Abstract
Purpose
We have developed a statistical prediction model for prostate cancer based on four kallikrein markers in blood: total, free, and intact prostate specific antigen (PSA) and kallikrein-related peptidase 2 (hK2). Although this model accurately predicts the result of biopsy in unscreened men, its properties for men with a history of PSA screening have not been fully characterized.
Experimental Design
1501 previously screened men with elevated PSA underwent initial biopsy during rounds 2 and 3 of the European Randomized Study of Prostate Cancer Screening, Rotterdam, with 388 cancers diagnosed. Biomarker levels were measured in serum samples taken before biopsy. The prediction model developed on the unscreened cohort was then applied and predictions compared to biopsy outcome.
Results
The previously developed four-kallikrein prediction model had much higher predictive accuracy than PSA and age alone (area-under-the-curve of 0.711 vs. 0.585 and 0.713 vs. 0.557 with and without digital rectal exam, respectively; both p<0.001). Similar statistically significant enhancements were seen for high-grade cancer. Applying the model with a cut-off of 20% cancer risk as the criterion for biopsy would reduce the biopsy rate by 362 for every 1000 men with elevated PSA. Although diagnosis would be delayed for 47 cancers, these would be predominately low stage and low grade (83% Gleason 6 T1c).
Conclusions
A panel of four kallikreins can help predict the result of initial biopsy in previously screened men with elevated PSA. Use of a statistical model based on the panel would substantially decrease rates of unnecessary biopsy.
Keywords: prostate cancer, biomarkers, predictive value of tests, prostate-specific antigen, cancer screening
Statement of Translational Relevance.
The work demonstrates that a panel of four kallikrein markers is a highly accurate predictor of prostate biopsy outcome in men with who have undergone prior prostate cancer screening. The study involved application of a previously developed model and thus involves strict separation of training and validation sets. We previously demonstrated the utility of the kallikrein panel for men with no prior screening. The current study shows that the panel can also be used for men undergoing repeat prostate cancer screening, as an important aid to determining whether an initial biopsy is warranted for men with an elevated PSA. Use of the panel could substantially reduce the number of biopsies given to men at low risk of harboring prostate cancer, thereby reducing the discomfort, risk of infection, anxiety and cost associated with biopsy and likely the number of screened detected cases needed to be treated to prevent one prostate cancer death.
Introduction
A cure for metastatic prostate cancer, like most other solid tumors, remains stubbornly elusive. Accordingly, screening and early detection remains the most promising avenue for reducing prostate cancer–specific death. Yet the recent reports from two large randomized trials of screening gave equivocal results (1-2). The PLCO study found no reduction in mortality from screening, although this could, at least in part, be attributed to widespread prostate specific antigen (PSA) testing at baseline, and during the trial among the men randomized to the control group (2). The European Randomized Screening Study of Prostate Cancer (ESRPC), which had less contamination of the control arm, showed a 20% reduction in cancer mortality after 9 years. An oft-cited figure is that 48 men would need to be treated after being diagnosed through screening in order to prevent one death (1).
In addition, recent data have demonstrated some of the shortcomings of PSA as a basis for biopsy decisions. The positive predictive value of an elevated PSA is in the 20–30% range (3), implying that a large number of men receive unnecessary biopsy. In addition, many of the cancers found by PSA constitute overdiagnosis, such that treatment—which is associated with important morbidities—has little if any benefit (4).
It is plausible that supplementing PSA with other markers during prostate cancer screening would reduce both unnecessary biopsy and overdetection. Using a data set from the Göteborg section of the ERSPC, we have reported that a panel of four kallikrein markers—total PSA, free PSA, intact PSA and kallikrein-related peptidase 2 (hK2)—was strongly predictive of biopsy outcome in men with elevated PSA at their first PSA test (5). We estimated that using the model to determine referral to biopsy would reduce biopsy rates by 573 per 1000 men with elevated PSA and would miss only a small number of cancers (42 per 1000). Moreover, most of the cancers missed were the low-grade, low-stage cancers most likely to constitute overdiagnosis. We subsequently replicated this result on an independent cohort of unscreened men biopsied in the first round of ERSPC Rotterdam, with very similar results (6).
It is reasonable to suppose that PSA screening history would affect the properties of predictive models for prostate cancer. Accordingly, we applied the kallikrein panel to men biopsied in subsequent rounds of ERSPC Göteborg, to address whether it retained its value in men with a recent PSA test. We found similar increments in predictive accuracy: use of the model to determine biopsy would lead to a sharp decrease in the number of biopsies and delay the diagnosis of only 1 high-grade cancer per 1000 men with elevated PSA (7). To determine whether this finding can be replicated, here we apply the predictive model from the kallikrein panel to men with a normal PSA at initial screening and who were subsequently biopsied in rounds 2 and 3 of the Rotterdam section of the ERPSC.
Methods
Patient cohort
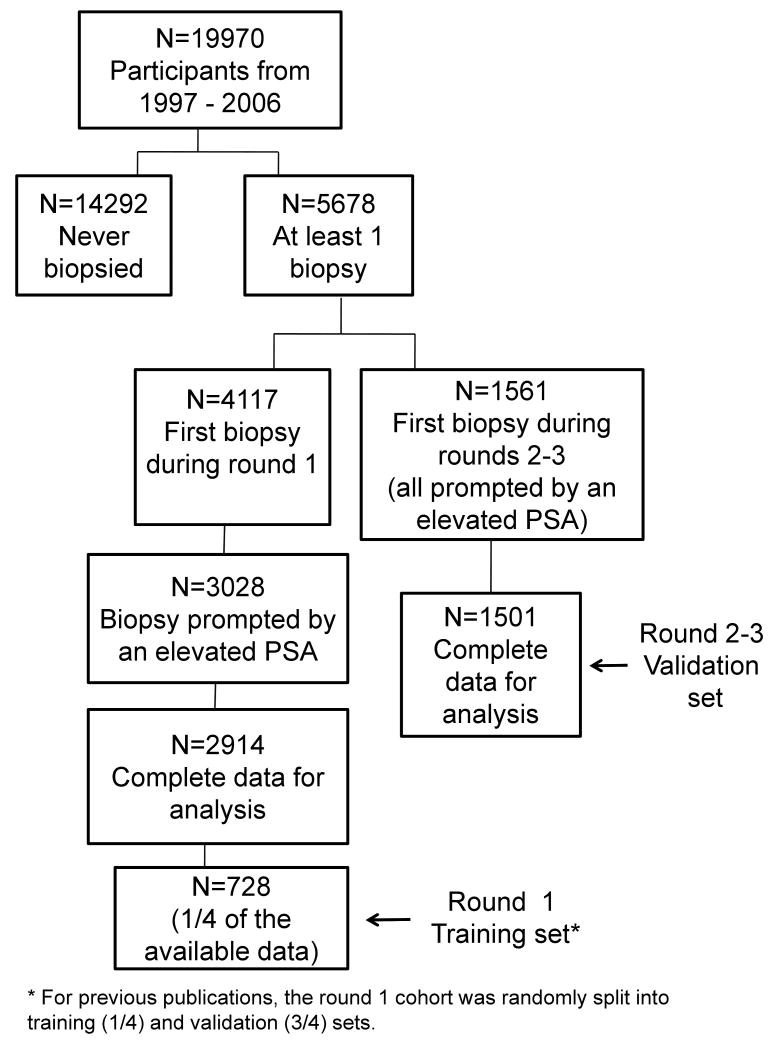
The cohort analyzed in the current analysis has been described previously (6, 8-9). In brief, we identified men with PSA < 3 ng/ml at initial screening, and who underwent a biopsy for the first time during screening rounds 2-3 of the Rotterdam section of the ERSPC. This group of men constitutes a cohort with recent PSA screening. We included only those participants whose biopsy was prompted by an elevated PSA. Following an initial PSA test (“round 1”), men not diagnosed with cancer and aged <75 were invited for up to two additional quadrennial screens (rounds 2 and 3). The current cohort includes screens conducted through 12/31/06. The rate at which participants in the screening arm continued to undergo subsequent screening was very high, with ≈80% of men participating in all 3 screening rounds, until a cancer diagnosis, or age 75. The use of materials and data was approved on the basis of the originally signed consent. Men with an elevated PSA level in serum (defined as ≥3 ng/mL) were invited to undergo subsequent clinical examination and were highly compliant (≈88%). This examination consisted of transrectal ultrasound guided laterally directed sextant biopsy and digital rectal examination (DRE). The flow of participants through the study is given in Figure 1.
Figure 1. Flow of participants.
Laboratory methods
Laboratory methods were as for our prior publication (6). Serum samples were retrieved from the archival serum bank in Rotterdam (where they had been stored frozen at -80°C after their initial processing within 3 hours from venipuncture) and shipped frozen on dry ice to Malmö, Sweden in 2005-2007. Analyses of free, total, and intact PSA and hK2 were performed in Dr. Lilja's laboratory at the Wallenberg Research Laboratories, Department of Laboratory Medicine, Lund University, University Hospital in Malmö, Sweden during 2005 and 2007. Free and total PSA were measured using the dual-label DELFIA Prostatus® total/free PSA Assay (Perkin-Elmer, Turku, Finland). Intact PSA and hK2 were measured by using F(ab′)2 fragments of the monoclonal capture antibodies in order to significantly reduce the frequency of non-specific assay interference (10). The intact PSA assay measures only free, uncomplexed intact PSA (i.e. not cleaved at Lys145-Lys146). All analyses were conducted blind to biopsy result.
Statistical considerations
Our overall hypothesis was that additional kallikreins (free PSA, intact PSA, and hK2) could enhance identification of prostate cancer diagnosis in recently screened men with elevated PSA, when compared to a base laboratory model (including age and total PSA), or a base clinical model (including age, total PSA, and DRE result). In our initial paper on the value of the kallikrein panel for previously screened men in the ERSPC Göteborg cohort (7), we came to three conclusions. First, PSA values in recently screened men referred for biopsy on the basis of an elevated PSA are extremely homogeneous; as a result, PSA is a poor predictor of biopsy outcome. Second, the additional kallikreins enhanced discrimination of prostate cancer in recently screened men. Third, the statistical model developed on unscreened men had reasonable discrimination, but poor calibration, when applied to recently screened men; accordingly, different statistical models to predict biopsy outcome are required depending on whether a man has undergone recent screening. The primary aim of the current paper was to test each of these three findings on an entirely independent data set.
Although it would have been ideal to test the statistical model developed to analyze the Göteborg data set, modifications to the assays used to measure intact PSA and hK2 in the Rotterdam cohort precluded such an analysis, as described previously (6). Accordingly, we created our statistical models using the Rotterdam round 1 training set, a random sample of one-fourth of the participants biopsied during round 1 for an elevated PSA (6). We used a slightly simplified model in comparison to our previous papers on this cohort, using only linear terms for intact PSA and hK2, and using non-linear terms (restricted cubic splines with knots at the tertiles) only for total PSA and free PSA. In addition, the equation was modified so that, in men with very high PSA, risk was based on PSA alone, to avoid unusual situations where men with very high PSA were given a low risk of cancer. The model was finalized before analysis of the data from the this cohort were conducted, such that the study involves strict separation of training and validation sets, with models developed and tested on independent cohorts.
We considered two separate base models. The base laboratory model included only information available to a laboratory: age and total PSA. The base clinical model also included the DRE result on the grounds that this would be available during a clinical consultation. We evaluated the increment in predictive accuracy when all additional kallikreins were added to the base models to create models that included four kallikreins. Predictive accuracy was given as the area under the receiver operating characteristic curve (AUC). High-grade cancer was defined as biopsy Gleason score 7 or higher. The AUC for high-grade cancer was calculated from the predicted probabilities of any cancer, that is, we did not build a separate model for the outcome of high-grade cancer. Confidence intervals and inference statistics for differences between AUCs were obtained using the method of Delong (11).
To characterize the clinical effects of the models, we used decision curve analysis (12). This method estimates a “net benefit” for prediction models by summing the benefits (true positives) and subtracting the harms (false positives), where the latter is weighted by a factor related to the relative harm of a missed cancer compared to an unnecessary biopsy. A model is of clinical value if it has the highest net benefit across the full range of threshold probabilities at which a patient would choose to be biopsied.
All statistics and graphs given here are for the independent validation set, based on the blood sample taken closest to the time of biopsy. Statistical analyses were conducted using Stata 10.1 (StataCorp LP, College Station TX).
Results
Characteristics of the 1501 participants biopsied for the first time during screening rounds 2-3 are given in Table 1. Overall, 388 cancers were diagnosed (26%), of which 91 were high grade (6%). The total PSA levels were very similar between participants with and without a cancer diagnosis (median 4.0 and 3.8 ng/ml, respectively). PSA was above 5 ng/ml for 22% (n=244) of those without a diagnosis, 21% (n=63) of those diagnosed with low-grade cancer, and 48% (n=44) of those diagnosed with high-grade cancer.
Table 1. Characteristics of participants biopsied for the first time during Rotterdam screening rounds 2-3. Values are frequency (%) or median (interquartile range).
| No cancer n=1113 |
Cancer n=388 |
|
|---|---|---|
| Biopsy round | ||
| Round 2 | 936 (84%) | 324 (84%) |
| Round 3 | 177 (16%) | 64 (16%) |
| Age | 67 (63, 71) | 67 (64, 71) |
| Total PSA, ng/mL | 3.80 (3.30, 4.80) | 4.00 (3.40, 5.20) |
| Free PSA, ng/mL | 0.84 (0.66, 1.10) | 0.75 (0.58, 1.02) |
| Intact PSA, ng/mL | 0.47 (0.36, 0.62) | 0.49 (0.37, 0.67) |
| hK2, ng/mL | 0.059 (0.041, 0.082) | 0.073 (0.051, 0.101) |
| Abnormal DRE | 194 (17%) | 121 (31%) |
| Biopsy Gleason grade | ||
| ≤6 | -- | 297 (77%) |
| 7 | -- | 78 (20%) |
| ≥ 8 | -- | 13 (3%) |
| Clinical stage | ||
| T1C | -- | 236 (61%) |
| T2 | -- | 138 (36%) |
| T3 | -- | 14 (4%) |
Models were developed with men biopsied during round 1 and independently validated for men with an initial biopsy during rounds 2-3; the predictive accuracies are given in Table 2. Both the laboratory and clinical base models had poor discriminative accuracy for prediction of any cancer (AUC 0.557 and 0.585, respectively). The addition of free PSA, intact PSA, and hK2 to these two models significantly increased discriminative accuracy (AUC 0.713 and 0.711, respectively; increment of 0.156 and 0.126, respectively; both p<0.001). For the outcome of high-grade cancer, the base laboratory model had a discriminative accuracy of 0.699 and 0.709 for the base clinical model. Nonetheless, the additional kallikreins significantly enhanced the AUC for high-grade cancer (AUC 0.793 for full laboratory model, enhancement 0.094, p=0.003; AUC 0.798 for full clinical model, enhancement 0.089, p=0.001).
Table 2. Area under the receiver operating characteristic curve for various models to predict biopsy outcome. Models were developed using men biopsied during round 1 and independently validated using men with an initial biopsy during rounds 2-3.
| Any Cancer | High-grade cancer | |||
|---|---|---|---|---|
| AUC (95% CI) | P value vs base | AUC (95% CI) | P value vs base | |
| Laboratory models | ||||
| Base: age, tPSA | 0.557 (0.524, 0.590) | -- | 0.699 (0.642, 0.755) | -- |
| Full: age, tPSA, fPSA, iPSA, hK2 | 0.713 (0.682, 0.743) | <0.001 | 0.793 (0.744, 0.842) | 0.003 |
| Clinical models | ||||
| Base: DRE, age, tPSA | 0.585 (0.551, 0.619) | -- | 0.709 (0.646, 0.771) | -- |
| Full: DRE, age, tPSA, fPSA, iPSA, hK2 | 0.711 (0.681, 0.741) | <0.001 | 0.798 (0.749, 0.847) | 0.001 |
iPSA: intact PSA; fPSA: free PSA
To put these results in a clinical context, we considered the scenario where a clinician would recommend biopsy to men with a ≥20% predicted probability of a positive biopsy (Table 3). Applying this rule with the laboratory full model would reduce the number of biopsies by 30%, while delaying the diagnosis of 32 low-grade cancers and only 4 high-grade cancers per 1000 men biopsied. Applying this rule with the clinical full model would reduce the number of biopsies by 36%, with a delayed diagnosis for 43 low-grade and 4 high-cancers per 1000 men biopsied. Of the 43 low-grade cancers with a delayed diagnosis, 39 would be T1C and 4 would be T2; of the 4 high-grade cancers with a delayed diagnosis, 3 would T1C and 1 a T2.
Table 3. Reduction in biopsies per 1000 men, when biopsying only those with a 20% or higher predicted probability of positive biopsy result.
| Biopsies | Cancers | High-grade cancers | ||||
|---|---|---|---|---|---|---|
| Performed | Reduced | Caught | Missed | Caught | Missed | |
| Laboratory full model | 702 | 298 | 222 | 36 | 57 | 4 |
| Clinical full model | 638 | 362 | 211 | 47 | 57 | 4 |
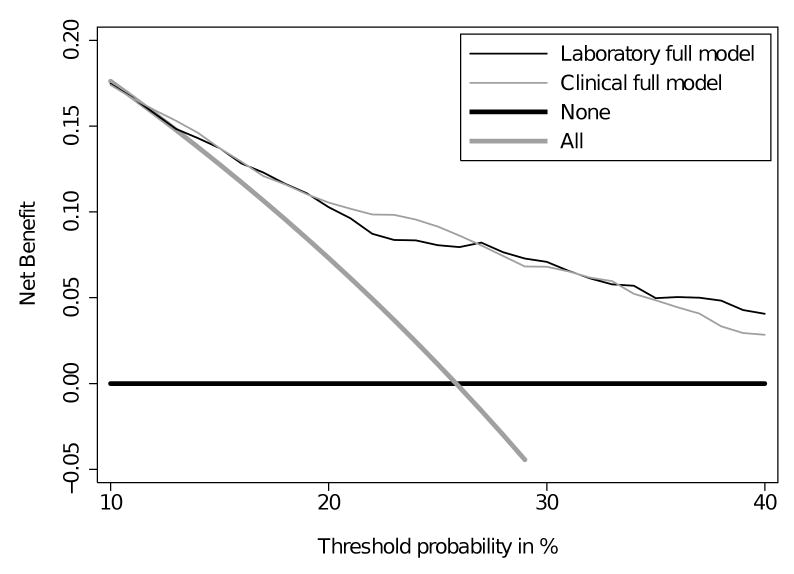
We have shown the benefit of the full models using a single cut-point of 20% as the threshold probability for biopsy. Since preferences will vary for the individual patient and clinician, we plotted a decision curve to explore how individual preferences might impact the results. Decision curve analysis (figure 2) demonstrates that the full models were superior to the base models and the strategies of biopsying all men or biopsying no men for the full range of relevant threshold probabilities (10-40%). This demonstrates that using the kallikrein model to determine biopsy would lead to superior clinical outcomes irrespective of a patient's or clinician's preference as to the relative harms and benefits of cancer detection versus unnecessary biopsy.
Figure 2. Decision curve analysis.
Clinical net benefit for various biopsy strategies plotted against the risk threshold at which a patient or clinician would opt for biopsy. The clinical net benefit is greater across the full range of reasonable risk thresholds for biopsying based on the kallikrein models than for the alternative strategies of biopsying all men with elevated PSA or biopsying no men.
To examine whether a separate statistical model is required for men with a history of recent screening, we created a model using the round 2–3 data. The predictive accuracies of the models were very similar to those given in Table 2 (AUC for any cancer: laboratory base model 0.575, laboratory full model 0.713, clinical base model 0.609, clinical full model 0.725). The models also led to similar clinical outcomes. For example, the clinical full model would reduce the number of biopsies by 44%, with a delayed diagnosis of 57 low-grade and 4 high-grade cancers per 1000 men biopsied. Therefore, we saw no evidence to suggest that a model developed among men without recent screening is not appropriate for men with a history of recent screening. Table 4 compares the incremental value of the different kallikreins for the Rotterdam and Göteborg cohorts. Overall, the updated assays used in Rotterdam appear to improve discrimination.
Table 4. Incremental enhancement in AUC for prediction of any cancer attributable to each of the additional kallikreins.
Results are presented separately for the Rotterdam cohort and Göteborg cohort (7), when models are developed among unscreened men and independently validated among recently screened men
| Rotterdam Cohort | Göteborg Cohort | |||
|---|---|---|---|---|
| AUC (95% CI) | Incremental enhancement | AUC (95% CI) | Incremental enhancement | |
| Laboratory models | ||||
| Full: age, tPSA, fPSA, iPSA, hK2 | 0.713 (0.682, 0.743) | -- | 0.656 (0.621, 0.691) | -- |
| Without fPSA | 0.576 (0.542, 0.610) | 0.137 | 0.620 (0.583, 0.657) | 0.036 |
| Without iPSA | 0.669 (0.637, 0.701) | 0.044 | 0.660 (0.625, 0.695) | -0.004 |
| Without hK2 | 0.701 (0.671, 0.732) | 0.012 | 0.592 (0.556, 0.629) | 0.064 |
| Clinical models | ||||
| Full: DRE, age, tPSA, fPSA, iPSA, hK2 | 0.711 (0.681, 0.741) | -- | 0.678 (0.643, 0.712) | -- |
| Without fPSA | 0.606 (0.573, 0.640) | 0.105 | 0.651 (0.614, 0.687) | 0.027 |
| Without iPSA | 0.679 (0.648, 0.710) | 0.032 | 0.683 (0.648, 0.718) | -0.005 |
| Without hK2 | 0.700 (0.670, 0.731) | 0.011 | 0.632 (0.596, 0.669) | 0.046 |
The ERSPC study used 6-core biopsies, and in general a more extensive biopsy leads to more cancers being detected. In our previous analyses of the kallikrein panel with ERSPC cohorts, we performed sensitivity analyses to address whether biopsy scheme would impact our results. In the sensitivity analyses, we assumed that men with a positive biopsy within 4 years of initial biopsy would have had cancer detected at initial biopsy if they had received an extended core biopsy. We considered performing a similar sensitivity analysis for this cohort; however, we found that there were only 30 such cases of cancers diagnosed with an additional 4 years of follow-up. Therefore, it is very unlikely that we would come to different conclusions had extended core biopsies been performed. Note also the number of aggressive cancers with a poor outcome missed by sextant biopsy has been shown to be extremely low(13).
Discussion
In this study, we used an independent cohort to test three previously published results. We replicated our finding that PSA is a poor predictor of initial biopsy outcome in men with a recent screening history and with elevated PSA, and that three additional kallikreins can dramatically improve discrimination and thereby reduce unnecessary biopsy in this group of men. However, we failed to replicate our finding that different statistical models for the kallikreins are required for recently screened men as compared to unscreened men.
There is accumulating evidence that prior screening affects the properties of PSA. A widely cited report by Stamey et al found that the association between PSA and cancer characteristics decreased over time as a result of PSA screening, leading to the conclusion that “the prostate specific antigen era … is over” (14). In three separate studies biopsy outcome of men with elevated PSA after repeat screening - our report here, our previous report on the Göteborg section of the ERSPC (7), and Eggener et al's paper from a US screening cohort (15) – the predictive accuracy of PSA been reported to have an AUC of close to 0.55, little better than a coin-flip. Moreover, in a study of a clinical cohort, Schwartz et al reported that PSA entirely lost its ability to predict biopsy outcome between 1993 and 2005, an effect that might be explained, at least in part, by the increased proportion of men with elevated PSA having undergone prior screening. The results of the Prostate Cancer Prevention Trial (PCPT) (16) stand in distinction to these findings: even though participants in the PCPT participated in yearly PSA tests, the predictive of value of PSA was maintained, with an AUC of 0.68. The clear difference in the PCPT is that men received a protocolled biopsy irrespective of PSA level. We can conclude that in a recently screened man with a PSA elevated above typical biopsy thresholds, such as 3 ng/ml, the degree of elevation does not markedly affect risk: a PSA, 6 ng/ml, does not constitute an importantly higher risk than a PSA of 4 ng/ml. However, PSAs above commonly used thresholds represent a higher risk than those below: a PSA of 4 ng/ml does represent a much higher risk than a PSA of 1 ng/ml(16).
Our finding that the kallikrein panel helps predict the result of biopsy is also highly consistent with the prior literature. We have now found that the kallikrein panel improves discrimination in unscreened men in both an initial study (5) and independent replication set (6), and also in previously screened men (7), with the current study replicating those findings. We have also investigated two of the three additional kallikreins, free PSA and hK2, for long-term prediction of clinically diagnosed prostate cancer, and found that they improve prediction over and above that of PSA alone in a cohort of 60-year-old men (17). Several other reports support our findings. A large number of reports have shown that free PSA (or free-to-total PSA ratio) improves discrimination of prostate cancer (18). It is likely that intact PSA may reflect prostate pathology in a manner quite similar to a proPSA assay that was highly predictive of cancer in a recently screened population (19). hK2 has been shown to be highly predictive of prostate cancer in several studies by independent authors (20-22). To our knowledge, we are the first group to combine all four markers into a single statistical model.
We hypothesize that there are two general explanations for why a new model for recently screened men was necessary with the Göteborg cohort but not with the Rotterdam cohort. First, the screening interval was longer for Rotterdam than for Göteborg (4 years versus 2 years). The longer the screening interval, the less homogenous PSA and other measurements will be at subsequent screenings. For example, 44% versus 30% of men biopsied in subsequent rounds of Rotterdam and Göteborg sections had PSA > 4 ng/ml, and 11% versus 5% had PSA > 6 ng/ml. In other words, due to the longer screening interval, the men biopsied in the first versus subsequent screening rounds were more similar in the Göteborg cohort than in the Rotterdam cohort, and the models have greater predictive performance as a result. Second, a newer version of the assays to measure intact PSA and hK2 was used for the Rotterdam measurements, which led to more precise measurements of both hK2 and intact PSA, which is a subform of free PSA(10). The improved measurements appear to have led to better enhancements in predictive accuracy associated with free PSA and intact PSA (table 4). For example, the addition of intact PSA enhanced the AUC by 0.044 in the Rotterdam cohort, but did not enhance the AUC at all in the Göteborg cohort. These enhancements also appear to have translated into improved calibration with the addition of these markers. In both Göteborg and Rotterdam, models lacking either intact PSA or free PSA were not well calibrated; with the addition of these markers in the Rotterdam cohort, the full models achieved good calibration.
Clear strengths of our study include the use of completely independent training and validation sets, the replication of a previously published model, and the very close concordance between our initial findings and those reported here. For example, we initially reported (7) that AUC for the detection of any cancer increased from 0.564 for PSA and age to 0.674 with the additional kallikreins; the comparable numbers here are 0.557 and 0.713. For prediction of high-grade cancer, our prior paper gave an AUC of 0.717 for the base model including DRE and 0.828 for the kallikrein panel plus DRE; here we report AUCs of 0.709 and 0.798, respectively.
An additional strength is our use of a decision analytic approach, clearly demonstrating that application of the kallikrein panel would improve clinical decision-making. Again, we find a strong concordance between our previously published results and those shown here. We initially reported that use of the kallikrein panel plus DRE, using a threshold probability of 20% for biopsy, would lead to 369 fewer biopsies per 1000 men with elevated PSA, but would lead to a failure to biopsy 66 men with low-grade cancer and 5 men with high-grade disease. The comparable figures reported here are 362, 43, and 4.
As previously pointed out (6), a major advantage of our approach is that it does not require novel clinical procedures, such as collection of urine after prostatic massage (23), or novel laboratory tests, such as mass spectroscopy (24). Indeed, the very same blood sample identifying a man as potentially in need of biopsy—that is, a PSA of 3 ng/ml or above—could be retested for the additional kallikreins. It would also be relatively straightforward to convert the current PSA assay to a multiplex kallikrein assay.
In a previous analysis of this cohort(8), we did not find that PSA velocity helped to predict biopsy outcome. Use of PSA velocity added only slightly to predictive accuracy, and once men with high PSA velocities, who had a lower risk of cancer, were excluded there was no benefit to PSA velocity. Hence, the kallikrein panel should be used on its own without inclusion of PSA velocity.
We recognize two limitations of our study. First, participants in this study were subject to sextant biopsy, as was current practice when the PCPT was designed. Contemporary biopsies more typically involve 10 or 12 cores. We see no reason why the additional yield of prostate cancers from a more extensive biopsy would materially impact the predictive value of a marker that is highly predictive of cancer detected on sextant biopsy. Moreover, in our prior paper we demonstrated that the increment in predictive accuracy associated with the full kallikrein panel was similar when simulating the results of an extended biopsy (5-7). The second limitation is that the samples available for testing had been stored for several years, and may have been thawed and refrozen before analysis. Previous reports have shown that long-term storage and repeated freezing and rethawing degrade kallikreins (25), affecting the predictive accuracy of the four-kallikrein panel. Yet this would act as a conservative bias, acting to decrease the apparent clinical value of our approach.
In summary, we have replicated, using an independent sample, a published finding that a panel of four kallikreins can help predict the result of initial biopsy in previously screened men with elevated PSA who have no history of negative biopsy. Our models can therefore be used to determine which men should be advised to have biopsy and which might be advised to continue screening, but defer biopsy until evidence of malignancy was stronger. As such, the models have the capacity to shift favorably the balance between the benefits and harms of PSA screening.
Acknowledgments
This research was funded by a P50-CA92629 SPORE from the National Cancer Institute (Pilot Project 7), Swedish Cancer Society project no. 0345, Swedish Research Council (Medicine) project no. 20095, European Union 6th Framework contract LSHC-CT-2004-503011 (P-Mark), Academy of Finland (Project 206690), Fundación Federico SA, funds from David H. Koch provided through the Prostate Cancer Foundation, and by The Sidney Kimmel Center for Prostate and Urologic Cancers. The ERSPC Rotterdam study was funded by grants of the Dutch Cancer Society, The Netherlands Organisation for Health Research and Development, 6th Framework Program of the EU: P-Mark, and by Beckman Coulter Hybritech Inc.
References
- 1.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, Crawford ED, Grubb RL, 3rd, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulmert D, Serio AM, O'Brien MF, et al. Long-term prediction of prostate cancer: prostate-specific antigen (PSA) velocity is predictive but does not improve the predictive accuracy of a single PSA measurement 15 years or more before cancer diagnosis in a large, representative, unscreened population. J Clin Oncol. 2008;26:835–41. doi: 10.1200/JCO.2007.13.1490. [DOI] [PubMed] [Google Scholar]
- 4.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986-2005. J Natl Cancer Inst. 2009;101:1325–9. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med. 2008;6:19. doi: 10.1186/1741-7015-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickers A, Cronin A, Roobol M, et al. Reducing unnecessary biopsy during prostate cancer screening using a four kallikrein panel: an independent replication. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.24.1968. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vickers AJ, Cronin AM, Aus G, et al. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated prostate specific antigen: data from the European Randomized Study of Prostate Cancer Screening in Gothenburg, Sweden. Cancer. 2010 doi: 10.1002/cncr.25010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vickers AJ, Wolters T, Savage CJ, et al. Prostate-Specific Antigen Velocity for Early Detection of Prostate Cancer: Result from a Large, Representative, Population-based Cohort. Eur Urol. 2009 doi: 10.1016/j.eururo.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roobol MJ, Kirkels WJ, Schroder FH. Features and preliminary results of the Dutch centre of the ERSPC (Rotterdam, the Netherlands) BJU Int. 2003;92 2:48–54. doi: 10.1111/j.1464-410x.2003.04390.x. [DOI] [PubMed] [Google Scholar]
- 10.Vaisanen V, Peltola MT, Lilja H, Nurmi M, Pettersson K. Intact free prostate-specific antigen and free and total human glandular kallikrein 2. Elimination of assay interference by enzymatic digestion of antibodies to F(ab′)2 fragments. Anal Chem. 2006;78:7809–15. doi: 10.1021/ac061201+. [DOI] [PubMed] [Google Scholar]
- 11.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 12.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder FH, van den Bergh RC, Wolters T, et al. Eleven-Year Outcome of Patients with Prostate Cancers Diagnosed During Screening After Initial Negative Sextant Biopsies. Eur Urol. 2009 doi: 10.1016/j.eururo.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Stamey TA, Caldwell M, McNeal JE, Nolley R, Hemenez M, Downs J. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol. 2004;172:1297–301. doi: 10.1097/01.ju.0000139993.51181.5d. [DOI] [PubMed] [Google Scholar]
- 15.Eggener SE, Yossepowitch O, Roehl KA, Loeb S, Yu X, Catalona WJ. Relationship of prostate-specific antigen velocity to histologic findings in a prostate cancer screening program. Urology. 2008;71:1016–9. doi: 10.1016/j.urology.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Thompson IM, Ankerst DP, Chi C, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 17.Vickers AJ, Ulmert D, Serio AM, et al. The predictive value of prostate cancer biomarkers depends on age and time to diagnosis: towards a biologically-based screening strategy. Int J Cancer. 2007;121:2212–7. doi: 10.1002/ijc.22956. [DOI] [PubMed] [Google Scholar]
- 18.Roddam AW, Duffy MJ, Hamdy FC, et al. Use of prostate-specific antigen (PSA) isoforms for the detection of prostate cancer in men with a PSA level of 2-10 ng/ml: systematic review and meta-analysis. Eur Urol. 2005;48:386–99. doi: 10.1016/j.eururo.2005.04.015. discussion 98-9. [DOI] [PubMed] [Google Scholar]
- 19.Mikolajczyk SD, Catalona WJ, Evans CL, et al. Proenzyme forms of prostate-specific antigen in serum improve the detection of prostate cancer. Clin Chem. 2004;50:1017–25. doi: 10.1373/clinchem.2003.026823. [DOI] [PubMed] [Google Scholar]
- 20.Nam RK, Zhang WW, Trachtenberg J, et al. Single nucleotide polymorphism of the human kallikrein-2 gene highly correlates with serum human kallikrein-2 levels and in combination enhances prostate cancer detection. J Clin Oncol. 2003;21:2312–9. doi: 10.1200/JCO.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Martin BJ, Cheli CD, Sterling K, et al. Prostate specific antigen isoforms and human glandular kallikrein 2--which offers the best screening performance in a predominantly black population? J Urol. 2006;175:104–7. doi: 10.1016/S0022-5347(05)00069-8. [DOI] [PubMed] [Google Scholar]
- 22.Martin BJ, Finlay JA, Sterling K, et al. Early detection of prostate cancer in African-American men through use of multiple biomarkers: human kallikrein 2 (hK2), prostate-specific antigen (PSA), and free PSA (fPSA) Prostate Cancer Prostatic Dis. 2004;7:132–7. doi: 10.1038/sj.pcan.4500706. [DOI] [PubMed] [Google Scholar]
- 23.Deras IL, Aubin SM, Blase A, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008;179:1587–92. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 24.Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–84. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulmert D, Becker C, Nilsson JA, et al. Reproducibility and accuracy of measurements of free and total prostate-specific antigen in serum vs plasma after long-term storage at -20 degrees C. Clin Chem. 2006;52:235–9. doi: 10.1373/clinchem.2005.050641. [DOI] [PubMed] [Google Scholar]