Abstract
Mutations in the FUS gene have recently been described as a cause of familial ALS, but their role in the pathogenesis of sporadic ALS is unclear. We undertook mutational screening of all coding exons of FUS in 228 sporadic ALS cases, and, as previous reports suggest that exon 15 represents a mutational hotspot, we sequenced this exon in an additional 1,295 sporadic cases. Six variants in six different cases were found, indicating that FUS mutations can underlie apparently sporadic ALS, but account for less than 1% of this form of disease.
Keywords: amyotrophic lateral sclerosis, sporadic disease, FUS, Italy, United States of America
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease leading to progressive paralysis and ultimately death from respiratory failure, typically within three years of symptom onset. Population-based epidemiological studies have shown that approximately 5% of cases are familial in nature, whereas the remaining 95% of cases are considered to be sporadic as they occur randomly throughout the community (Chiò, et al., 2008). Although several genetic defects underlying familial ALS have been identified (Valdmanis and Rouleau, 2008), the etiology of sporadic ALS remains poorly understood. Mutations in the fusion (involved in t(12:16) in malignant liposarcoma) FUS gene on chromosome 16p11.2 have recently been described as a cause of familial ALS (Chiò, et al., 2009,Kwiatkowski, et al., 2009,Vance, et al., 2009). However, the importance of this gene in the pathogenesis of the more common sporadic form of the disease remains unclear. We undertook mutational screening of a large cohort of sporadic cases to determine the occurrence of FUS mutations in this disease subtype.
2. Methods
2.1. Subjects
DNA samples were obtained from 436 Italian sporadic ALS patients and 1,087 US sporadic ALS patients. Of the US samples, 538 are publicly available as pre-compiled plates of DNA from the NINDS DNA Repository at the Coriell Institute for Medical Research (NDPT025 to NDPT030, www.coriell.org). All cases were diagnosed as having definite, probable or probable laboratory-supported ALS according to El Escorial criteria (Brooks, 1994), and none of them had a known family history of ALS. Demographics and clinical features of the samples are shown in Supplemental Table 1. Control samples were obtained from 368 neurologically normal Italian individuals (consisting of 280 previously published controls (Chiò, et al., 2009) and 88 additional Italian control samples) and 338 neurologically normal US individuals (pre-compiled panels NDPT006, NDPT019, NDPT021, NDPT022 that are publicly available, www.corriell.org). An additional 273 samples that are part of the Human Genome Diversity Panel (HGDP) (Cann, et al., 2002) were included in the mutational analysis as controls to evaluate the genetic variability of FUS in non-Caucasian populations. These samples originated from sixteen different geographical regions, namely Algeria (n = 10), Cambodia (n = 9), China (n = 47), Democratic Republic of Congo (n = 10), France (n = 29), Israel (n = 5), Italy (n = 17), Japan (n = 10), Kenya (n = 11), Namibia (n = 5), Nigeria (n = 20), Orkney Islands (n = 6), Pakistan (n = 32), Russia (n = 29), Senegal (n = 20), Siberia (n = 5) and South Africa (n = 8). Written informed consent for genetic analysis was obtained from each individual, and appropriate institutional review board approval was obtained concerning human subjects.
2.2 Genetic analysis
Genomic DNA was extracted from peripheral blood using standard methods. All fifteen exons and flanking introns of the FUS gene (NM_004960.2) were sequenced in 159 Italian and 69 US sporadic cases using the Big-Dye Terminator v3.1 sequencing kit (Applied Biosystems Inc., Foster City, CA, USA) run on an ABI 3730xl genetic analyzer, and analyzed using Sequencher software version 4.2 (Gene Codes Corp., Ann Arbor, MI, USA). As published studies suggest that pathogenic mutations are concentrated in exon 15 (Chiò, et al., 2009,Kwiatkowski, et al., 2009,Vance, et al., 2009), we sequenced this exon in an additional 1,295 sporadic cases.
3. Results
In our screening of 1,523 ALS cases, we found six cases carrying six distinct variants in the FUS gene (Table 1). Of these, c.1561C>T (leading to a p.R521C change in the amino acid sequence, n = 1 case) and c.1562G>A (p.R521H, n = 1) have been previously described, whereas c.198T>C (p.Y66Y, n = 1), c.1520G>A (p.G507D, n = 1), c.1552A>G (p.R518G, n = 1) and c.1575G>T (p.P525P, n = 1) were novel. The four non-synonymous variants are likely to be pathogenic, as they were not present in a large number of population-matched control subjects, and have not been previously reported in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Table 1.
Likely pathogenic FUS mutations identified in US (n = 1,087) and Italian ALS cases (n = 436).
| Mutation | Age at onset | Gender | Site of onset | Country of origin |
|---|---|---|---|---|
| c.198T>C (p.Y66Y) | 69 | Male | Limb | Italy |
| c.1520G>A (p.G507D) | 41 | Male | Limb | Italy |
| c.1552A>G (p.R518G) | 46 | Male | Limb | US* |
| c.1561C>T (p.R521C) | 34 | Male | Limb | Italy |
| c.1562G>A (p.R521H) | 37 | Male | Limb | Italy |
| c.1575G>T (p.P525P) | 55 | Female | Bulbar | US |
Coriell/NINDS DNA Repository ID for this sample is ND14136
It is unclear whether the two synonymous variants (c.198T>C, p.Y66Y; c.1575G>T, p.P525P) are truly pathogenic or merely represent benign population polymorphisms. Neither was present in 987 controls or in dbSNP. However, these variants were found in single cases, and, as would be expected for sporadic disease, it was not possible to demonstrate segregation of the variant with disease.
Clinical phenotypes of the six mutation carriers are listed in Table 1. All of the cases, except one, were male, initially manifested limb weakness, and symptom onset was before 50 years of age in more than half of the cases carrying a FUS mutation.
Two thirds of the observed mutations (4/6 mutations) were found in exon 15 of the FUS gene. Furthermore, there were no variants detected in exon 15 in either a large number of control subjects or in the 273 non-Caucasian HGDP samples. A list of variants found in cases and in controls is provided in Supplemental Table 2. Of note, the synonymous variant c.G1566A (p.R522R) was present in four ALS cases, and was not present in either 987 controls or in dbSNP. However, a second cousin (i.e. sharing a great-grandparent) of one of these cases, who had also been diagnosed with ALS, did not carry this variant, the variant has been described in control samples (Corrado, et al., 2009), and there was no difference in either FUS mRNA expression or protein level between patients carrying and not carrying the c.1566G>A variant (data not shown).
4. Discussion
In our study, FUS mutations account for less than 1% of sporadic ALS cases. It is possible that the true rate may be higher as Sanger sequencing may have missed duplications or deletions of the gene, and as our mutational screening was not designed to detect non-coding SNPs that alter disease risk. However, the chromosome 16p11.2 locus has not being identified by any of the genome-wide association studies of sporadic ALS to date, suggesting that common variants in this region are not risk factors for sporadic disease. FUS mutations were also reported to be rare in smaller cohorts of sporadic ALS patients (Belzil, et al., 2009,Corrado, et al., 2009).
Our data potentially expand the type of mutations that can disrupt the FUS gene to include synonymous mutations. The potential of synonymous mutations to cause human disease by altering mRNA splicing is increasingly recognized (Ars, et al., 2000,Bourbon, et al., 2007,Pagani, et al., 2005). However, screening of additional case and control cohorts is required to confirm the pathogenicity of the two identified synonymous mutations, and to assess their biological effect.
It is unclear whether sporadic cases carrying FUS mutations should be considered to be truly sporadic or as “apparently” sporadic cases. Correct classification of a case as familial or truly sporadic ALS is complicated by a wide variety of factors including poor diagnosis in the past, lack of knowledge of family history, mutation carriers in previous generations dying of other diseases prior to developing motor neuron degeneration, and even varying manifestations of motor and cognitive dysfunction among mutation carriers within the same family (Traynor and Singleton, 2009). Genome-wide data offer an alternative method to overcome these issues and provide further impetus to make raw genotyping data publicly available.
A strikingly uniform phenotype was observed in our mutation carrying patients, namely limb-onset disease in young individuals (Table 1). This is consistent with previous reports (Chiò, et al., 2009,Kwiatkowski, et al., 2009,Vance, et al., 2009), and mutational screening of FUS in such ALS cases may be warranted, even if they appear to be sporadic in nature. FUS mutations also appear to be more common in the Italian ALS population (4 mutations out of 436 screened cases) compared to the US population (2 mutations out of 1,087 screen cases), suggesting that mutational screening of this gene could be prioritized in Italians and patients with Italian heritage.
In keeping with previous reports, our study suggests exon 15 of the FUS gene may be a mutational hotspot. This may reflect bias arising from our study design, as exon 15 was screened in six times as many cases, compared to samples in which the entire coding region was sequenced. However, we did not find polymorphisms in exon 15 in our large series of controls taken from two Caucasian populations and from non-Caucasian populations, there are no SNPs listed in dbSNP for this exon, it is highly conserved across species, and the majority of FUS mutations described to date have been found in the last exon. These data suggest that understanding how mutations disrupt the RNA binding domain encoded by exon 15 may be key to unraveling the role of FUS in motor neuron degeneration.
Supplementary Material
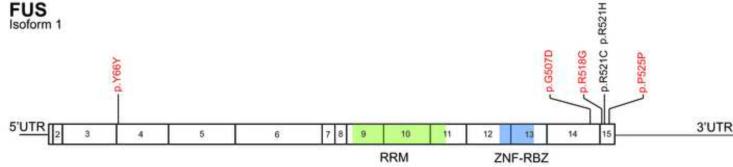
Figure 1. Distribution of FUS mutations detected in sporadic ALS patients*.
*Fifteen exons of FUS are numbered. Novel mutations are indicated in red, whereas previously described mutations are in black. RRM, RNA-recognition motif; ZNF-RBZ, zinc finger-RNA binding zone.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (Z01-AG000949-02). The work was also supported by Ministero della Salute, Ricerca Sanitaria Finalizzata 2007 (to AC, GR and GM); Fon- dazione Vialli e Mauro for ALS, Torino (to AC and GM); and Regione Piemonte, Progetti Finalizzati (to GR).
Appendix A. Other members of the ITALSGEN Consortium
Claudia Ricci (Siena), Cristina Moglia (Turin), Stefania Cammarosano (Turin), Roberto Mutani (Turin), Maura Brunetti (Torino), Irene Ossola (Torino), Laura Papetti (Milan), Marco Luigetti (Rome), Vincenzo La Bella (Palermo), Piera Paladino (Palermo), and Gabriele Siciliano (Pisa).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement The authors have no conflicts of interests.
References
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9:237–47. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, Hince P, Desjarlais A, Bouchard JP, Lacomblez L, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–9. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon M, Sun XM, Soutar AK. A rare polymorphism in the low density lipoprotein (LDL) gene that affects mRNA splicing. Atherosclerosis. 2007;195:e17–20. doi: 10.1016/j.atherosclerosis.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–2. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Mandrioli J, Salvi F, Spataro R, Schymick J, Traynor BJ, La Bella V. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Traynor BJ, Lombardo F, Fimognari M, Calvo A, Ghiglione P, Mutani R, Restagno G. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–7. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Soraru G, Carlomagno Y, Ghezzi S, Pensato V, Colombrita C, Gagliardi S, Cozzi L, Orsetti V, Mancuso M, Siciliano G, Mazzini L, Comi GP, Gellera C, Ceroni M, D'Alfonso S, Silani V. Mutations of FUS Gene in Sporadic Amyotrophic Lateral Sclerosis. J Med Genet. 2009 doi: 10.1136/jmg.2009.071027. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102:6368–72. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor BJ, Singleton AB. What's the FUS! Lancet Neurol. 2009;8:418–9. doi: 10.1016/S1474-4422(09)70088-2. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Rouleau GA. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–52. doi: 10.1212/01.wnl.0000296811.19811.db. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.