derived GRp170 may have antitumor activity by inducing innate and adaptive immune responses (32).
Defective autophagy and keratin homeostasis
The keratin cytoskeleton acts as a signaling platform and provides cytoprotection in epithelial cells (33). Keratin 8 and 18-deficient mice exhibit susceptibility to toxic agents and Fas-induced apoptosis, whereas K8/K18 variants predispose humans to end-stage liver disease, acute liver failure, and liver fibrosis in patients with chronic hepatitis C (34). On the other hand, keratin overexpression correlates with extent of epithelial cell injury (35) and is a common finding even in non-epithelial tumor cells, such as malignant melanoma (36). In most cases, it is not known whether keratin accumulation is a cause or result of neoplastic transformation. In a mouse cancer model, transgenic K8 expression in the epidermis altered epidermal cell differentiation and favored transition to malignancy (37), indicating that keratin may be an active player in tumorigenesis. Keratin-containing aggregates have also been associated with liver disease, as Mallory bodies (MBs) or Mallory-Denk bodies (MDBs), consisting of ubiquitinated K8/K18, chaperones and p62, are found in alcoholic and non-alcoholic steatohepatitis (38) and a subset of liver tumors (39). Recent studies implicated defective autophagy in MDB formation (40), as activation of autophagy by rapamycin was shown to eliminate mouse MDBs and block their proteasome inhibitor-mediated formation (41). The role of MDBs in liver disease pathogenesis remains unclear, but their presence is a histological and potential progression marker in liver disease (42).
Our studies define the important role of autophagy in keratin homeostasis in the mammary gland, similarly to what was already described for the liver (41), and indicate that keratins may be more than simple mammary epithelial state markers. Although MDBs have not been described in the mammary gland or breast tumors, abnormal keratin accumulation in mammary tumors may be a histological marker of defective autophagy status, elevated ER and oxidative stress, and possibly more aggressive disease. Whether defective autophagy-associated keratin upregulation in breast tumors has prognostic and/or treatment implications for breast cancer patients remains to be investigated in relevant animal models and human breast cancer specimens.
Autophagy deficiency signature in breast cancer
Our work indicates that high levels of ER chaperones, p62, phospho-K8, and keratins 8, 17 and 19 may constitute an autophagy deficiency signature in mammary epithelial cells, as monoallelic beclin 1 deletion resulted in concurrent upregulation of these proteins in mammary tumor cells in vitro and in vivo, and in mammary tissues. The association between defective autophagy, ER stress and breast cancer was discussed earlier. Higher p62 expression in beclin 1+/− mammary tumor cells is not surprising, given the well-established association between defective autophagy and abnormal p62 accumulation in neurons and hepatocytes (7), and kidney epithelial cells and tumors (5). Indeed, high p62 levels appear to be a reliable autophagy deficiency marker independently of tissue or autophagy defect type. In breast cancer, p62 expression also correlates with grade, distant metastasis, and EGF receptor expression (43), suggesting that defective autophagy may be associated with more aggressive breast malignancies.
Accumulation of keratins, particularly post-translationally modified keratin products, is relevant to breast cancer, as ubiquitin-immunoreactive K8/18 degradation products are detected in breast tumors and correlate with cancer aggressiveness (44). Furthermore, K17 is upregulated in wound-activated skin epithelial cells (45), which may explain why beclin 1+/− iMMECs, which are susceptible to metabolic stress and DNA damage (10), accumulate K17 under stress. K17 also regulates protein synthesis and epithelial cell growth through binding to the adaptor protein 14-3-3σ (46), indicating that it may be involved in defective autophagy-mediated mammary tumorigenesis. Finally, K19 is associated with aggressive breast cancer, as K19 mRNA-positive circulating tumor cell (CTC) detection before adjuvant chemotherapy predicts poor clinical outcome in breast cancer patients (47).
K8 becomes phosphorylated at Ser73 by stress-activated kinases and participates in shear stress-mediated keratin filament disassembly (48) and ubiquitin-proteasome-mediated keratin degradation (49) in alveolar epithelial cells. Our studies revealed that phospho(Ser73)-K8 also plays a role in autophagy- and p62-mediated keratin remodeling in mammary epithelial cells under metabolic stress and that strong (3+) phospho(Ser73)-K8 staining by IHC inversely correlate with absolute Beclin 1 expression in breast cancer. Thus, although phospho(Ser73)-K8 expression is a known general epithelial cell stress indicator, high (3+ staining) phospho(Ser73)-K8 levels may be further indicative of autophagy deficiency in breast tumors, and possibly in other human malignancies. Identification of an autophagy deficiency marker, and possibly an autophagy deficiency signature, in breast cancer may have prognostic and therapeutic implications for cancer patients, as the role of defective autophagy in breast cancer pathogenesis, prognosis and treatment responsiveness becomes more precisely defined. Ultimately, successfully recognizing the functional status of autophagy in human tumors will lead to personalized cancer treatment and hopefully improved clinical outcomes (23).
Supplementary Material
Acknowledgments
We thank Dr. Komatsu for providing atg7F/F mice, and Drs. Heintz, Yue and Jin for beclin 1+/− mice. We thank Dr. Zimmermann for GRp170-antibody, Dr. Kane-Goldsmith for help with confocal microscopy and Rajesh Patel for technical support with electron microscopy. We thank Dr. Omary for reviewing the manuscript and providing insightful feedback. We thank Applied Biomics, Inc., Hayward, CA, for technical help with 2-DIGE and proteomic analyses, and the CINJ Tissue Analytical Services and Biomedical Imaging core facilities for their assistance. This work was supported by grants from NIH (1K99CA133181), Damon Runyon Cancer Research Foundation (Clinical Investigator Award) and New Jersey Commission on Cancer Research to V. K.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7(12):961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival andrestricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763(12):1767–75. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21(13):1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62--more than just a scaffold. FEBS Lett. 2007;581(2):175–9. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karantza-Wadsworth V, White E. A mouse mammary epithelial cell model to identify molecular mechanisms regulating breast cancer progression. Methods Enzymol. 2008;446:61–76. doi: 10.1016/S0076-6879(08)01604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto N, Ezaki J, Komatsu M, et al. Comprehensive proteomics analysis of autophagy-deficient mouse liver. Biochem Biophys Res Commun. 2008;368(3):643–9. doi: 10.1016/j.bbrc.2008.01.112. [DOI] [PubMed] [Google Scholar]
- 15.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12(1):39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 16.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–68. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 18.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii T, Itoh K, Takahashi S, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275(21):16023–9. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 20.Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci. 2006;31(7):383–94. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Ku NO, Azhar S, Omary MB. Keratin 8 phosphorylation by p38 kinase regulates cellular keratin filament reorganization: modulation by a keratin 1-like disease causing mutation. J Biol Chem. 2002;277(13):10775–82. doi: 10.1074/jbc.M107623200. [DOI] [PubMed] [Google Scholar]
- 22.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 23.Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding WX, Ni HM, Gao W, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakaki K, Wu J, Kaufman RJ. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283(22):15370–80. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding WX, Ni HM, Gao W, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282(7):4702–10. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 27.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281(40):30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4(12):966–77. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez PM, Tabbara SO, Jacobs LK, et al. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat. 2000;59(1):15–26. doi: 10.1023/a:1006332011207. [DOI] [PubMed] [Google Scholar]
- 30.Dong D, Ni M, Li J, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 31.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9(12):2277–93. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 32.Wang XY, Kazim L, Repasky EA, Subjeck JR. Immunization with tumor-derived ER chaperone grp170 elicits tumor-specific CD8+ T-cell responses and reduces pulmonary metastatic disease. Int J Cancer. 2003;105(2):226–31. doi: 10.1002/ijc.11058. [DOI] [PubMed] [Google Scholar]
- 33.Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313(10):2021–32. doi: 10.1016/j.yexcr.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Strnad P, Stumptner C, Zatloukal K, Denk H. Intermediate filament cytoskeleton of the liver in health and disease. Histochem Cell Biol. 2008;129(6):735–49. doi: 10.1007/s00418-008-0431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toivola DM, Nakamichi I, Strnad P, et al. Keratin overexpression levels correlate with the extent of spontaneous pancreatic injury. Am J Pathol. 2008;172(4):882–92. doi: 10.2353/ajpath.2008.070830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seftor EA, Meltzer PS, Kirschmann DA, et al. Molecular determinants of human uveal melanoma invasion and metastasis. Clin Exp Metastasis. 2002;19(3):233–46. doi: 10.1023/a:1015591624171. [DOI] [PubMed] [Google Scholar]
- 37.Casanova ML, Bravo A, Martinez-Palacio J, et al. Epidermal abnormalities and increased malignancy of skin tumors in human epidermal keratin 8-expressing transgenic mice. FASEB J. 2004;18(13):1556–8. doi: 10.1096/fj.04-1683fje. [DOI] [PubMed] [Google Scholar]
- 38.Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: Lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782(12):764–74. doi: 10.1016/j.bbadis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Denk H, Stumptner C, Fuchsbichler A, et al. Are the Mallory bodies and intracellular hyaline bodies in neoplastic and non-neoplastic hepatocytes related? J Pathol. 2006;208(5):653–61. doi: 10.1002/path.1946. [DOI] [PubMed] [Google Scholar]
- 40.Harada M, Strnad P, Toivola DM, Omary MB. Autophagy modulates keratin-containing inclusion formation and apoptosis in cell culture in a context-dependent fashion. Exp Cell Res. 2008;314(8):1753–64. doi: 10.1016/j.yexcr.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47(6):2026–35. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 42.Zatloukal K, French SW, Stumptner C, et al. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313(10):2033–49. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 43.Rolland P, Madjd Z, Durrant L, Ellis IO, Layfield R, Spendlove I. The ubiquitin-binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr Relat Cancer. 2007;14(1):73–80. doi: 10.1677/erc.1.01312. [DOI] [PubMed] [Google Scholar]
- 44.Iwaya K, Ogawa H, Mukai Y, Iwamatsu A, Mukai K. Ubiquitin-immunoreactive degradation products of cytokeratin 8/18 correlate with aggressive breast cancer. Cancer Sci. 2003;94(10):864–70. doi: 10.1111/j.1349-7006.2003.tb01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276(5309):75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 46.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441(7091):362–5. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 47.Ignatiadis M, Xenidis N, Perraki M, et al. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J Clin Oncol. 2007;25(33):5194–202. doi: 10.1200/JCO.2007.11.7762. [DOI] [PubMed] [Google Scholar]
- 48.Ridge KM, Linz L, Flitney FW, et al. Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J Biol Chem. 2005;280(34):30400–5. doi: 10.1074/jbc.M504239200. [DOI] [PubMed] [Google Scholar]
- 49.Jaitovich A, Mehta S, Na N, Ciechanover A, Goldman RD, Ridge KM. Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J Biol Chem. 2008;283(37):25348–55. doi: 10.1074/jbc.M801635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.