Abstract
A longstanding research goal has been to develop a self-sustained chemical system that is capable of undergoing Darwinian evolution. The notion of primitive RNA-based life suggests this goal might be achieved by constructing an RNA enzyme that catalyzes the replication of RNA molecules, including the RNA enzyme itself. This reaction recently was demonstrated in a cross-catalytic system involving two RNA enzymes that catalyze each other’s synthesis from a total of four component substrates. The cross-replicating RNA enzymes undergo self-sustained exponential amplification at a constant temperature in the absence of proteins or other biological materials. Amplification occurs with a doubling time of 30–60 min, and can be continued indefinitely. Small populations of cross-replicating RNA enzymes can be made to compete for limited resources within a common environment. The molecules reproduce with high fidelity, but occasionally give rise to recombinants that also can replicate. Over the course of many “generations” of selective amplification, novel variants arise and grow to dominate the population based on their relative fitness under the chosen reaction conditions. This is the first example, outside of biology, of evolutionary adaptation in a molecular genetic system.
The last time the Cold Spring Harbor Symposium focused on evolution was in 1987, on the topic “The Evolution of Catalytic Function”. I was happy to have attended that meeting. Being a postdoctoral fellow at that time, I felt obliged to write out my introductory remarks, which I have saved to this day. In my introduction I said: “I choose to interpret ‘evolution of catalytic function’ in the prospective sense, by which I mean the potential to evolve novel catalysts in the laboratory”. I also said: “In the laboratory we focus on the problem of replication and on trying to copy genetic information without the aid of an external catalyst” (Joyce 1987).
I was hardly the first person to have had such thoughts. In that same meeting, Jeremy Knowles said, quoting from his paper in the 1987 Symposium volume: “We outline the first steps of an attempt to monitor the improvement in catalytic efficiency of an enzyme as its gene is mutagenized at random and more efficient catalysts are selected for” (Hermes et al. 1987). Knowles described what were some of the first directed evolution experiments, in which he randomly mutagenized the gene for triose phosphate isomerase and screened for variant enzymes with improved catalytic efficiency. Twenty years before that, Francis Crick discussed the possibility of replication of RNA genomes without the aid of an external catalyst. He said: “Possibly the first ‘enzyme’ was an RNA molecule with RNA replicase properties” (Crick 1968). In Crick’s view, RNA was the Ur enzyme — the first enzyme to be capable of bringing about its own replication, thereby providing the basis for Darwinian evolution.
Since the time of the 1987 Symposium, the technology of directed evolution has advanced tremendously, for both proteins and RNA. My own laboratory has focused on the in vitro evolution of RNA enzymes, especially those relevant to the replication of genetic information. The technology itself has become so powerful, and yet so routine, that it can be practiced by any biochemist or molecular biologist. It is straightforward to amplify RNA molecules by a combination of reverse transcription, PCR amplification, and forward transcription. Once can impose selection constraints on the RNA molecules such that if they meet those constraints (for example, binding a target ligand or performing some catalytic function), then they become eligible for amplification. And one can introduce random mutations, usually at the level of double-stranded DNA, through mutagenic or recombinagenic PCR procedures. Taken together, the ability to amplify, select, and mutate populations of RNA molecules gives one the opportunity to carry out the Darwinian evolution of RNA-based catalytic function (Joyce 1989; Beaudry and Joyce 1992).
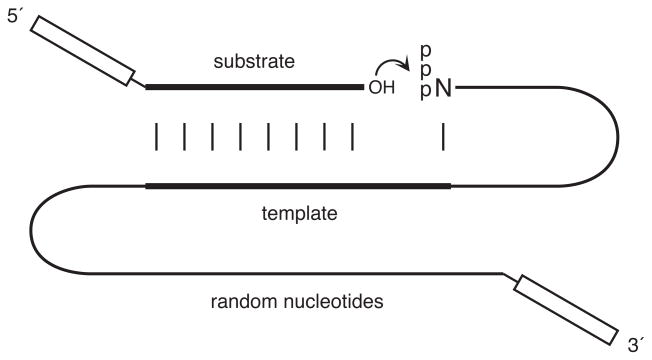
One of the first examples of the directed evolution of RNA enzymes concerned the same function that Francis Crick had talked about in 1968: the ability of RNA to catalyze the RNA-templated joining of RNA molecules (Bartel and Szostak 1993). This is fundamentally the same chemistry that is brought about by RNA-dependent RNA polymerase proteins. In order to discover RNA enzymes that catalyze this reaction, one can go searching in random sequence space. One can attach random sequence polynucleotides to an RNA template-substrate complex, and install primer binding sites at the 3′ end of the random sequence region and at the 5′ end of substrate (Figure 1). Then, through selective RT-PCR, one can amplify only those molecules that have catalyzed the joining of the substrate to themselves. The first application of this selection scheme, and the first case in which enzymatic function was derived starting from random sequence RNAs, was the work of David Bartel and Jack Szostak (1993) that resulted in the “class I” RNA ligase enzyme. It is a robust enzyme, with a kcat of 14 min−1 and Km of 9 μM, obtained from a starting population of ~1015 random sequence 220mers. This work demonstrates that Crick’s notion of RNA-catalyzed RNA replication, together with Knowles’ approach to the directed evolution of catalytic function, are experimentally viable.
Figure 1.
Scheme for selective amplification of RNA molecules that catalyze the RNA-templated joining of RNA. The putative catalytic domain consists of random-sequence nucleotides that are attached to a template region, which is complementary to the 3′ end of an oligonucleotide substrate and to the 5′ end of the population of RNAs. Any RNA molecule that catalyzes ligation of the substrate to itself (curved arrow) will contain two primer binding sites (boxed regions) that are necessary for reverse transcription and PCR amplification.
Continuous in Vitro Evolution
More recently, but still more than 10 years ago, a new technology for the directed evolution of RNA was devised in our laboratory; what we termed “continuous in vitro evolution” (Wright and Joyce 1997). This method first was applied to the class I RNA ligase, which was challenged to attach an oligonucleotide substrate to the 5′ end of the RNA enzyme. The substrate had the sequence of the T7 RNA polymerase promoter, containing mostly deoxynucleotides, but also a few ribonucleotides at its 3′ end. RNA enzymes that reacted with this substrate became reversed transcribed, in the same mixture, to yield double-stranded RNA-DNA molecules that contained a functional promoter element. The reaction mixture also included T7 RNA polymerase, which generated multiple copies of “progeny” RNA enzymes per reacted parental molecule. These progeny in turn could catalyze additional ligation reactions, and so on, resulting in the exponential amplification of functional RNAs. This cycle of events could be continued indefinitely, so long as one maintained a supply of the promoter-containing substrate and other reagents, usually accomplished through a serial transfer procedure.
The continuous in vitro evolution of RNA enzymes is analogous to the continuous culture of bacterial or eukaryotic cells, except that our culture medium is purely biochemical, containing two polymerase proteins (reverse transcriptase and T7 RNA polymerase), the four NTPs and dNTPs, salts, and buffer. This system enables longitudinal studies of the Darwinian evolution of RNA enzymes, analogous to the work of Richard Lenski and colleagues concerning the long-term experimental evolution of E. coli (Elena et al. 1996; Blount et al. 2008).
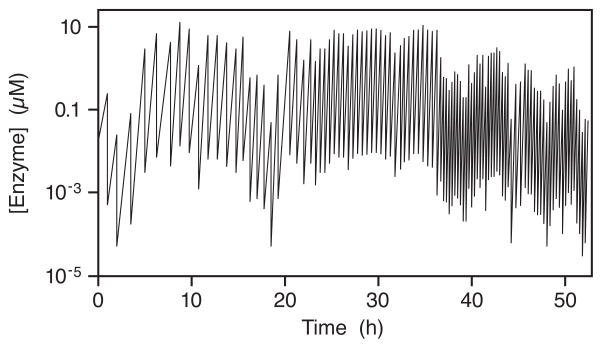
One way to track the evolving population of RNA enzymes is to measure the concentration of RNA before and after each transfer throughout the course of a serial transfer experiment. This produces what we term “zigzag plots”, reflecting repeated rounds of growth and dilution (Figure 2). In the first of many such continuous in vitro evolution experiments, we carried out 300 successive rounds of ~1,000-fold growth and 1,000-fold dilution, achieving an overall amplification of ~10300-fold in 52 h (Wright and Joyce 1997). The evolving population not only withstood this extreme dilution schedule, but also exhibited progressive improvement in its catalytic function. The most fit enzymes grew preferentially to dominate the population, and had the opportunity to give rise to novel variants with even higher catalytic efficiency. The starting class I ligase enzyme exhibited a catalytic efficiency (kcat/Km) of 8 × 102 M−1 min−1, whereas the evolved enzyme exhibited a catalytic efficiency of 1 × 107 M−1 min−1 (measured in the presence of 15 mM MgCl2 at pH 8.5 and 37 °C). This improvement of ~104-fold was attributable to 30 acquired mutations that improved both the kcat and Km of the ligase enzyme.
Figure 2.
Time course of continuous evolution of the class I RNA ligase enzyme in a serial transfer experiment involving 100 successive rounds of ~1,000-fold growth and 1,000-fold dilution. The concentration of RNA enzymes was measured before and after each transfer (zigzag line). The time between transfers was decreased as tolerated, initially 1 h and eventually 15 min (figure based on Wright and Joyce 1997).
Continuous in vitro evolution, although a powerful method for witnessing the evolution of catalytic function in real time (Paegel and Joyce 2008), suffers from the fact that behind the curtain lurk two informational macromolecules: reverse transcriptase and T7 RNA polymerase, which themselves are not subject to evolution within the system. Reverse transcriptase, derived from a retrovirus, and T7 RNA polymerase, derived from a bacteriophage, are the products of biological evolution, and not what I had in mind at the 1987 Symposium when I discussed the imperative to “copy genetic information without the aid of an external catalyst” (Joyce 1987). Instead what one wants is what Francis Crick talked about: an RNA enzyme that is “capable of bringing about its own replication” (Crick 1968). One wants a system in which the evolving RNA molecules adopt a structure that confers the ability to catalyze the amplification of RNA molecules, including the production of new copies of the enzymes themselves. Mutations will occur as a matter of course, and selection would be based on the differential replication rate of various RNA molecules in the population. In this way, the Darwinian evolution of RNA could be a self-sustaining process.
Self-sustained Replication of RNA
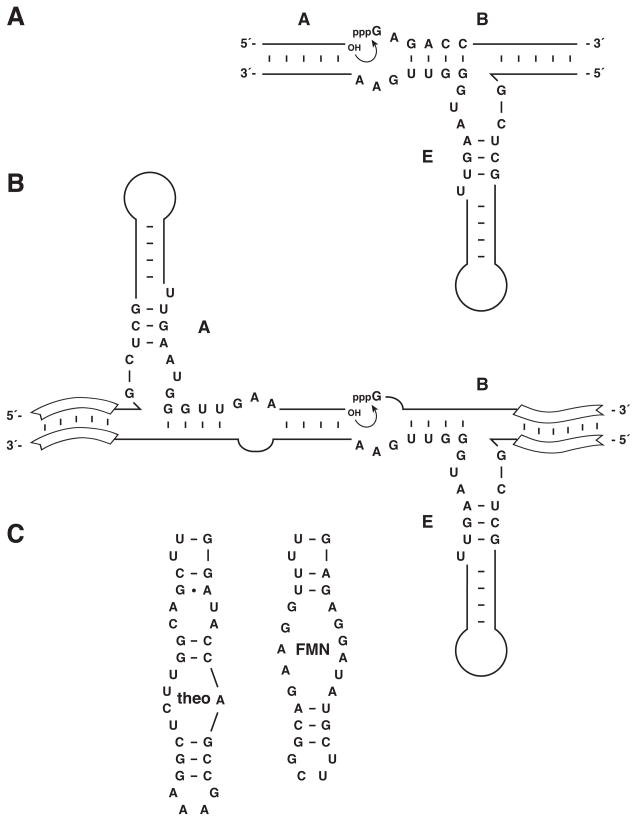
In recent years we have made substantial progress in developing RNA enzymes that catalyze their own replication. This work involves a different RNA ligase, the “R3C” RNA enzyme, which also was obtained by directed evolution starting from a large population of random sequence RNAs (Rogers and Joyce 2001). Like the class I ligase, the R3C ligase catalyzes the joining of two RNA substrates, one bearing a 3′-hydroxyl and the other bearing a 5′-triphosphate, forming a 3′,5′-phosphodiester and releasing inorganic pyrophosphate. The R3C ligase has a simple three-way junction architecture, consisting of three stem-loops that are joined at a central location that contains the catalytic domain of the enzyme (Figure 3A). Nucleotides within the catalytic domain are highly conserved in sequence, but those within the pendant stem-loops are generic, so long as they form a stable duplex structure.
Figure 3.
Sequence and secondary structure of various forms of the R3C ligase enzyme. A, The enzyme (E) adopts a three-way junction structure upon binding two oligonucleotide substrates (A and B), which become ligated (curved arrow) to form the product. Conserved nucleotides that are essential for catalytic function are shown. B, The self-replicating or cross-replicating enzyme ligates two substrates to yield a new copy of the enzyme or its cross-catalytic partner, respectively. Open boxes indicate regions of Watson-Crick pairing between enzyme and substrates that can have any complementary sequence. C, The central stem-loop of the enzyme can be replaced by an aptamer domain, configured such that binding of the corresponding ligand is required to stabilize the active structure of the enzyme. The aptamer domains for theophylline (theo) and flavin mononucleotide (FMN) are shown.
Two of the stem-loop regions within the R3C ligase are involved in binding the RNA substrates. Because these regions are generic in sequence, they can be designed to accommodate substrates whose sequences are identical to that of the enzyme itself. The two substrates (A and B) can be made to correspond to the 5′ and 3′ portions of the enzyme (E), so that when the substrates become ligated they form another copy of the enzyme (Figure 3B). In this way, at least in a formal sense, one can carry out the self-replication of an RNA enzyme (Paul and Joyce 2002). The reaction does indeed proceed autocatalytically, but is not very efficient and does not reach a high maximum extent. For example, if one employs 1 μM starting concentration of ligase enzyme and 2 μM each of the two RNA substrates, there is an initial exponential burst that consumes ~5% of the substrates in 20 min, followed by a slow linear phase that proceeds at a rate of <0.01% min−1. In absence of any starting enzyme there is no exponential burst, consistent with the autocatalytic nature of the system. However, even under optimal conditions, an incubation time of 17 h is required to produce as many new enzyme molecules as the number that were present at the outset (Paul and Joyce 2002). Reaching this breakeven point, and doing so many times over, is critical for achieving self-sustained replication of RNA.
Taking a lesson from the semi-conservative nature of nucleic acid replication in biology, the next step was to devise two ligase enzymes: a plus-strand enzyme that directs the synthesis of a minus-strand enzyme, which in turn directs the synthesis of a new plus-strand enzyme (Kim and Joyce 2004). This approach causes replication to proceed in a cross-catalytic manner, with two enzymes (E and E′) catalyzing each other’s synthesis from a total of four component substrates (A′ + B′ → E′ and A + B → E, respectively). Compared to self-replication, cross-replication places fewer design constraints on the sequences of the replicating molecules. The self-replicating enzyme must be fully palindromic (in the molecular biology sense), while the cross-replicating enzymes need only have short regions of complementarity between the replicating partners. Furthermore, the extensive self-complemementarity of the self-replicating enzyme is the chief reason for its limited extent of growth in the exponential phase of the reaction (Paul and Joyce 2002). This is because the two substrate molecules are complementary to each other (as well as to the parent), and therefore have a tendency to form a non-productive substrate-substrate complex. The initial exponential phase consumes the readily available substrate molecules, and the subsequent linear phase reflects the slow dissociation of substrate molecules from the non-productive complexes. Importantly, the step of product release is not rate limiting, freeing the newly-synthesized enzyme molecules to enter another round of replication.
Initial attempts to carry out cross-catalytic replication were an improvement compared to self-replication, but still disappointing with regard to the goal of reaching the breakeven point. Employing a starting concentration of 1 μM each of E and E′ and 2 μM each of the four RNA substrates, the exponential phase consumed ~25% of the substrates in 6 h (Kim and Joyce 2004). Under optimized reaction conditions and employing long incubation times it would be possible to limp past the breakeven mark, but this is hardly sufficient for sustained replication. One needs to think in terms of achieving 10- to 100-fold breakeven so that, like for protein-mediated continuous in vitro evolution, one can carry out serial transfer experiments that allow replication to proceed indefinitely.
It thus became necessary to return to directed evolution methods to improve the rate and maximum extent of the cross-replicating RNA enzymes. This was done by evolving each enzyme separately, but seeking solutions that would apply to both members of the cross-replication pair. A quench-flow apparatus was used to select molecules that could react in times as short as 10 milliseconds. The resulting E and E′ enzymes exhibited a 38- and 12-fold improvement in catalytic rate, respectively, and reacted to a maximum extent of ~90% in the initial fast phase. These optimized molecules were found to be capable of undergoing self-sustained replication, achieving 100-fold amplification in 5 h at a constant temperature of 42 °C (Lincoln and Joyce 2009).
A serial transfer experiment was carried out employing a starting concentration of 0.1 μM each of E and E′ and 5 μM each of the four RNA substrates, in the presence of 25 mM MgCl2 and 50 mM EPPS (pH 8.5), but with no proteins or other biological molecules. Following 5 h incubation at 42 °C, 4% of the reaction mixture was transferred to a new reaction vessel that contained a fresh supply of the substrates, but only those enzymes that were carried over in the transfer. This procedure was repeated for six rounds, resulting in an overall amplification of >108-fold in 30 h (Lincoln and Joyce 2009). The corresponding zigzag plot was highly regular, each round consisting of ~25-fold amplification of both E and E′ followed by 25-fold dilution (Figure 4A). This process can indeed be continued indefinitely.
Figure 4.
Self-sustained cross-replication of the R3C RNA ligase enzyme in a serial transfer experiment. The concentrations of E (black) and E′ (gray) were measured before and after each transfer. A, A single cross-replicator was propagated for six successive rounds of ~25-fold growth and 25-fold dilution. B, A starting population of 12 different cross-replicators were propagated for 20 successive rounds of ~20-fold growth and 20-fold dilution, with the opportunity for recombination throughout (figure based on Jackson and Joyce 2009).
An Artificial Genetic System
Immortality can be rather dreary if it does not allow for the possibility of variation. What one wants is not a single replicating entity, but rather a heterogeneous populations of replicators that can undergo mutation and selection. The cross-replicating RNA enzymes provide the opportunity to construct an artificial genetic system based on the transmission of sequence information from parent to progeny molecules. The replicating enzymes contain two “alleles”, represented by the two regions of base-pairing interactions between E and E′. Each allele encodes a corresponding trait, represented by the catalytic domain that is covalently linked to the allele.
In principle, the cross-replicating RNAs have the potential to transmit 26 bits of genetic information via the 15 base pairs (two bits per base pair) that comprise the two alleles. One of the alleles contains seven base pairs (16,384 possible variants), and the other contains eight base pairs (65,536 possible variants). However, not all sequences will be discriminated with high fidelity, especially at the extreme 5′ and 3′ ends of the molecule, thus reducing the information capacity of the system. Significantly, there is the opportunity for combinatorial diversity through recombination of the two alleles. This can occur due to occasional incorporation of a mismatched substrate, which results in a recombinant enzyme that also can cross-replicate. Recombinants can give rise to other recombinants, as well as revert back to non-recombinants. Over the course of many “generations” of selective amplification, novel replicators can arise through recombination and can grow to dominate the population, exhibiting Darwinian behavior in a non-biological system.
As a test case, we constructed a model population of 12 different pairs of cross-replicating RNAs (Lincoln and Joyce 2009). Each pair had a different genetic sequence in the two allelic regions, which encoded different functional sequences in the corresponding catalytic domains of the E and E′ molecules. A coding relationship was established between a particular genetic allele and its associated phenotypic trait, implemented through the chemical synthesis of the various RNA molecules. Together, the 12 pairs of cross-replicators have the potential to give rise to 132 pairs of recombinants, which may be more or less fit than their progenitors. A serial transfer experiment was carried out, starting with ~0.1 μM each of the 12 different E and E′ molecules and 5 μM each of the various A, B, A′, and B′ molecules. The population was subjected to 20 successive rounds of ~20-fold amplification and 20-fold dilution (~1026-fold overall amplification) in 100 h. In this case the zigzag plot was not uniform, as novel variants arose and competed with existing members of the population, resulting in the preferential survivial of the most efficient replicators (Figure 4B).
After 20 rounds (86 doublings) of evolution, 100 individuals were cloned from the population and sequenced. The great majority of these (93%) were recombinants that were not present at the start of the experiment. Three such recombinants dominated the population, together accounting for one-third of all clones. These three recombinants all contained the A5 allele, together with the A2′, A3′, or A4′ allele. Overall, the A5 and A3′ alleles were the most enriched, while the A8, A11 and A11′ alleles were the most depleted among the evolved population of replicators (Lincoln and Joyce 2009).
What was the basis for the selective advantage of the dominant individuals? In the presence of their cognate substrates alone, the three dominant recombinants are less efficient replicators compared to the most efficient of the 12 starting replicators. The most efficient recombinant (A5-A3′) has an exponential growth rate of 0.68 h-1, while the most efficient starting replicator (A1-A1′) has a growth rate of 0.75 h−1. However, in the presence of the complete set of 48 substrates, the A5-A3′ recombinant amplifies more efficiently (0.33 h−1) compared to the A1-A1′ starting replicator (0.10 h−1). Furthermore, when the A5-A3′ recombinant is supplied with just the eight substrates that correspond to the enriched set of alleles, it has an exponential growth rate of 0.84 h−1, the highest measured in the study.
It appears that the three dominant recombinants form a clique, not only replicating themselves efficiently, but also giving rise to each other through preferred mutational pathways. An analysis of predicted ΔG values for each combination of matched and mismatched substrates suggests that the most likely recombination events involve exchange of the A2′ and A3′ alleles and exchange of the A3′ and A4′ alleles, favoring the interconversion of the three dominant replicators (Lincoln and Joyce 2009).
Replication Contingent on Other Functions
Although replication efficiency is the ultimate measure of fitness, other traits may confer selective advantage to biological organisms through their indirect effect on fecundity. So too in an artificial genetic system it is possible to make reproductive fitness contingent on the execution of some other function. The cross-replicating RNA enzymes contain three generic stem-loops, two that are committed to substrate binding, and a third that can contain a functional domain (Figure 3C). The functional domain might be an RNA aptamer that binds a specific ligand or a catalyst that has some function other than replication. The activity of this added functional domain must somehow relate to replication so that molecules that are better able to execute the secondary function will enjoy a replicative advantage.
It is straightforward to install an aptamer domain within the central stem-loop of the replicating enzymes, configured so that the enzymes undergo exponential amplification in the presence, but not the absence, of the corresponding ligand. Such constructs are termed “aptazymes”, and have been developed in the laboratory for simple RNA enzymes (Tang and Breaker 1997), and have been discovered within naturally-occurring “riboswitches” (Winker et al. 2004). We installed aptamers that specifically recognize either theophylline (Jenison et al. 1994) or FMN (Burgstaller et al. 1994) in either one or both members of a cross-replicating pair, causing exponential amplification to be dependent on the presence of one or both ligands (Lam and Joyce 2009). In the absence of the ligand the aptamer is unstructured and cannot support the active structure of the enzyme, while in the presence of the ligand the aptamer adopts a well-defined structure that stabilizes and therefore activates the adjacent catalytic domain.
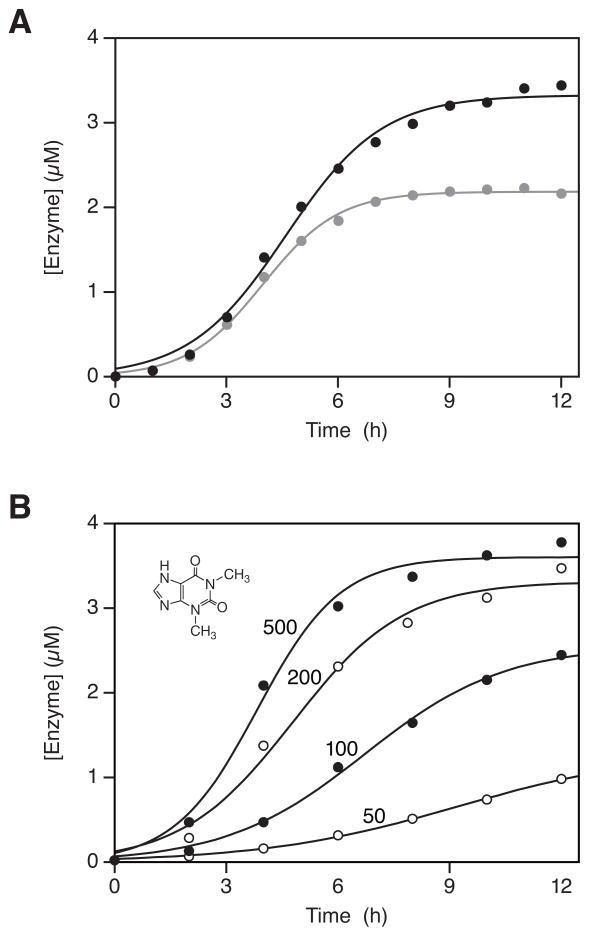
Cross-replicating enzymes that contained the theophylline aptamer exhibited exponential growth in the presence of theophylline, with growth leveling off as the supply of substrates became depleted (Figure 5A). In the absence of theophylline, or in the presence of the closely related molecule caffeine (which differs from theophylline by the presence of a single methyl group at the N7 position), no growth was detected (Lam and Joyce 2009). All-or-none, ligand-dependent, isothermal exponential amplification is highly unusual. The closest parallel is the isothermal exponential amplification of nucleic acids (Guatelli et al. 1990; Walker et al. 1992; Notomi et al. 2000), which can be highly specific for a particular target, but only applies to nucleic acid targets.
Figure 5.
Ligand-dependent exponential amplification of cross-replicating RNA enzymes that contain the theophylline aptamer (see Figure 3C). A, Amplification of E (black) and E′ (gray) occurs in the presence of 5 mM theophylline, but not 5 mM caffeine (figure based on Lam and Joyce 2009). B, The exponential growth rate depends on the concentration of theophylline, which was either 50, 100, 200, or 500 μM. The chemical structure of theophylline is shown.
The exponential growth rate of the cross-replicating aptazymes depends on the concentration of the ligand relative to the Kd of the aptamer domain (Figure 5B). This provides a way to measure the concentration of ligand in an unknown sample, analogous to quantitative PCR, but for a broad range of ligands (Lam and Joyce 2009). It also provides a means for the replicating molecules to sense their local environment, and to reflect this behavior in their reproductive fitness. Cross-replication can be made dependent on two different ligands by installing a different aptamer domain in the two members of a cross-replicating pair. This was done by installing the theophylline aptamer in E and the FMN aptamer in E′ (or vice versa). In the presence of just one ligand, linear growth was observed. This is because only one of the two enzymes was active, but still able to operate with multiple turnover. In the presence of both ligands, however, both enzymes were active and exponential amplification occurred (Lam and Joyce 2009). In principle, multiple aptamer domains could be installed in series within E or E′, resulting in more complex ligand-dependent behavior. Such tandem aptazymes have been constructed previously in the laboratory (Jose et al. 2001), and tandem riboswitches have been found to occur in nature (Sudarsan et al. 2006).
Is it Alive?
No. The artificial genetic system based on RNA enzymes that catalyze their own replication has many of the properties of a living system, but lacks the ability to bring about inventive Darwinian evolution. The molecules can undergo self-sustained replication with exponential growth. “Self-sustained” in this context refers to their ability to operate without the aid of an external catalyst. All of the genetic information that is necessary for the system to replicate and evolve is part of the system that is undergoing replication and evolution. Genetic information within the system is represented by the two regions of base-pairing interactions between the E and E′ enzymes, and that information is inherited through the process of cross-replication. The system is informational because many such genetic sequences can be represented, each of which can be maintained in a heritable fashion. Furthermore, this genetic information encodes complex phenotypic traits, reflected in the catalytic and ligand-recognition properties of the associated functional domain.
The opportunity exists for mutation through recombination within the artificial genetic system, and the resulting recombinants also are capable of propagating genetic information. However, the sequence space available to the system is meager, limited to the n × m combinations of the two genetic alleles. Sequence space in biology is far more generous due to the 4n possible combinations for a nucleic acid genome of length n. In the artificial genetic system that we have demonstrated, n and m were chosen to be 12 and 12, resulting in 144 possible cross-replicating pairs (Lincoln and Joyce 2009). In principle, n and m each could be on the order of 104–105, giving 108–1010 possible combinations. However, not all of these potential genotypes would be discriminated with high fidelity. In addition, it would be difficult for any replicator to find its corresponding substrates among a mixture of tens of thousands of potential substrates. Complexities on the order of 103 × 103 are likely to be the maximum that can be achieved, unless one resorts to methods outside the system to reduce substrate diversity in a selective manner, for example, by employing deconstructive PCR methods to convert the population of newly-formed enzymes to a daughter population of substrates (Lincoln and Joyce 2009).
Even with a complexity of 12 × 12, it was possible to carry out Darwinian evolution in the artificial genetic system, seen as the emergence of novel variants and survival of the fittest in response to a particular set of environmental conditions. Fitness can be made to reflect not just the replicative function, but also other functions that are linked to replication, such as ligand recognition. What the system cannot do, and the chief reason why it cannot be considered alive even in a molecular reductionist sense, is invent novel function within the system. There are evolved entities still lurking behind the curtain — not polymerase enzymes borrowed from biology, but the R3C catalytic motif and various aptamer motifs that were obtained by directed evolution conducted outside the system. Once placed within the synthetic genetic system these preexisting motifs can be further evolved, but how could functional motifs be invented within the system?
A living system must not only be capable of undergoing Darwinian evolution in a self-sustained manner, but also have a broad inventive capability that enables the discovery of adaptive solutions to a variety of challenges imposed by the environment. The cross-replicating system based on the R3C ligase may indeed have the capacity for inventive Darwinian evolution, but this will depend on the degree of complexity that can be implemented through a simple n × m genetics. One can imagine many thousands of replicators, each with a particular genetic sequence encoding a different randomly chosen sequence within the corresponding functional domain. A diverse population of such replicating RNAs may provide the basis for the discovery of novel function, although the extent to which such inventive capability can lead to the emergence of complex and interesting behaviors remains to be seen.
Acknowledgments
This work was supported by NASA grant NNX07AJ23G, NIH grant R01GM065130, and NSF grant MCB-0614614. I am grateful to Roslind Varghese for preparing a transcript of my lecture at the 2009 Symposium, which was the basis for this manuscript.
References
- Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Beaudry AA, Joyce GF. Directed evolution of an RNA enzyme. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]
- Burgstaller P, Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew Chemie. 1994;33:1084–1087. [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Elena SF, Cooper VS, Lenski RE. Punctuated evolution caused by selection of rare beneficial mutations. Science. 1996;272:1802–1804. doi: 10.1126/science.272.5269.1802. [DOI] [PubMed] [Google Scholar]
- Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes JD, Blacklow SC, Knowles JR. The development of enzyme catalytic efficiency: an experimental approach. Cold Spring Harbor Symp Quant Biol. 1987;52:597–602. doi: 10.1101/sqb.1987.052.01.068. [DOI] [PubMed] [Google Scholar]
- Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Jose AM, Soukup GA, Breaker RR. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 2001;7:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce GF. Nonenzymatic template-directed synthesis of informational macromolecules. Cold Spring Harbor Symp Quant Biol. 1987;52:41–51. doi: 10.1101/sqb.1987.052.01.008. [DOI] [PubMed] [Google Scholar]
- Joyce GF. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Kim DE, Joyce GF. Cross-catalytic replication of an RNA ligase ribozyme. Chem Biol. 2004;11:1505–1512. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Lam BJ, Joyce GF. Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nat Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2008;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paegel BM, Joyce GF. Darwinian evolution on a chip. PLoS Biol. 2008;6:900–906. doi: 10.1371/journal.pbio.0060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc Natl Acad Sci USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1693. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]