Abstract
This study used microarray analysis to examine age-related changes in gene expression 6 and 12 hr following a single estradiol injection in ovariectomized mice. Estradiol-responsive gene expression at the 6 hr time point was reduced in aged (18 mo) animals compared to young (4 mo) and middle-aged (MA, 12 mo) mice. Examination of gene clustering within biological and functional pathways indicated that young and MA mice exhibited increased expression of genes for cellular components of the synapse and decreased expression of genes related to oxidative phosphorylation and mitochondrial dysfunction. At the 12 hr time point, estradiol-responsive gene expression increased in aged animals and decreased in young and MA mice compared to the 6 hr time point. Gene clustering analysis indicated that aged mice exhibited increased expression of genes for signaling pathways that are rapidly influenced by estradiol. The age differences in gene expression for rapid signaling pathways may relate to disparity in basal pathway activity and estradiol mediated activation of rapid signaling cascades.
Keywords: Estrogen, Hippocampus, Microarray, Signaling
Introduction
In humans, age-related impairments in hippocampal-dependent memory begin in middle-age and cognitive weakening continues with advancing age (Foster, 2006; Small et al., 1999). Estrogen treatment in women (Sherwin, 2006), nonhuman primates (Lacreuse et al., 2002; Rapp et al., 2003), and rodents (Aenlle et al., 2009; Foster et al., 2003; Markham et al., 2002) has been shown to protect against cognitive decline. However, it is becoming apparent that estradiol treatment initiated late in life is less effective (Adams et al., 2001; Daniel et al., 2006; Foster et al., 2003; Sherwin and Henry, 2008).
The mechanism for differential estradiol effects across the lifespan is unclear. In younger animals, estradiol has numerous effects on the hippocampus that could provide a mechanism for improved cognition. For example, estradiol can rapidly activate signaling pathways for neuroprotection (Guerra et al., 2004; Jover-Mengual et al., 2007; Kuroki et al., 2001; Sarkar et al., 2008; Wu et al., 2005) and synaptogenesis (Akama and McEwen, 2003; Mukai et al., 2007) and estradiol effects on neuroprotection and synaptogenesis may be impaired in aged animals (Adams et al., 2001; Brinton, 2008; Miranda et al., 1999; Yildirim et al., 2008) suggesting a possible breakdown in estrogen signaling.
To determine whether the age differences in synaptogenesis and neuroprotection result from a weakening of the signaling pathways, we investigated differences in gene expression following an acute estradiol treatment. In vitro (Carroll et al., 2006; Schnoes et al., 2008) and in vivo studies (Fertuck et al., 2003; Naciff et al., 2007; Pechenino and Frick, 2009) have provided evidence for distinct temporal patterns of estrogen-mediated gene expression. In general, genes related to the regulation of transcription are altered within the first 2 hrs of treatment. Protein changes associated with this early transcription contribute to the amplification in the number of altered genes occurring between 4-12 hr. Furthermore, this second wave of altered genes expression, between 4-12 hr, includes genes related to the functional effects of estrogen treatment for specific cell systems. Therefore, 17β-estradiol was injected in ovariectomized mice and estradiol-responsive genes were identified by transcript profiling at 6 and 12 hr after treatment. Pathway analysis of estradiol-responsive genes identified age-related differences in functional pathways related to oxidative phosphorylation, synaptic plasticity, and estrogen responsive signaling cascades.
Materials and Methods
Subjects
Procedures involving animal subjects have been reviewed and approved by the Institutional Animal Care and Use Committee at the University of Florida and were in accordance with guidelines established by the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. Initially 85 female C57/BL6 mice were obtained from National Institute of Aging for gene array analysis, with one gene chip per animal. However, quality controls for gene arrays indicate that 5 chips were outliers and the data for these animals was removed from further analysis Therefore, a total of 80 female mice (young: n = 26, 4 months; middle-aged: n = 26, 12 months; aged: n = 28, 18 months) were employed in this study. Animals were housed 3-5 per cage and maintained on 12:12 light:dark cycle (lights on at 6 am). Following one-week habituation, mice were anesthetized (2 mg ketamine and 0.2 mg xylazine per 20 gm of body weight) and ovaries were removed through a small midline incision on the abdomen. All mice received ad lib access to food (Purina mouse chow, St Louis, MO) and water, until the surgery when they were placed on Casein based chow (Cincinnati Lab Supply, Cincinnati, OH), which is low in phytoestrogens found in soy based chow.
Hormone administration
Briefly, a single injection of 17β-estradiol (Sigma Chemical Co, St Louis, MO) or mineral oil was initiated 10 days after ovariectomy (OVX) at 10 pm or 4 am. To control for time of day effects, all animal were sacrificed between 10 - 11 am, ~4 hr after lights on and either 6 hrs (injection at 4 am) or 12 hrs (injection at 10 pm) following the injection of estradiol or oil. Estradiol was dissolved in light mineral oil (Fisher Scientific, Pittsburgh, PA) to concentration of 0.1 mg/ml. Oil or estradiol (5μg) in oil was injected subcutaneously at the nape of the neck in volumes of 0.05 ml. The groups included: young receiving oil and sacrificed 6 hr (n = 5) and 12 hr (n = 3) later; young receiving estradiol and sacrificed 6 hr (n = 10) and 12 hr (n = 8) later; middle-aged receiving oil and sacrificed 6 hr (n = 5) and 12 hr (n = 6) later; middle-aged receiving estradiol and sacrificed 6 hr (n = 9) and 12 hr (n = 6) later; aged receiving oil and sacrificed 6 hr (n = 5) and 12 hr (n = 7) later; aged receiving estradiol and sacrificed 6 hr (n = 9) and 12 hr (n = 7) later. To determine effectiveness of estradiol treatment, uteri were excised at the time of sacrifice and weighed immediately. An analysis of variance (ANOVA) was used to compare main effects on uterine weight.
At the time of sacrifice, each animal was anesthetized with CO2 and decapitated. The brain was quickly removed and placed in ice-cold artificial cerebral spinal fluid. Both hippocampus were removed, frozen in liquid nitrogen, and stored at -80°C. RNA was isolated from each sample using Qiagen RNeasy Lipid Tissue Mini Kit (Qiagen, Germantown, MD). RNA concentration was determined using spectrophometer and a subset of samples was examined using Agilent 2100 Bioanalyzer (Santa Clara, CA). Microarray analysis was performed for individual animals (one chip per animal).
Microarray Hybridization and signal detection
An amount of 5μg of total RNA was synthesized to cRNA using Affymetrix amplification kit following the manufacture’s protocol. Hybridization of cRNA was carried out by the Interdisciplinary Center for Biotechnology Research Microarray Core, University of Florida. Hybridization of Affymetrix Mouse 430 2.0 Arrays occurred for 17 hours at 60°C in accordance with manufacture’s instructions and arrays were scanned using an Affymetrix Microarray scanner. Images were analyzed using Affymetrix Gene Chip Operating System software (GCOS version 1.1) and scaled to 500. Hybridization signal intensities between GeneChips were normalized using dChip’s (Li and Wong, 2001) model-based expression index with the PM-only model. The model was used to set thresholds for identify outlier probe sets. Arrays with a large number of outlier probe sets (> 5% of total) were removed from further analysis. Data was then transferred into Microsoft excel for further analysis. Probe sets were annotated using Affymetrix® NetAffx™ (12/2007).
Real Time-PCR
Real time PCR (RT-PCR) was performed to verify microarray results. RNA from each group was treated with Turbo DNAfree (Ambion, Austin TX) to remove any remaining genomic DNA. RNA was then converted to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Primers and probes for WDFY1, GABRA2, NNT, PPARGC1A, AFT4, ENTPD4 and GAPDH were purchased from Applied Biosystems. Briefly, 3μg of total RNA from a single animal was incubated with appropriate reagents at 25°C for 10 min and then heated to 37°C for 120 min using 7300 Fast Real-Time PCR System (Applied Biosystems). For relative quantification of RNA, 100ng in 2.5μl of cDNA was added to 12.5μl of Taqman® Universal PCR Master Mix (2X), 1.25μl of 20X Gene Expression Assay Mix, a probe specific primer mixture (Table 1), and 8.75μl of nuclease-free water for a total volume of 25μl. Thermal cycler conditions were set at 2 min at 50°C, 10 min at 95°C and cycles 15s at 95°C and 1 min at 60°C for 40 cycles. The point at which the fluorescence crosses the threshold (Ct) was determined using 7300 Real-Time PCR System and SDS Software 1.3.1 analysis software (Applied Biosystems). Each sample was in triplicate and normalized to corresponding GAPDH values (ΔCtsample) and then compared to normalized young oil (ΔCtreference). The mean normalized values were compared using ΔΔCt method as described by Applied Biosystems to derive fold change (Aenlle et al., 2009), where ΔΔCt(ΔCtsample) (ΔCtreference).
Table 1. Context Sequence of Genes for RT-PCR Analysis.
| Gene Symbol | Assay ID | Context Sequence |
|---|---|---|
| WDFY1 | Mm00840455_m1 | GGGGTGTGATGGAATTTCACGTTT |
| GABRA2 | Mm01211683_m1 | CGGGAAGAGTGTAGTCAATGACAAG |
| NNT | Mm01298455_m1 | GCCAACATCTCTGGTTATAAGGCTG |
| PPARGC1A | Mm00447183_m1 | CGCAACATGCTCAAGCCAAACCAAC |
| ATF4 | Mm00515324_m1 | GCCATGGCGCTCTTCACGAAATCCA |
| ENTPD4 | Mm00491888_m1 | TTCCTGCCCTTGAGAGACATCCGGC |
| GAPDH | Mm99999915_g1 | GAACGGATTTGGCCGTATTGGGCGC |
Statistical analysis
Probe set filtering and initial statistical analysis was performed according to our previously published work (Aenlle et al., 2009; Blalock et al., 2003). Briefly, the number of present calls for each probe was determined across all chips and the probed was removed if fewer than 80% of the chips exhibited a present call for the probe. For all studies, differential expression was determined using two-tailed t-tests with the alpha level set at 0.025 in accordance with our previous studies (Aenlle et al., 2009; Blalock et al., 2003). The probes sets that exhibited an increase or decrease in expression following treatment were submitted to Ingenuity Pathway Analysis’s (IPA; Ingenuity Systems). With alpha set at p<0.025 we were able to obtain >800 molecules for generating networks, in accordance with IPA best practices for pathway analysis. The IPA program uses a right-tailed Fisher’s Exact Test to compute the likelihood that the relationship between the list of submitted genes and a set of genes representing a given pathway is due to chance. A similar procedure was employed for determining overrepresentation of genes related to synaptic structure using the Expression Analysis Systematic Explorer (EASE) through the NIHDAVID Bioinformatics Resources (Hosack et al., 2003).
Results
For each age group, all animals treated with oil (young = 8, MA = 11, aged = 12) were used as controls to determine effects of treatment for age matched animals sacrificed 6 hr (young = 10, MA = 9, aged = 9) or 12 hr (young = 8, MA = 6, aged = 7) after a single estradiol injection. To determine the effectiveness of the estradiol treatment uterine weight was compared across groups (young oil 20 ± 5 mg; young estradiol 6 hr 54 ± 3 mg; young estradiol 12 hr 50 ± 3 mg; middle-aged oil 30 ± 6 mg; middle-aged estradiol 6 hr 52 ± 3 mg; middle-aged estradiol 12 hr 43 ± 10 mg; aged oil 34 ± 4 mg; aged estradiol 6 hr 56 ± 3 mg; aged estradiol 12 hr 56 ± 4 mg). An ANOVA indicated an overall treatment effect(p < 0.0001) in the absence of an age differences and post hoc FLSD tests indicated a significant increase in uterine weight at 6 hr (p < 0.0001) and 12 hr (p < 0.0001) following treatment relative to oil treated controls.
Age differences in estradiol-responsive genes for synaptogenesis, and neuroprotection 6 hr after treatment
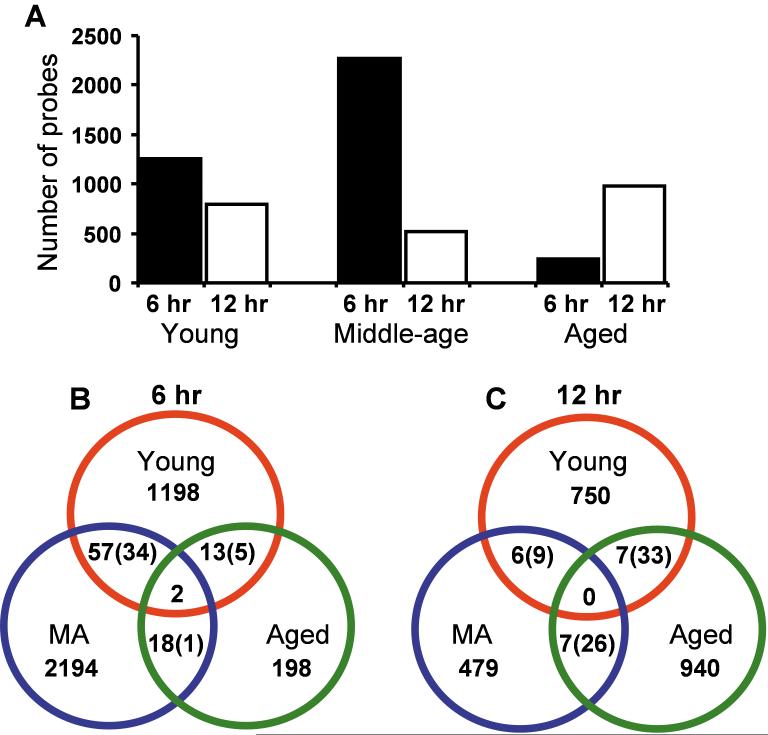
Figure 1 illustrates the number of estradiol-responsive probes for the 6 and 12 hr time points. At the 6 hr time point the MA mice exhibited the greatest shift in gene expression with approximately twice as many probes exhibiting altered expression relative to young animals and approximately a ten fold increase in the number of altered probes relative to aged mice. Age-related differences in the pattern of estradiol-responsive gene expression were also apparent. For probes that were observed to change expression at 6 hrs, young and MA mice exhibited increased expression for ~60% of the probes, while the majority (64%) of estradiol-responsive probes were decreased in aged animals. A few probes were altered in the same direction across the different age groups; however, in some cases estradiol effects were in the opposite direction (Fig 1B).
Figure 1.
Estradiol-responsive gene expression is altered over the course of aging. A) Illustration of the number of probes that were increased or decreased by estradiol treatment in young (Young), middle-aged (MA) and aged (Aged) animals at 6 hr (filled bars) or 12 hr (open bars) after a single estradiol injection. MA animals exhibited over two times the number of altered probes at 6 hr relative to the other two age groups. The number of altered probes decreased at 12 hr relative to 6 hr for young and MA animals. In contrast, aged animals exhibited approximately a five fold increase in the number of estradiol-responsive probes during this time period. B&C) Venn diagrams of the number of differentially expressed probes in response to estradiol treatment at B) 6 hr and C) 12 hr. The numbers in parentheses represent probes that changes in opposite directions.
To examine markers of synaptic components, estradiol-responsive genes were grouped according to age and whether the genes increased or decreased expression. The gene groups were submitted to DAVID Bioinformatics Resources to determine overrepresentation of genes related to the gene ontology classification for synapse cellular components (GO: 0045202). The results indicate that estradiol treatment was associated with increased expression of synaptic genes only for young (17 genes, p < 0.0005) and MA animals (26 genes, p < 0.00005) at the 6 hr time point. Four of the genes (ENAH, GRIA4, PJA2, GRIP1, GRIA1) were increased in both age groups (Table 2). A significant clustering was not observed for synaptic component genes that decreased expression (young: 4 genes; MA: 6 genes). Furthermore, aged animals did not exhibit altered expression, increasing or decreasing, for genes related to synaptic components.
Table 2. Synaptic Component Genes Increased at 6 hr in Young and MA Mice.
The Affymetrix probe identifier, gene symbol, gene description, t-test p-value and fold change are provided for genes of synaptic components that increase 6 hr following treatment in young and MA mice.
| Affymetrix | Symbol | Description | p-value | Fold |
|---|---|---|---|---|
| Young | ||||
| 1458298_at | CADPS | Ca2+ dependent activator protein for secretion | 1.58E-02 | 1.20 |
| 1443876_at | CAMK2A | Calcium/calmodulin-dependent protein kinase II alpha | 1.00E-02 | 1.19 |
| 1423286_at | CBLN1 | Cerebellin 1 precursor protein | 1.33E-02 | 1.53 |
| 1433607_at | CBLN4 | Cerebellin 4 precursor protein | 9.86E-03 | 1.38 |
| 1433451_at | CDK5R1 | Cyclin-dependent kinase 5, regulatory subunit (p35) 1 | 1.49E-03 | 1.26 |
| 1422887_a_at | CTBP2 | c-terminal binding protein 2 | 9.81E-03 | 1.20 |
| 1442223_at | ENAH | Enabled homolog (drosophila) | 9.46E-03 | 1.20 |
| 1455444_at | GABRA2 | Gamma-aminobutyric acid receptor, subunit alpha 2 | 1.06E-06 | 2.75 |
| 1434098_at | GLRA2 | Glycine receptor, alpha 2 subunit | 1.29E-02 | 1.34 |
| 1458285_at | GRIA1 | Glutamate receptor, ionotropic, ampa1 (alpha 1) | 5.61E-03 | 1.34 |
| 1440891_at | GRIA4 | Glutamate receptor, ionotropic, ampa4 (alpha 4) | 1.55E-02 | 1.48 |
| 1436575_at | GRIN3A | Glutamate receptor ionotropic, nmda3a | 1.35E-02 | 1.20 |
| 1435951_at | GRIP1 | Glutamate receptor interacting protein 1 | 4.60E-03 | 1.20 |
| 1437363_at | HOMER1 | Homer homolog 1 (drosophila) | 1.94E-02 | 1.19 |
| 1417376_a_at | IGSF4A | Immunoglobulin superfamily, member 4a | 1.19E-02 | 1.22 |
| 1450435_at | L1CAM | L1 cell adhesion molecule | 1.18E-02 | 1.15 |
| 1452328_s_at | PJA2 | Praja 2, ring-h2 motif containing | 1.78E-02 | 1.41 |
| Middle-age | ||||
| 1439220_at | ANK3 | Ankyrin 3, epithelial | 1.93E-02 | 2.01 |
| 1458525_at | APP | Amyloid beta (a4) precursor protein | 2.94E-03 | 2.03 |
| 1445798_at | DLGH1 | Discs, large homolog 1 (drosophila) | 1.95E-02 | 1.74 |
| 1446585_at | DLGH2 | Discs, large homolog 2 (drosophila) | 1.32E-02 | 1.37 |
| 1429768_at | DTNA | Dystrobrevin alpha | 5.52E-03 | 1.27 |
| 1445329_at | DTNB | Dystrobrevin, beta | 5.79E-03 | 1.66 |
| 1446426_at | ENAH | Enabled homolog (drosophila) | 4.17E-03 | 1.85 |
| 1454022_at | EPHB2 | Eph receptor b2 | 9.43E-03 | 1.58 |
| 1458285_at | GRIA1 | Glutamate receptor, ionotropic, ampa1 (alpha 1) | 1.36E-02 | 1.37 |
| 1453098_at | GRIA2 | Glutamate receptor, ionotropic, ampa2 (alpha 2) | 1.27E-02 | 1.75 |
| 1443285_at | GRIA4 | Glutamate receptor, ionotropic, ampa4 (alpha 4) | 4.53E-03 | 2.08 |
| 1440602_at | GRIK2 | Glutamate receptor, ionotropic, kainate 2 (beta 2) | 6.52E-03 | 2.57 |
| 1421350_a_at | GRIP1 | Glutamate receptor interacting protein 1 | 1.69E-02 | 1.57 |
| 1458861_at | GRM7 | Glutamate receptor, metabotropic 7 | 2.12E-02 | 1.42 |
| 1440637_at | ITSN1 | Intersectin 1 (sh3 domain protein 1a) | 1.58E-02 | 1.75 |
| 1424848_at | KCNMA1 | Potassium large conductance calcium-activated channel | 9.27E-03 | 2.99 |
| 1440807_at | MAGI2 | Membrane associated guanylate kinase | 1.97E-03 | 2.90 |
| 1420171_s_at | MYH9 | Myosin, heavy polypeptide 9, non-muscle | 1.25E-02 | 1.34 |
| 1422520_at | NEF3 | Neurofilament 3, medium | 2.28E-02 | 1.13 |
| 1447216_at | NRXN1 | Neurexin I | 1.37E-02 | 2.10 |
| 1457212_at | NRXN3 | Neurexin III | 9.69E-04 | 2.11 |
| 1444126_at | PJA2 | Praja 2, ring-h2 motif containing | 1.52E-04 | 2.07 |
| 1442620_at | PSD3 | Pleckstrin and sec7 domain containing 3 | 3.20E-04 | 2.12 |
| 1438282_at | SYT1 | Synaptotagmin I | 2.99E-03 | 2.02 |
| 1429729_at | SYT11 | Synaptotagmin 11 | 1.22E-02 | 1.88 |
| 1459009_at | UTRN | Utrophin | 2.27E-02 | 1.44 |
Estrogen responsive genes were submitted to IPA to determine whether expression changes were associated with gene-enrichment for signaling pathways. Table 3 shows the pathways that exhibited significant (p < 0.01) overrepresentation. For genes that increased expression at the 6 hr time point, only young animals exhibited overrepresentation in specific signaling pathways including PPAR/RAR signaling, which has been linked to neuroprotection (Martin et al., 2006; Rosa et al., 2008; Sanguino et al., 2006; Santos et al., 2005). Interestingly, while significant gene enrichment was not observed for the PPAR/RAR pathway in MA mice, three of the six genes that increased in MA mice were common for the young group (CLOCK, GNAQ, NCOR1). For genes that decreased expression at the 6 hr time point, young and MA animals exhibited clustering of genes for oxidative phosphorylation and mitochondrial dysfunction (Table 4), with two genes NDUFV1 and NDUFV2 decreased in both age groups.
Table 3. Estradiol-Responsive Pathways 6 hr Post Treatment.
Pathways with overrepresentation of genes that were observed to increase or decrease expression 6 hr following treatment. The p-value is calculated from a right-tailed Fisher’s Exact Test. The number of altered genes is also provided.
| Increasing | p-value | Genes | Decreasing | p-value | Genes |
|---|---|---|---|---|---|
| Young | |||||
| PPAR/RARa activation | 5.E-03 | 15 | Oxidative phosphorylation | 5.E-05 | 17 |
| Glutamate receptor signaling | 5.E-03 | 8 | Mytochondrial dysfunction | 1.E-03 | 14 |
| Circadian rhythm signaling | 1.E-02 | 4 | |||
| Middle-age | |||||
| None | Oxidative phosphorylation | 1.E-05 | 24 | ||
| Mytochondrial dysfunction | 5.E-03 | 18 | |||
| Protein ubiquination pathway | 1.E-02 | 25 | |||
| Aged | |||||
| None | None |
Table 4. Oxidative Phosphorylation and Mitochondrial Dysfunction Genes Altered at 6 hr in Young and Middle-Aged Mice.
The Affymetrix probe identifier, gene symbol, gene description, t-test p-value and fold change are provided for genes of in the oxidative phosphorylation and mitochondrial dysfunction pathways that were decrease 6 hr following treatment in young and MA mice.
| Affymetrix | Symbol | Description | p-value | Fold |
|---|---|---|---|---|
| Young | ||||
| 1417607_at | COX6A2 | cytochrome c oxidase subunit VIa polypeptide 2 | 4.53E-03 | -1.37 |
| 1424364_a_at | UCRC | ubiquinol-cytochrome c reductase complex (7.2 kD) | 1.86E-02 | -1.31 |
| 1416057_at | NDUFB11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa | 1.98E-02 | -1.28 |
| 1417286_at | NDUFA5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa | 9.06E-03 | -1.27 |
| 1454716_x_at | COX5B | cytochrome c oxidase subunit Vb | 2.13E-02 | -1.25 |
| 1437680_x_at | GLRX2 | glutaredoxin 2 | 7.65E-03 | -1.22 |
| 1423676_at | ATP5H (includes EG:10476) | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | 2.48E-02 | -1.17 |
| 1453229_s_at | HCG 25371 | hCG25371 | 7.61E-03 | -1.16 |
| 1428360_x_at | NDUFA7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa | 1.42E-02 | -1.16 |
| 1416526_a_at | PARK7 | Parkinson disease (autosomal recessive, early onset) 7 | 3.80E-03 | -1.16 |
| 1416495_s_at | NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) | 2.01E-03 | -1.14 |
| 1428322_a_at | NDUFB10 (includes EG:4716) | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa | 3.85E-03 | -1.14 |
| 1455283_x_at | NDUFS8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) | 1.12E-02 | -1.14 |
| 1415980_at | ATP5G2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 1.45E-02 | -1.13 |
| 1428075_at | NDUFB4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa | 3.76E-03 | -1.13 |
| 1426689_s_at | SDHA | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 1.11E-02 | -1.11 |
| 1428179_at | NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | 9.55E-03 | -1.10 |
| 1415966_a_at | NDUFV1 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | 1.02E-02 | -1.07 |
| 1449622_s_at | ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | 1.66E-02 | -1.07 |
| Middle-age | ||||
| 1429329_at | COX10 | COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) | 7.36E-04 | -1.38 |
| 1419544_at | ATP6V1C1 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit c1 | 1.98E-02 | -1.32 |
| 1426742_at | ATP5F1 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 9.93E-03 | -1.30 |
| 1415967_at | NDUFV1 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | 3.37E-03 | -1.30 |
| 1428782_a_at | UQCRC1 | ubiquinol-cytochrome c reductase core protein I | 3.35E-03 | -1.29 |
| 1455640_a_at | TXN2 | thioredoxin 2 | 7.96E-03 | -1.29 |
| 1423711_at | NDUFAF1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 | 1.42E-03 | -1.27 |
| 1417799_at | ATP6V1G2 | ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 | 1.13E-02 | -1.25 |
| 1423737_at | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) | 5.67E-04 | -1.24 |
| 1451312_at | NDUFS7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 4.13E-03 | -1.23 |
| 1424488_a_at | PPA2 | pyrophosphatase (inorganic) 2 | 4.49E-04 | -1.23 |
| 1448331_at | NDUFB7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa | 8.45E-03 | -1.22 |
| 1450968_at | UQCRFS1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 1.08E-02 | -1.22 |
| 1437013_x_at | ATP6V0B | ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b | 4.57E-03 | -1.21 |
| 1432264_x_at | COX7A2L | cytochrome c oxidase subunit VIIa polypeptide 2 like | 1.44E-02 | -1.21 |
| 1448153_at | COX5A | cytochrome c oxidase subunit Va | 2.60E-05 | -1.20 |
| 1448292_at | UQCR | ubiquinol-cytochrome c reductase 6.4kDa subunit | 2.33E-02 | -1.20 |
| 1448286_atHSD17B10 | hydroxysteroid (17-beta) dehydrogenase 10 | 4.13E-03 | -1.19 | |
| 1448589_at | NDUFB5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa | 1.29E-03 | -1.18 |
| 1451096_at | NDUFS2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 4.95E-03 | -1.18 |
| 1428631_a_at | ubiquinol-cytochrome c reductasecore protein II | 2.34E-02 | -1.17 | |
| 1428179_at | NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | 1.60E-03 | -1.16 |
| 1416663_at | NDUFA9 (includes EG:4704) | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 2.01E-03 | -1.16 |
| 1416952_at | ATP6V1D | ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D | 1.41E-02 | -1.16 |
| 1448203_at | ATP5L | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit G | 7.34E-03 | -1.15 |
| 1415671_at | ATP6V0D1 | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 | 1.19E-03 | -1.13 |
| 1428075_at | NDUFB4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa | 1.56E-02 | -1.12 |
Age differences in estradiol-responsive genes 12 hr after treatment
The number of probes influenced by treatment decreased from the 6 to 12 hr time points for young and MA mice. In contrast, aged animals exhibited approximately a five fold increase in the number of estradiol-responsive probes at 12 hr relative to the 6 hr time point (Fig 1A). Most of the probes (67%) for aged animals exhibited decreased expression at 12 hr. When estradiol-responsive genes were compared across age groups, common probes were usually altered in the opposite direction in aged animals compared to the other two groups (Fig 1C) and include a number of genes involved in the regulation of transcription (Table 5).
Table 5. Genes from aged mice which exhibited expression opposite young or MA mice at 12 hr.
Genes from aged mice which exhibited expression opposite young or MA mice at 12 hr. The Affymetrix probe identifier, gene symbol, gene description, t-test p-value and fold change are provided for genes in young (Y), MA, and aged (A) mice 12 hr following treatment. The p-values are all < 0.025 for aged mice and for either young or MA mice.
| p-value | Fold Change | |||||||
|---|---|---|---|---|---|---|---|---|
| Affy ID | Gene | Protein | Y | MA | A | Y | MA | A |
| 1452369_at | MAGI1 | MEMBRANE ASSOCIATED GUANYLATE KINASE, WW AND PDZ DOMAIN CONTAINING 1 | 6.3E-04 | 4.4E-01 | 3.8E-03 | -1.29 | -1.10 | 1.51 |
| 1434008_at | SCN4B | SODIUM CHANNEL, TYPE IV, BETA | 2.2E-02 | 3.9E-01 | 3.9E-03 | -1.26 | -1.16 | 1.47 |
| 1419008_at | NPY5R | NEUROPEPTIDE Y RECEPTOR Y5 | 9.2E-01 | 2.1E-02 | 5.3E-05 | 1.01 | -1.52 | 1.45 |
| 1426495_at | 2410042D21RIK | RIKEN CDNA 2410042D21 GENE | 5.2E-01 | 1.2E-02 | 1.9E-03 | 1.08 | -1.51 | 1.41 |
| 1448795_a_at | TBRG4 | TRANSFORMING GROWTH FACTOR BETA REGULATED GENE 4 | 6.8E-01 | 1.6E-02 | 5.3E-03 | -1.05 | -1.34 | 1.33 |
| 1427329_a_at | IGH-6 | IMMUNOGLOBULIN HEAVY CHAIN 6 (HEAVY CHAIN OF IGM) | 5.0E-02 | 2.3E-02 | 1.3E-02 | -1.41 | -1.27 | 1.32 |
| 1455277_at | HHIP | HEDGEHOG-INTERACTING PROTEIN | 8.4E-05 | 7.1E-01 | 1.1E-02 | -1.45 | 1.06 | 1.29 |
| 1426582_at | ATF2 | ACTIVATING TRANSCRIPTION FACTOR 2 | 1.4E-01 | 2.1E-02 | 2.4E-02 | 1.27 | -1.45 | 1.28 |
| 1429249_at | 4833424O15RIK | RIKEN CDNA 4833424O15 GENE | 7.1E-01 | 1.9E-02 | 5.9E-03 | 1.04 | -1.58 | 1.27 |
| 1456904_at | EST | 8.0E-01 | 1.0E-02 | 2.2E-02 | 1.02 | -1.18 | 1.26 | |
| 1426806_at | OBFC2A | OLIGONUCLEOTIDE BINDING FOLD CONTAINING 2A | 2.6E-01 | 1.9E-02 | 1.9E-02 | 1.15 | -1.51 | 1.25 |
| 1428429_at | RGMB | RGM DOMAIN FAMILY, MEMBER B | 2.9E-01 | 2.2E-02 | 1.3E-02 | 1.13 | -1.43 | 1.25 |
| 1435165_at | CNTN2 | CONTACTIN 2 | 2.1E-02 | 4.4E-01 | 1.1E-02 | -1.23 | -1.09 | 1.25 |
| 1416286_at | RGS4 | REGULATOR OF G-PROTEIN SIGNALING 4 | 4.9E-01 | 2.3E-02 | 5.9E-04 | 1.05 | -1.13 | 1.23 |
| 1433719_at | SLC9A9 | SOLUTE CARRIER FAMILY 9 (SODIUM/HYDROGEN EXCHANGER), ISOFORM 9 | 2.3E-02 | 4.4E-01 | 3.5E-03 | -1.18 | -1.07 | 1.21 |
| 1455734_at | CRBN | CEREBLON | 2.1E-01 | 2.4E-02 | 2.3E-03 | 1.09 | -1.15 | 1.20 |
| 1436056_at | KIF13B | KINESIN FAMILY MEMBER 13B | 7.6E-03 | 6.6E-01 | 2.4E-02 | -1.23 | 1.04 | 1.20 |
| 1419184_a_at | FHL2 | FOUR AND A HALF LIM DOMAINS 2 | 4.7E-03 | 5.6E-01 | 1.7E-02 | -1.17 | -1.03 | 1.20 |
| 1456967_at | TRIM66 | KIAA0298 HYPOTHETICAL PROTEIN (HUMAN) | 1.2E-02 | 7.4E-01 | 1.8E-02 | -1.24 | 1.04 | 1.19 |
| 1448752_at | CAR2 | CARBONIC ANHYDRASE 2 | 7.9E-01 | 2.4E-02 | 9.7E-03 | 1.02 | -1.25 | 1.16 |
| 1449164_at | CD68 | CD68 ANTIGEN | 9.2E-01 | 1.3E-02 | 7.7E-03 | -1.00 | -1.33 | 1.15 |
| 1428903_at | 3110037I16RIK | RIKEN CDNA 3110037I16 GENE | 9.8E-03 | 9.8E-01 | 1.2E-02 | -1.13 | -1.00 | 1.14 |
| 1423332_at | SDCBP | SYNDECAN BINDING PROTEIN | 7.4E-01 | 1.8E-02 | 1.4E-02 | 1.03 | -1.13 | 1.13 |
| 1429087_at | 1110054O05RIK | RIKEN CDNA 1110054O05 GENE | 2.3E-02 | 4.7E-02 | 2.0E-02 | -1.20 | -1.18 | 1.13 |
| 1429227_x_at | NAP1L1 | NUCLEOSOME ASSEMBLY PROTEIN-1 | 2.1E-01 | 1.5E-02 | 2.4E-03 | 1.09 | -1.25 | 1.13 |
| 1424801_at | ENAH | ENABLED HOMOLOG (DROSOPHILA) | 3.8E-01 | 1.9E-02 | 2.5E-02 | 1.05 | -1.20 | 1.12 |
| 1416458_at | ARF2 | ADP-RIBOSYLATION FACTOR 2 | 9.6E-01 | 1.4E-02 | 2.0E-02 | -1.00 | -1.26 | 1.12 |
| 1434440_at | GNAI1 | GUANINE NUCLEOTIDE BINDING PROTEIN, ALPHA INHIBITING 1 | 6.2E-01 | 1.4E-02 | 2.3E-02 | 1.05 | -1.10 | 1.12 |
| 1424594_at | LGALS7 | LECTIN, GALACTOSE BINDING, SOLUBLE 7 | 1.0E-02 | 3.1E-01 | 6.3E-03 | -1.12 | -1.05 | 1.12 |
| 1455403_at | MANEA | MANNOSIDASE, ENDO-ALPHA | 1.8E-02 | 9.3E-01 | 2.3E-02 | -1.12 | 1.01 | 1.11 |
| 1434612_s_at | SBNO1 | SNO, STRAWBERRY NOTCH HOMOLOG 1 (DROSOPHILA) | 8.1E-01 | 2.2E-02 | 2.4E-02 | 1.01 | -1.17 | 1.11 |
| 1448963_at | NFYC | NUCLEAR TRANSCRIPTION FACTOR-Y GAMMA | 1.8E-02 | 4.5E-01 | 1.9E-03 | -1.10 | -1.05 | 1.11 |
| 1455011_at | STARD4 | RIKEN CDNA 4632419C16 GENE | 9.2E-03 | 4.4E-03 | 2.1E-02 | -1.23 | -1.17 | 1.10 |
| 1417364_at | EEF1G | EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 GAMMA | 8.8E-01 | 2.4E-02 | 1.2E-02 | -1.01 | -1.13 | 1.09 |
| 1452159_at | 2310001A20RIK | RIKEN CDNA 2310001A20 GENE | 2.1E-04 | 2.4E-01 | 2.2E-02 | 1.17 | -1.10 | -1.10 |
| 1417252_at | NT5C | 5′,3′-NUCLEOTIDASE, CYTOSOLIC | 1.4E-03 | 1.2E-01 | 2.9E-03 | 1.32 | -1.17 | -1.12 |
| 1429048_at | BLOC1S2 | BIOGENESIS OF LYSOSOME-RELATED ORGANELLES COMPLEX-1, SUBUNIT 2 | 2.1E-02 | 2.4E-02 | 1.5E-02 | 1.24 | -1.28 | -1.14 |
| 1434521_at | RFXDC2 | REGULATORY FACTOR X DOMAIN CONTAINING 2 HOMOLOG (HUMAN) | 7.8E-01 | 9.3E-03 | 7.9E-03 | -1.01 | 1.20 | -1.15 |
| 1459874_s_at | MTMR4 | MYOTUBULARIN RELATED PROTEIN 4 | 4.9E-03 | 1.9E-01 | 1.3E-02 | 1.22 | 1.14 | -1.16 |
| 1434745_at | CCND2 | CYCLIN D2 | 1.1E-03 | 2.3E-01 | 1.5E-02 | 1.23 | -1.09 | -1.16 |
| 1456748_a_at | NIPSNAP1 | 4-NITROPHENYLPHOSPHAT ASE DOMAIN AND NON-NEURONAL SNAP25-LIKE PROTEIN H... | 1.3E-02 | 6.0E-01 | 2.2E-02 | 1.21 | 1.04 | -1.17 |
| 1426858_at | INHBB | INHIBIN BETA-B | 1.5E-02 | 5.6E-01 | 2.2E-02 | 1.20 | -1.05 | -1.17 |
| 1441003_at | ERCC4 | EXCISION REPAIR CROSS-COMPLEMENTING RODENT REPAIR DEFICIENCY, COMPLEME... | 2.8E-01 | 2.9E-03 | 2.3E-02 | -1.14 | 1.48 | -1.19 |
| 1431890_a_at | MLLT3 | DNA SEGMENT, CHR 4, ERATO DOI 321, EXPRESSED | 6.5E-03 | 8.2E-01 | 2.5E-04 | 1.14 | 1.02 | -1.20 |
| 1455940_x_at | WDR6 | WD REPEAT DOMAIN 6 | 2.4E-02 | 3.6E-01 | 2.0E-02 | 1.16 | 1.14 | -1.20 |
| 1447320_x_at | RPO1-3 | RNA POLYMERASE 1-3 | 7.7E-03 | 1.4E-01 | 1.3E-02 | 1.29 | -1.20 | -1.20 |
| 1436443_a_at | KDELC1 | KDEL (LYS-ASP-GLU-LEU) CONTAINING 1 | 7.4E-03 | 9.5E-01 | 1.6E-02 | 1.31 | -1.01 | -1.21 |
| 1448694_at | JUN | JUN ONCOGENE | 3.4E-01 | 1.4E-02 | 2.8E-03 | -1.05 | 1.13 | -1.22 |
| 1436114_at | Rnf165 | Ring finger protein 165 | 2.3E-02 | 2.4E-01 | 1.3E-03 | 1.19 | 1.11 | -1.25 |
| 1455039_a_at | SIN3B | TRANSCRIPTIONAL REGULATOR, SIN3B (YEAST) | 1.8E-02 | 6.7E-01 | 4.5E-03 | 1.23 | 1.04 | -1.30 |
| 1441727_s_at | ZFP467 | HYPOTHETICAL PROTEIN, MNCB-3350 | 1.6E-02 | 3.1E-01 | 5.6E-03 | 1.34 | 1.10 | -1.33 |
| 1456573_x_at | NNT | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE | 2.0E-03 | 5.8E-01 | 1.7E-02 | 1.77 | 1.09 | -1.33 |
| 1434210_s_at | LRIG1 | LEUCINE-RICH REPEATS AND IMMUNOGLOBULIN-LIKE DOMAINS 1 | 2.1E-03 | 9.9E-01 | 2.1E-02 | 1.42 | -1.00 | -1.34 |
| 1438157_s_at | NFKBIA | NUCLEAR FACTOR OF KAPPA LIGHT CHAIN GENE ENHANCER IN B-CELLS INHIBITOR | 1.4E-02 | 3.0E-01 | 2.4E-03 | 1.29 | 1.14 | -1.35 |
| 1439422_a_at | C1QDC2 | C1Q DOMAIN CONTAINING 2 | 2.4E-02 | 4.1E-01 | 3.2E-03 | 1.25 | -1.06 | -1.37 |
| 1429372_at | SOX11 | SRY-BOX CONTAINING GENE 11 | 3.5E-03 | 6.7E-01 | 1.3E-02 | 1.51 | 1.06 | -1.39 |
| 1446464_at | PSME4 | PROTEASOME (PROSOME, MACROPAIN) ACTIVATOR SUBUNIT 4 | 9.3E-01 | 5.4E-03 | 1.5E-02 | -1.02 | 2.11 | -1.58 |
| 1454869_at | WDR40B | WD REPEAT DOMAIN 40B | 8.2E-01 | 2.1E-02 | 1.3E-03 | -1.07 | 1.59 | -1.74 |
To examine overrepresentation in functional categories, the list of significantly altered genes was submitted to DAVID Bioinformatics Resources for examination of synaptic components. For all age groups, overrepresentation of synaptic component genes was not observed. Data were then submitted to IPA for determination of over representation in functional pathways (p < 0.01). No significant clustering was observed for genes that exhibited decreased expression, regardless of age group (Table 6). In the case of increased expression, only aged animals exhibited gene enrichment which was largely focused on signaling pathways that are rapidly influenced by estrogen including α-adrenergic signaling (Aydin et al., 2008; Bowman et al., 2002; Favit et al., 1991; Heikkinen et al., 2002), Ca2+ signaling (Brewer et al., 2006; Foster, 2005; Zhao and Brinton, 2007), synaptic plasticity (Cordoba Montoya and Carrer, 1997; Smith and McMahon, 2005; Warren et al., 1995), and IGF-1 signaling (Azcoitia et al., 1999; Donahue et al., 2006; Perez-Martin et al., 2003). Several of the genes interact with multiple signaling pathways (Table 7). Finally, the PPAR pathway was increased at 6 hr in young and 12 hr in aged mice; however, only one gene, CHUK, was common for young 6 hr and aged 12 hr groups.
Table 6. Estradiol-Responsive Pathways 12 hr Post Treatment.
Pathways with overrepresentation of genes with altered expression 12 hr following treatment. The p-value is calculated from a right-tailed Fisher’s Exact Test. The number of altered genes is also provided.
| Increasing | p-value | Genes | Decreasing | p-value | Genes |
|---|---|---|---|---|---|
| Young | |||||
| None | None | ||||
| MA | |||||
| None | None | ||||
| Aged | |||||
| α-Adrenergic signaling | 5.E-05 | 13 | None | ||
| Calcium signaling | 1.E-04 | 16 | |||
| Long-term depression signaling | 5.E-04 | 13 | |||
| Long-term potentiation signaling | 1.E-03 | 12 | |||
| G-protein coupled receptor signaling | 5.E-03 | 13 | |||
| cAMP-mediated signaling | 1.E-02 | 11 | |||
| IGF-1 signaling | 1.E-02 | 10 | |||
| PPAR signaling | 1.E-02 | 7 | |||
| Neuregulin signaling | 1.E-02 | 8 |
Table 7. Signaling Genes Altered at 12 hr in Aged Mice.
The Affymetrix probe identifier, gene symbol, gene description, t-test p-value and fold increase are provided for genes increased 12 hr following treatment in signaling pathways in aged mice. An x indicates that the gene is a member of the pathway.
| Signaling Pathways | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affymetrix | Symbol | Description | p-value | Fold | α-Adrenergic | Calcium | LTD | LTP | G-Protein | cAMP | IGF | PPAR | Neuregulin |
| 1426585_s_at | MAPK1 | mitogen-activated protein kinase 1 | 1.47E-02 | 1.14 | x | x | x | x | x | x | x | x | x |
| 1416351_at | MAP2K1 | mitogen-activated protein kinase kinase 1 | 1.08E-02 | 1.11 | x | x | x | x | x | x | x | x | |
| 1453419_at | MRAS | muscle RAS oncogene homolog | 1.27E-02 | 1.25 | x | x | x | x | x | x | x | ||
| 1452032_at | PRKAR1A | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 4.08E-05 | 1.12 | x | x | x | x | x | x | |||
| 1440132_s_at | PRKAR1B | protein kinase, cAMP-dependent, regulatory, type I, beta | 5.23E-03 | 1.14 | x | x | x | x | x | x | |||
| 1460419_a_at | PRKCB | protein kinase C, beta | 2.03E-02 | 1.16 | x | x | x | x | x | ||||
| 1418754_at | ADCY8 | adenylate cyclase 8 (brain) | 1.26E-02 | 1.24 | x | x | x | x | x | ||||
| 1426582_at | ATF2 | activating transcription factor 2 | 2.39E-02 | 1.28 | x | x | x | x | |||||
| 1433592_at | CALM1 | calmodulin 1 (phosphorylase kinase, delta) | 3.37E-03 | 1.18 | x | x | x | x | |||||
| 1434440_at | GNAI1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 2.30E-02 | 1.12 | x | x | x | x | |||||
| 1450186_s_at | GNAS | GNAS complex locus | 1.96E-02 | 1.14 | x | x | x | x | |||||
| 1417279_at | ITPR1 | inositol 1,4,5-triphosphate receptor, type 1 | 1.33E-02 | 1.28 | x | x | x | x | |||||
| 1426645_at | HSP90AA1 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | 5.62E-03 | 1.36 | x | x | |||||||
| 1422103_a_at | STAT5B | signal transducer and activator of transcription 5B | 2.07E-02 | 1.16 | x | x | |||||||
| 1417091_at | CHUK | conserved helix-loop-helix ubiquitous kinase | 1.42E-03 | 1.26 | x | x | |||||||
| 1421622_a_at | RAPGEF4 | Rap guanine nucleotide exchange factor (GEF) 4 | 2.09E-02 | 1.49 | x | x | |||||||
| 1416286_at | RGS4 | regulator of G-protein signaling 4 | 5.89E-04 | 1.23 | x | x | |||||||
| 1450202_at | GRIN1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | 4.12E-03 | 1.35 | x | x | |||||||
| 1452533_at | RYR3 | ryanodine receptor 3 | 1.02E-03 | 1.36 | x | x | |||||||
| 1440962_at | SLC8A3 | solute carrier family 8 (sodium/calcium exchanger), member 3 | 3.51E-03 | 1.26 | x | x | |||||||
| 1450655_at | PTEN | phosphatase and tensin homolog | 2.15E-02 | 1.28 | x | ||||||||
| 1419073_at | TMEFF2 | transmembrane protein with EGF-like and two follistatin-like domains 2 | 1.08E-02 | 1.16 | x | ||||||||
| 1418099_at | TNFRSF1B | tumor necrosis factor receptor superfamily, member 1B | 5.29E-03 | 1.26 | x | ||||||||
| 1419036_at | CSNK2A1 | casein kinase 2, alpha 1 polypeptide | 4.59E-03 | 1.11 | x | ||||||||
| 1421992_a_at | IGFBP4 | insulin-like growth factor binding protein 4 | 1.76E-02 | 1.32 | x | ||||||||
| 1422313_a_at | IGFBP5 | insulin-like growth factor binding protein 5 | 1.65E-03 | 1.57 | x | ||||||||
| 1417933_at | IGFBP6 | insulin-like growth factor binding protein 6 | 8.90E-03 | 1.35 | x | ||||||||
| 1450431_a_at | NEDD4 | neural precursor cell expressed, developmentally down-regulated 4 | 1.33E-02 | 1.10 | x | ||||||||
| 1452046_a_at | PPP1CC | protein phosphatase 1, catalytic subunit, gamma isoform | 1.95E-02 | 1.15 | x | ||||||||
| 1420534_at | GUCY1A3 | guanylate cyclase 1, soluble, alpha 3 | 8.46E-03 | 1.56 | x | ||||||||
| 1420871_at | GUCY1B3 | guanylate cyclase 1, soluble, beta 3 | 3.69E-03 | 1.43 | x | ||||||||
| 1453260_a_at | PPP2R2A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | 2.23E-02 | 1.23 | x | ||||||||
| 1452788_at | PPP2R5E | protein phosphatase 2, regulatory subunit B’, epsilon isoform | 1.44E-02 | 1.87 | x | ||||||||
| 1417943_at | GNG4 | guanine nucleotide binding protein (G protein), gamma 4 | 4.75E-04 | 1.30 | x | ||||||||
| 1452363_a_at | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 1.76E-03 | 1.15 | x | ||||||||
| 1417606_a_at | CALR | calreticulin | 2.45E-02 | 1.16 | x | ||||||||
| 1434572_at | HDAC9 | histone deacetylase 9 | 7.05E-04 | 1.35 | x | ||||||||
| 1424852_at | MEF2C | myocyte enhancer factor 2C | 6.59E-03 | 1.27 | x | ||||||||
| 1450243_a_at | RCAN2 | regulator of calcineurin 2 | 1.57E-02 | 1.50 | x | ||||||||
| 1423721_at | TPM1 | tropomyosin 1 (alpha) | 1.41E-02 | 1.13 | x | ||||||||
The increase in the number of altered genes at 12 hr for aged animals suggests that gene changes observed in younger mice may have been delayed in older animals. To examine this possibility we employed the gene expression data for 6 hr in young and MA mice and compared it to gene expression in aged mice at 6 and 12 hr to determine the number of genes that changed in the same direction. As illustrated in Fig 1B for age mice, the number of genes that changes in the same direction at 6 hr was 13 compared to young and 18 compared to MA. When we used the gene expression from age mice at 12 hr, we expected to observe an increase of ~4 fold, since the number of genes for the aged group increase from 198 to 940. However, relative to the young 6 hr group we saw a small increase from 13 to 19 and relative to MA animals the number of genes decreased from 18 to 4.
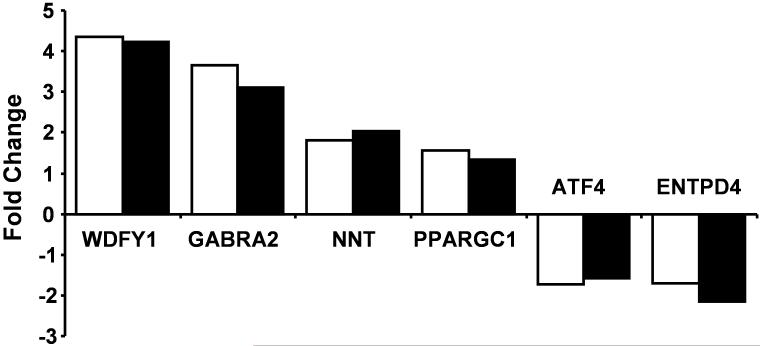
Four genes that increased (WDFY1, GABRA2, NNT, PPARGC1) and two that decreased (ATF4, ENTPD4) in young mice treated with estradiol were selected for validation of microarray results using RT-PCR. These genes were selected because they exhibited altered expression in the same direction at 6 hr and 12 hr after treatment at p < 0.025, except for PPARGC1 which was increased at 12 hr for p < 0.05. RNA isolated from oil treated mice (n = 3) was used as the control to calculate the fold change for mice (n = 3) treated 6 hr earlier with estradiol. Similarly, the fold change was calculated for values on the microarray for young oil and estradiol (6 hr) treated mice. Figure 2 illustrates that the direction and extent of altered transcription was similar for the microarray and RT-PCR.
Figure 2.
Validation of estradiol treatment effects in young animals at 6 hr for six genes using RT-PCR. The bars represent the mean fold change in gene expression for young mice 6 hr following estradiol treatment compared to the mean of age-matched oil treated animals using RT-PCR (open bars) and for microarray measures (filled bars).
Discussion
Age differences in estradiol-responsive gene signatures 6 hr after treatment
The current study examined altered gene expression in the hippocampus following estradiol treatment over the course of aging. The results reveal that aged animals were less responsive to estradiol treatment examined 6 hr after an acute treatment. In young and MA animals, estradiol treatment reduced expression for genes involved in oxidative phosphorylation and mitochondrial dysfunction. The decreased expression may represent feedback regulation due estradiol effects on oxidative phosphorylation. Altered oxidative phosphorylation is a major outcome of estradiol treatment in young animals and recent work indicates that in the brain, estradiol can enhance mitochondrial efficiency and decrease oxidative stress (Irwin et al., 2008; Massart et al., 2002; Nilsen et al., 2007; Stirone et al., 2005; Zheng and Ramirez, 1999). It is unclear whether estradiol influences oxidative phosphorylation to the same extent in aged animals. This point is important since previous research indicates that regulation of mitochondrial function and oxidative phosphorylation may constitute a corner stone for estrogen’s neuroprotective effects (Simpkins and Dykens, 2008). Thus it will be important for future studies to determine whether acute estradiol effects on oxidative phosphorylation are reduced with advanced age.
Age differences in genes that increased expression were also apparent. Estradiol treatment is associated with an increase in dendritic spines in the hippocampus of young adult rats (Gould et al., 1990; Woolley et al., 1990; Woolley and McEwen, 1992; Woolley and McEwen, 1993; Woolley et al., 1996), and this process is impaired in older rats (Adams et al., 2001; Miranda et al., 1999; Yildirim et al., 2008). We observed that young and MA, but not aged mice, exhibited an increase in expression of genes related to the synapse 6 hr after acute estradiol treatment. Young mice exhibited an increase in genes related to PPAR signaling. Although, MA did not exhibit a significant number of genes in this pathway, for the 6 genes that increased, 3 were common to young and MA animals. Aged animals exhibited increased expression of genes related to the PPAR pathway at 12 hr post treatment suggesting that the interaction of estrogen and PPAR signaling is maintained in advanced age. The ability of estrogen to increase gene expression of this pathway may be important for hippocampal aging since PPAR signaling has been implicated in the progression of Alzheimer’s disease (Dupuy et al., 2001) and neuroprotection from inflammation (Kapadia et al., 2008; Vegeto et al., 2008). Furthermore, age and sex specific changes have been noted for hippocampal PPAR signaling (Sanguino et al., 2006), suggesting that older females may be at greater risk.
Age differences in estradiol-responsive gene signatures 12 hr after treatment
In contrast to young and MA mice, which exhibited a decline in the number of altered gene between 6 and 12 hr, the number of genes with altered expression increased during this time in aged animals. For aged mice at the 12 hr time point after an acute injection, gene expression increased in signaling pathways that are rapidly influenced by estradiol including; Ca2+ signaling (Brewer et al., 2006; Foster, 2005; Zhao and Brinton, 2007), cAMP signaling (Gu and Moss, 1996), IGF-1 signaling (Azcoitia et al., 1999), and synaptic plasticity (Foy et al., 2008; Sharrow et al., 2002). The rapid activation of these pathways by estrogen is due to membrane interactions and not the result of classic transcriptional regulation. However, there is some indication for a reciprocal interaction between rapid membrane effects of estrogen on Ca2+, G-protein coupled receptor, and trophic factor signaling and estrogenic modulation of genes in these pathways (Foster, 2005).
Mechanisms for age-related differences in estradiol-responsive gene signatures
In the case of increased expression of genes for rapid signaling cascades, it may be important that these same signaling cascades decline during aging (Foster, 2005). Thus, aged cells may be differentially sensitive to estradiol influences due to age-related changes in baseline function and differential activation of rapid of signaling cascades by estradiol. Estradiol effects on Ca2+ signaling provides a prime example. Estradiol rapidly influences Ca2+ signaling (Sarkar et al., 2008; Wu et al., 2005). In turn, Ca2+ signaling can regulate gene expression through “non-classical” transcriptional regulation, independent of estrogen nuclear receptor mechanisms (Bading et al., 1993; Foster, 2005). Aged hippocampal neurons exhibit altered Ca2+ homeostasis and estrogen has effects on Ca2+-dependent processes, which are opposite that observed during aging (Foster, 2007). For example, the Ca2+-dependent afterhyperpolarization is increased with age and estradiol reduces the afterhyperpolarization (Kumar and Foster, 2002). Furthermore, estradiol pretreatment may have a greater effect on Ca2+ regulation in aged cells (Brewer et al., 2006; Brewer et al., 2009). Together the results indicate that age differences in gene expression for rapid signaling pathways may relate to disparity in basal pathway activity and estrogen mediated activation of rapid signaling cascades.
In addition, to age-related changes in rapid signaling cascades, it is likely that changes in estrogen receptors contribute to differences in gene expression. In brain regions like the hippocampus that express both estrogen receptor alpha (ERα) and beta (ERβ), the magnitude and direction of gene regulation will depend on the relative expression of each receptor and the interaction of receptors (Gonzales et al., 2008; Gottfried-Blackmore et al., 2007). While it is unclear how estrogen receptor expression changes in the hippocampus of mice, aging female mice exhibit a decrease in the transcription and expression of ERβ in the cortex (Sharma and Thakur, 2006; Thakur and Sharma, 2007). In contrast, an age-related shift in the hippocampal expression of ERα splice variants may reduce the sensitivity to estrogen treatment in women (Ishunina et al., 2007). In the hippocampus of rats, expression of both ERα and ERβ declines during aging (Mehra et al., 2005) and the loss of ERα is associated with the decreased responsiveness of hippocampal synapses to estradiol (Adams et al., 2002). Indeed, previous work indicates an important role for ERα in the estrogen-mediated increase in synaptic markers (Jelks et al., 2007; Morissette et al., 2008; Mukai et al., 2007). However, several of these studies report a similar, though usually blunted effect of ERβ activation (Jelks et al., 2007; Morissette et al., 2008; Patrone et al., 2000), suggesting that ERβ may be less active but have similar effects on transcription (Lindberg et al., 2003).
An age-related change in ERα and ERβ or a decline in rapid signaling pathways could have reduced or delayed estradiol induced signaling and gene regulation. In the current study there appears to be a delay in the expression of PPAR genes. However, only one gene was common for young 6 hr and aged 12 hr groups. Although, gene enrichment was observed for the PPAR pathway in young and aged mice, IPA analysis indicated that many more pathways were differentially influenced between the two age groups. Similarly, synaptic component genes increased for young, but not for aged mice. Finally, examination of all genes indicated little correspondence in the gene changes between young 6 hr and aged 12 hr groups. Together, the results indicate that delayed activation is not responsible for most of the age differences in altered gene expression between 6 and 12 hr. The longer-term effects of estradiol on gene transcription in aged animals remain to be determined.
It is possible that gene changes observed in aged animals could act as a priming response for successive estradiol induced changes beyond the 12 hr time point. For example, estradiol application to hippocampal slices rapidly increases ERK/MAPK activation and NMDA receptor function (Bi et al., 2003) and the magnitude of LTP (Foy et al., 2008) in young, but not aged animals. In contrast, in vivo priming with estradiol 48 hr prior to sacrifice can enhance LTP in slices from young and aged animals (Smith and McMahon, 2005; Yun et al., 2007). Previous works suggests that both estrogen receptors and rapid signaling cascades are involved in the estradiol-mediated spine growth and synaptogenesis (Akama and McEwen, 2003; Lee et al., 2004; Mukai et al., 2007; Murphy and Segal, 1996; Murphy and Segal, 1997; Yildirim et al., 2008; Znamensky et al., 2003). Thus, while young mice exhibit a rapid increase in the expression of the synaptic marker synaptophysin, an increase in synaptophysin can also be observed in aged mice following treatment with estradiol over several days (Frick et al., 2002; Spencer et al., 2008). Similarly, behavioral studies suggest that a single estradiol injection delivered after training can improve memory in young and MA animals, but not aged animals (Frick, 2009), consistent with the idea that aged animals are less responsive to a single injection. Estradiol treatment for several days prior to training reliably improves memory in middle-aged animals (Foster, 2005; Frick, 2009); however, the effects in aged animals can vary across species. Treatment prior to training improves memory in mice (Frick et al., 2002; Heikkinen et al., 2004; Vaucher et al., 2002), and is less effective in aged rats (Foster et al., 2003; Savonenko and Markowska, 2003; Talboom et al., 2008). Similar differences in responsiveness are noted for estradiol effects on synaptic markers, which can be increased in aged mice (Frick et al., 2002; Spencer et al., 2008), but not in aged rats (Adams et al., 2001; Miranda et al., 1999; Yildirim et al., 2008). The difference in rats and mice may be due to differences in the expression of estrogen receptors during aging as noted above. Regardless, the results indicate that aged animals are less responsive to a single injection of estradiol, however; depending on the species, estradiol priming may rescue estrogen responsiveness. It would be enlightening to determine whether an increase in the expression of estrogen receptors or an enhancement of rapid signaling would ameliorate age-related differences in gene changes, synaptic plasticity and memory following estradiol treatment.
Acknowledgement
This work was supported by NIA AG02499, AG14979, NIMH 059891 and the Evelyn F. McKnight Brain Research Foundation.
References
- Adams MM, Fink SE, Shah RA, Janssen WG, Hayashi S, Milner TA, McEwen BS, Morrison JH. Estrogen and aging affect the subcellular distribution of estrogen receptor-alpha in the hippocampus of female rats. J Neurosci. 2002;22(9):3608–14. doi: 10.1523/JNEUROSCI.22-09-03608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MM, Oung T, Morrison JH, Gore AC. Length of postovariectomy interval and age, but not estrogen replacement, regulate N-methyl-D-aspartate receptor mRNA levels in the hippocampus of female rats. Exp Neurol. 2001;170(2):345–56. doi: 10.1006/exnr.2001.7716. [DOI] [PubMed] [Google Scholar]
- Aenlle KK, Kumar A, Cui L, Jackson TC, Foster TC. Estrogen effects on cognition and hippocampal transcription in middle-aged mice. Neurobiol Aging. 2009;30(6):932–45. doi: 10.1016/j.neurobiolaging.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama KT, McEwen BS. Estrogen stimulates postsynaptic density-95 rapid protein synthesis via the Akt/protein kinase B pathway. J Neurosci. 2003;23(6):2333–9. doi: 10.1523/JNEUROSCI.23-06-02333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin M, Yilmaz B, Alcin E, Nedzvetsky VS, Sahin Z, Tuzcu M. Effects of letrozole on hippocampal and cortical catecholaminergic neurotransmitter levels, neural cell adhesion molecule expression and spatial learning and memory in female rats. Neuroscience. 2008;151(1):186–94. doi: 10.1016/j.neuroscience.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Azcoitia I, Sierra A, Garcia-Segura LM. Neuroprotective effects of estradiol in the adult rat hippocampus: interaction with insulin-like growth factor-I signalling. J Neurosci Res. 1999;58(6):815–22. doi: 10.1002/(sici)1097-4547(19991215)58:6<815::aid-jnr8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260(5105):181–6. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Bi R, Foy MR, Thompson RF, Baudry M. Effects of estrogen, age, and calpain on MAP kinase and NMDA receptors in female rat brain. Neurobiol Aging. 2003;24(7):977–83. doi: 10.1016/s0197-4580(03)00012-5. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23(9):3807–19. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RE, Ferguson D, Luine VN. Effects of chronic restraint stress and estradiol on open field activity, spatial memory, and monoaminergic neurotransmitters in ovariectomized rats. Neuroscience. 2002;113(2):401–10. doi: 10.1016/s0306-4522(02)00156-2. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Reichensperger JD, Brinton RD. Prevention of age-related dysregulation of calcium dynamics by estrogen in neurons. Neurobiol Aging. 2006;27(2):306–17. doi: 10.1016/j.neurobiolaging.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Brewer LD, Dowling AL, Curran-Rauhut MA, Landfield PW, Porter NM, Blalock EM. Estradiol reverses a calcium-related biomarker of brain aging in female rats. J Neurosci. 2009;29(19):6058–67. doi: 10.1523/JNEUROSCI.5253-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton RD. The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends Neurosci. 2008;31(10):529–37. doi: 10.1016/j.tins.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. others. [DOI] [PubMed] [Google Scholar]
- Cordoba Montoya DA, Carrer HF. Estrogen facilitates induction of long term potentiation in the hippocampus of awake rats. Brain Res. 1997;778(2):430–8. doi: 10.1016/s0006-8993(97)01206-7. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Hulst JL, Berbling JL. Estradiol replacement enhances working memory in middle-aged rats when initiated immediately after ovariectomy but not after a long-term period of ovarian hormone deprivation. Endocrinology. 2006;147(1):607–14. doi: 10.1210/en.2005-0998. [DOI] [PubMed] [Google Scholar]
- Donahue CP, Kosik KS, Shors TJ. Growth hormone is produced within the hippocampus where it responds to age, sex, and stress. Proc Natl Acad Sci U S A. 2006;103(15):6031–6. doi: 10.1073/pnas.0507776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AM, Mas E, Ritchie K, Descomps B, Badiou S, Cristol JP, Touchon J. The relationship between apolipoprotein E4 and lipid metabolism is impaired in Alzheimer’s disease. Gerontology. 2001;47(4):213–8. doi: 10.1159/000052801. [DOI] [PubMed] [Google Scholar]
- Favit A, Fiore L, Nicoletti F, Canonico PL. Estrogen modulates stimulation of inositol phospholipid hydrolysis by norepinephrine in rat brain slices. Brain Res. 1991;555(1):65–9. doi: 10.1016/0006-8993(91)90860-x. [DOI] [PubMed] [Google Scholar]
- Fertuck KC, Eckel JE, Gennings C, Zacharewski TR. Identification of temporal patterns of gene expression in the uteri of immature, ovariectomized mice following exposure to ethynylestradiol. Physiol Genomics. 2003;15(2):127–41. doi: 10.1152/physiolgenomics.00058.2003. [DOI] [PubMed] [Google Scholar]
- Foster TC. Interaction of rapid signal transduction cascades and gene expression in mediating estrogen effects on memory over the life span. Front Neuroendocrinol. 2005;26(2):51–64. doi: 10.1016/j.yfrne.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Foster TC. Biological markers of age-related memory deficits: treatment of senescent physiology. CNS Drugs. 2006;20(2):153–66. doi: 10.2165/00023210-200620020-00006. [DOI] [PubMed] [Google Scholar]
- Foster TC. Calcium homeostasis and modulation of synaptic plasticity in the aged brain. Aging Cell. 2007;6(3):319–25. doi: 10.1111/j.1474-9726.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Foster TC, Sharrow KM, Kumar A, Masse J. Interaction of age and chronic estradiol replacement on memory and markers of brain aging. Neurobiol Aging. 2003;24(6):839–52. doi: 10.1016/s0197-4580(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Foy MR, Baudry M, Foy JG, Thompson RF. 17beta-estradiol modifies stress-induced and age-related changes in hippocampal synaptic plasticity. Behav Neurosci. 2008;122(2):301–9. doi: 10.1037/0735-7044.122.2.301. [DOI] [PubMed] [Google Scholar]
- Frick KM. Estrogens and age-related memory decline in rodents: what have we learned and where do we go from here? Horm Behav. 2009;55(1):2–23. doi: 10.1016/j.yhbeh.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick KM, Fernandez SM, Bulinski SC. Estrogen replacement improves spatial reference memory and increases hippocampal synaptophysin in aged female mice. Neuroscience. 2002;115(2):547–58. doi: 10.1016/s0306-4522(02)00377-9. [DOI] [PubMed] [Google Scholar]
- Gonzales KL, Tetel MJ, Wagner CK. Estrogen receptor (ER) beta modulates ERalpha responses to estrogens in the developing rat ventromedial nucleus of the hypothalamus. Endocrinology. 2008;149(9):4615–21. doi: 10.1210/en.2008-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried-Blackmore A, Croft G, McEwen BS, Bulloch K. Transcriptional activity of estrogen receptors ERalpha and ERbeta in the EtC.1 cerebellar granule cell line. Brain Res. 2007;1186:41–7. doi: 10.1016/j.brainres.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Gould E, Woolley CS, Frankfurt M, McEwen BS. Gonadal steroids regulate dendritic spine density in hippocampal pyramidal cells in adulthood. J Neurosci. 1990;10(4):1286–91. doi: 10.1523/JNEUROSCI.10-04-01286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Moss RL. 17 beta-Estradiol potentiates kainate-induced currents via activation of the cAMP cascade. J Neurosci. 1996;16(11):3620–9. doi: 10.1523/JNEUROSCI.16-11-03620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra B, Diaz M, Alonso R, Marin R. Plasma membrane oestrogen receptor mediates neuroprotection against beta-amyloid toxicity through activation of Raf-1/MEK/ERK cascade in septal-derived cholinergic SN56 cells. J Neurochem. 2004;91(1):99–109. doi: 10.1111/j.1471-4159.2004.02695.x. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Puolivali J, Liu L, Rissanen A, Tanila H. Effects of ovariectomy and estrogen treatment on learning and hippocampal neurotransmitters in mice. Horm Behav. 2002;41(1):22–32. doi: 10.1006/hbeh.2001.1738. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Puolivali J, Tanila H. Effects of long-term ovariectomy and estrogen treatment on maze learning in aged mice. Exp Gerontol. 2004;39(9):1277–83. doi: 10.1016/j.exger.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin RW, Yao J, Hamilton RT, Cadenas E, Brinton RD, Nilsen J. Progesterone and estrogen regulate oxidative metabolism in brain mitochondria. Endocrinology. 2008;149(6):3167–75. doi: 10.1210/en.2007-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishunina TA, Fischer DF, Swaab DF. Estrogen receptor alpha and its splice variants in the hippocampus in aging and Alzheimer’s disease. Neurobiol Aging. 2007;28(11):1670–81. doi: 10.1016/j.neurobiolaging.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Jelks KB, Wylie R, Floyd CL, McAllister AK, Wise P. Estradiol targets synaptic proteins to induce glutamatergic synapse formation in cultured hippocampal neurons: critical role of estrogen receptor-alpha. J Neurosci. 2007;27(26):6903–13. doi: 10.1523/JNEUROSCI.0909-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover-Mengual T, Zukin RS, Etgen AM. MAPK signaling is critical to estradiol protection of CA1 neurons in global ischemia. Endocrinology. 2007;148(3):1131–43. doi: 10.1210/en.2006-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia R, Yi JH, Vemuganti R. Mechanisms of anti-inflammatory and neuroprotective actions of PPAR-gamma agonists. Front Biosci. 2008;13:1813–26. doi: 10.2741/2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Foster TC. 17beta-estradiol benzoate decreases the AHP amplitude in CA1 pyramidal neurons. J Neurophysiol. 2002;88(2):621–6. doi: 10.1152/jn.2002.88.2.621. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Fukushima K, Kanda Y, Mizuno K, Watanabe Y. Neuroprotection by estrogen via extracellular signal-regulated kinase against quinolinic acid-induced cell death in the rat hippocampus. Eur J Neurosci. 2001;13(3):472–6. doi: 10.1046/j.0953-816x.2000.01409.x. [DOI] [PubMed] [Google Scholar]
- Lacreuse A, Wilson ME, Herndon JG. Estradiol, but not raloxifene, improves aspects of spatial working memory in aged ovariectomized rhesus monkeys. Neurobiol Aging. 2002;23(4):589–600. doi: 10.1016/s0197-4580(02)00002-7. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Campomanes CR, Sikat PT, Greenfield AT, Allen PB, McEwen BS. Estrogen induces phosphorylation of cyclic AMP response element binding (pCREB) in primary hippocampal cells in a time-dependent manner. Neuroscience. 2004;124(3):549–60. doi: 10.1016/j.neuroscience.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98(1):31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg MK, Moverare S, Skrtic S, Gao H, Dahlman-Wright K, Gustafsson JA, Ohlsson C. Estrogen receptor (ER)-beta reduces ERalpha-regulated gene transcription, supporting a “ying yang” relationship between ERalpha and ERbeta in mice. Mol Endocrinol. 2003;17(2):203–8. doi: 10.1210/me.2002-0206. [DOI] [PubMed] [Google Scholar]
- Markham JA, Pych JC, Juraska JM. Ovarian hormone replacement to aged ovariectomized female rats benefits acquisition of the morris water maze. Horm Behav. 2002;42(3):284–93. doi: 10.1006/hbeh.2002.1819. [DOI] [PubMed] [Google Scholar]
- Martin B, Mattson MP, Maudsley S. Caloric restriction and intermittent fasting: two potential diets for successful brain aging. Ageing Res Rev. 2006;5(3):332–53. doi: 10.1016/j.arr.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart F, Paolini S, Piscitelli E, Brandi ML, Solaini G. Dose-dependent inhibition of mitochondrial ATP synthase by 17 beta-estradiol. Gynecol Endocrinol. 2002;16(5):373–7. [PubMed] [Google Scholar]
- Mehra RD, Sharma K, Nyakas C, Vij U. Estrogen receptor alpha and beta immunoreactive neurons in normal adult and aged female rat hippocampus: a qualitative and quantitative study. Brain Res. 2005;1056(1):22–35. doi: 10.1016/j.brainres.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Miranda P, Williams CL, Einstein G. Granule cells in aging rats are sexually dimorphic in their response to estradiol. J Neurosci. 1999;19(9):3316–25. doi: 10.1523/JNEUROSCI.19-09-03316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette M, Le Saux M, Di Paolo T. Effect of oestrogen receptor alpha and beta agonists on brain N-methyl-D-aspartate receptors. J Neuroendocrinol. 2008;20(8):1006–14. doi: 10.1111/j.1365-2826.2008.01754.x. [DOI] [PubMed] [Google Scholar]
- Mukai H, Tsurugizawa T, Murakami G, Kominami S, Ishii H, Ogiue-Ikeda M, Takata N, Tanabe N, Furukawa A, Hojo Y. Rapid modulation of long-term depression and spinogenesis via synaptic estrogen receptors in hippocampal principal neurons. J Neurochem. 2007;100(4):950–67. doi: 10.1111/j.1471-4159.2006.04264.x. others. [DOI] [PubMed] [Google Scholar]
- Murphy DD, Segal M. Regulation of dendritic spine density in cultured rat hippocampal neurons by steroid hormones. J Neurosci. 1996;16(13):4059–68. doi: 10.1523/JNEUROSCI.16-13-04059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DD, Segal M. Morphological plasticity of dendritic spines in central neurons is mediated by activation of cAMP response element binding protein. Proc Natl Acad Sci U S A. 1997;94(4):1482–7. doi: 10.1073/pnas.94.4.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naciff JM, Overmann GJ, Torontali SM, Carr GJ, Khambatta ZS, Tiesman JP, Richardson BD, Daston GP. Uterine temporal response to acute exposure to 17alpha-ethinyl estradiol in the immature rat. Toxicol Sci. 2007;97(2):467–90. doi: 10.1093/toxsci/kfm046. [DOI] [PubMed] [Google Scholar]
- Nilsen J, Irwin RW, Gallaher TK, Brinton RD. Estradiol in vivo regulation of brain mitochondrial proteome. J Neurosci. 2007;27(51):14069–77. doi: 10.1523/JNEUROSCI.4391-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrone C, Pollio G, Vegeto E, Enmark E, de Curtis I, Gustafsson JA, Maggi A. Estradiol induces differential neuronal phenotypes by activating estrogen receptor alpha or beta. Endocrinology. 2000;141(5):1839–45. doi: 10.1210/endo.141.5.7443. [DOI] [PubMed] [Google Scholar]
- Pechenino AS, Frick KM. The effects of acute 17beta-estradiol treatment on gene expression in the young female mouse hippocampus. Neurobiol Learn Mem. 2009;91(3):315–22. doi: 10.1016/j.nlm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martin M, Azcoitia I, Trejo JL, Sierra A, Garcia-Segura LM. An antagonist of estrogen receptors blocks the induction of adult neurogenesis by insulin-like growth factor-I in the dentate gyrus of adult female rat. Eur J Neurosci. 2003;18(4):923–30. doi: 10.1046/j.1460-9568.2003.02830.x. [DOI] [PubMed] [Google Scholar]
- Rapp PR, Morrison JH, Roberts JA. Cyclic estrogen replacement improves cognitive function in aged ovariectomized rhesus monkeys. J Neurosci. 2003;23(13):5708–14. doi: 10.1523/JNEUROSCI.23-13-05708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa AO, Egea J, Martinez A, Garcia AG, Lopez MG. Neuroprotective effect of the new thiadiazolidinone NP00111 against oxygen-glucose deprivation in rat hippocampal slices: implication of ERK1/2 and PPARgamma receptors. Exp Neurol. 2008;212(1):93–9. doi: 10.1016/j.expneurol.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Sanguino E, Roglans N, Rodriguez-Calvo R, Alegret M, Sanchez RM, Vazquez-Carrera M, Laguna JC. Ageing introduces a complex pattern of changes in several rat brain transcription factors depending on gender and anatomical localization. Exp Gerontol. 2006;41(4):372–9. doi: 10.1016/j.exger.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Santos MJ, Quintanilla RA, Toro A, Grandy R, Dinamarca MC, Godoy JA, Inestrosa NC. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. J Biol Chem. 2005;280(49):41057–68. doi: 10.1074/jbc.M505160200. [DOI] [PubMed] [Google Scholar]
- Sarkar SN, Huang RQ, Logan SM, Yi KD, Dillon GH, Simpkins JW. Estrogens directly potentiate neuronal L-type Ca2+ channels. Proc Natl Acad Sci U S A. 2008;105(39):15148–53. doi: 10.1073/pnas.0802379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savonenko AV, Markowska AL. The cognitive effects of ovariectomy and estrogen replacement are modulated by aging. Neuroscience. 2003;119(3):821–30. doi: 10.1016/s0306-4522(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Schnoes KK, Jaffe IZ, Iyer L, Dabreo A, Aronovitz M, Newfell B, Hansen U, Rosano G, Mendelsohn ME. Rapid recruitment of temporally distinct vascular gene sets by estrogen. Mol Endocrinol. 2008;22(11):2544–56. doi: 10.1210/me.2008-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PK, Thakur MK. Expression of estrogen receptor (ER) alpha and beta in mouse cerebral cortex: effect of age, sex and gonadal steroids. Neurobiol Aging. 2006;27(6):880–7. doi: 10.1016/j.neurobiolaging.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Sharrow KM, Kumar A, Foster TC. Calcineurin as a potential contributor in estradiol regulation of hippocampal synaptic function. Neuroscience. 2002;113(1):89–97. doi: 10.1016/s0306-4522(02)00151-3. [DOI] [PubMed] [Google Scholar]
- Sherwin BB. Estrogen and cognitive aging in women. Neuroscience. 2006;138(3):1021–6. doi: 10.1016/j.neuroscience.2005.07.051. [DOI] [PubMed] [Google Scholar]
- Sherwin BB, Henry JF. Brain aging modulates the neuroprotective effects of estrogen on selective aspects of cognition in women: a critical review. Front Neuroendocrinol. 2008;29(1):88–113. doi: 10.1016/j.yfrne.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Simpkins JW, Dykens JA. Mitochondrial mechanisms of estrogen neuroprotection. Brain Res Rev. 2008;57(2):421–30. doi: 10.1016/j.brainresrev.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Small SA, Stern Y, Tang M, Mayeux R. Selective decline in memory function among healthy elderly. Neurology. 1999;52(7):1392–6. doi: 10.1212/wnl.52.7.1392. [DOI] [PubMed] [Google Scholar]
- Smith CC, McMahon LL. Estrogen-induced increase in the magnitude of long-term potentiation occurs only when the ratio of NMDA transmission to AMPA transmission is increased. J Neurosci. 2005;25(34):7780–91. doi: 10.1523/JNEUROSCI.0762-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JL, Waters EM, Romeo RD, Wood GE, Milner TA, McEwen BS. Uncovering the mechanisms of estrogen effects on hippocampal function. Front Neuroendocrinol. 2008;29(2):219–37. doi: 10.1016/j.yfrne.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirone C, Duckles SP, Krause DN, Procaccio V. Estrogen increases mitochondrial efficiency and reduces oxidative stress in cerebral blood vessels. Mol Pharmacol. 2005;68(4):959–65. doi: 10.1124/mol.105.014662. [DOI] [PubMed] [Google Scholar]
- Talboom JS, Williams BJ, Baxley ER, West SG, Bimonte-Nelson HA. Higher levels of estradiol replacement correlate with better spatial memory in surgically menopausal young and middle-aged rats. Neurobiol Learn Mem. 2008;90(1):155–63. doi: 10.1016/j.nlm.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur MK, Sharma PK. Transcription of estrogen receptor alpha and beta in mouse cerebral cortex: effect of age, sex, 17beta-estradiol and testosterone. Neurochem Int. 2007;50(2):314–21. doi: 10.1016/j.neuint.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Vaucher E, Reymond I, Najaffe R, Kar S, Quirion R, Miller MM, Franklin KB. Estrogen effects on object memory and cholinergic receptors in young and old female mice. Neurobiol Aging. 2002;23(1):87–95. doi: 10.1016/s0197-4580(01)00250-0. [DOI] [PubMed] [Google Scholar]
- Vegeto E, Benedusi V, Maggi A. Estrogen anti-inflammatory activity in brain: a therapeutic opportunity for menopause and neurodegenerative diseases. Front Neuroendocrinol. 2008;29(4):507–19. doi: 10.1016/j.yfrne.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SG, Humphreys AG, Juraska JM, Greenough WT. LTP varies across the estrous cycle: enhanced synaptic plasticity in proestrus rats. Brain Res. 1995;703(1-2):26–30. doi: 10.1016/0006-8993(95)01059-9. [DOI] [PubMed] [Google Scholar]
- Woolley CS, Gould E, Frankfurt M, McEwen BS. Naturally occurring fluctuation in dendritic spine density on adult hippocampal pyramidal neurons. J Neurosci. 1990;10(12):4035–9. doi: 10.1523/JNEUROSCI.10-12-04035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley CS, McEwen BS. Estradiol mediates fluctuation in hippocampal synapse density during the estrous cycle in the adult rat. J Neurosci. 1992;12(7):2549–54. doi: 10.1523/JNEUROSCI.12-07-02549.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley CS, McEwen BS. Roles of estradiol and progesterone in regulation of hippocampal dendritic spine density during the estrous cycle in the rat. J Comp Neurol. 1993;336(2):293–306. doi: 10.1002/cne.903360210. [DOI] [PubMed] [Google Scholar]
- Woolley CS, Wenzel HJ, Schwartzkroin PA. Estradiol increases the frequency of multiple synapse boutons in the hippocampal CA1 region of the adult female rat. J Comp Neurol. 1996;373(1):108–17. doi: 10.1002/(SICI)1096-9861(19960909)373:1<108::AID-CNE9>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wu TW, Wang JM, Chen S, Brinton RD. 17Beta-estradiol induced Ca2+ influx via L-type calcium channels activates the Src/ERK/cyclic-AMP response element binding protein signal pathway and BCL-2 expression in rat hippocampal neurons: a potential initiation mechanism for estrogen-induced neuroprotection. Neuroscience. 2005;135(1):59–72. doi: 10.1016/j.neuroscience.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Yildirim M, Janssen WG, Tabori NE, Adams MM, Yuen GS, Akama KT, McEwen BS, Milner TA, Morrison JH. Estrogen and aging affect synaptic distribution of phosphorylated LIM kinase (pLIMK) in CA1 region of female rat hippocampus. Neuroscience. 2008;152(2):360–370. doi: 10.1016/j.neuroscience.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SH, Park KA, Kwon S, Woolley CS, Sullivan PM, Pasternak JF, Trommer BL. Estradiol enhances long term potentiation in hippocampal slices from aged apoE4-TR mice. Hippocampus. 2007;17(12):1153–7. doi: 10.1002/hipo.20357. [DOI] [PubMed] [Google Scholar]
- Zhao L, Brinton RD. Estrogen receptor alpha and beta differentially regulate intracellular Ca(2+) dynamics leading to ERK phosphorylation and estrogen neuroprotection in hippocampal neurons. Brain Res. 2007;1172:48–59. doi: 10.1016/j.brainres.2007.06.092. [DOI] [PubMed] [Google Scholar]
- Zheng J, Ramirez VD. Rapid inhibition of rat brain mitochondrial proton F0F1-ATPase activity by estrogens: comparison with Na+, K+ -ATPase of porcine cortex. Eur J Pharmacol. 1999;368(1):95–102. doi: 10.1016/s0014-2999(99)00012-6. [DOI] [PubMed] [Google Scholar]
- Znamensky V, Akama KT, McEwen BS, Milner TA. Estrogen levels regulate the subcellular distribution of phosphorylated Akt in hippocampal CA1 dendrites. J Neurosci. 2003;23(6):2340–7. doi: 10.1523/JNEUROSCI.23-06-02340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]