Abstract
We applied an established and commercially available serum proteomic classifier for survival after treatment with erlotinib (VeriStrat®) in a blinded manner to pre-treatment sera obtained from recurrent advanced NSCLC patients before treatment with the combination of erlotinib plus bevacizumab. We found that VeriStrat® could classify these patients into two groups with significantly better or worse outcomes and may enable rational selection of patients more likely to benefit from this costly and potentially toxic regimen.
Keywords: Lung Cancer, erlotinib, bevacizumab, serum biomarker
Introduction
Inhibition of the epidermal growth factor (EGFR) pathway with erlotinib improves survival compared to placebo for patients with advanced lung cancer [1], and antibodies against VEGF improve survival when combined with chemotherapy [2, 3]. Response rates and progression-free survival (PFS) in unselected patients treated with both erlotinib and bevacizumab are much higher than those in unselected patients treated with erlotinib alone, suggesting significant activity for this combination [4]. However, many patients are exposed to toxicities without evident clinical benefit and these patients may be better served by earlier access to alternative therapies. This has encouraged an intense search for biomarkers of clinical significance for this and other targeted therapies. In the Iressa Non-small cell lung cancer Trial Evaluating REsponse and Survival against Taxotere (INTEREST) trial, in which the patients were randomized between gefitinib and docetaxel, no statistically significant prediction of survival benefit was seen for any of the biomarkers tested, including EGFR expression and mutation, EGFR gene amplification, or ras mutation [5]. These biomarkers are also present only in a small minority of patients with NSCLC, and require the availability of a significant amount of fresh tumor tissue for analysis; thus none are adequate to practically stratify patients who can derive survival benefit from erlotinib-based therapy in a western population. Furthermore, there are no validated biomarkers for benefit from bevacizumab therapy [6, 7]. Better predictive tools are thus needed to guide and optimize treatment decisions to maximize treatment benefits while minimizing cost and toxicity.
Recently, we reported a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) proteomic signature (VeriStrat®), comprised of 8 protein features, that was able to classify patients for improved PFS and overall survival (OS) after treatment with EGFR-tyrosine kinase inhibitor (TKI) therapy but not with chemotherapy [8]. This signature was validated in two independent cohorts treated with gefitinib or erlotinib. In this study, we tested whether VeriStrat® could also predict outcome in an independent multi-institutional cohort of patients treated with erlotinib in combination with bevacizumab.
Materials and methods
Mass spectrometry was performed on 35 available pre-treatment serum samples from an open-labeled, phase I/II study (n=40) in which the patients were treated with erlotinib in combination with bevacizumab. All patients included in this study were diagnosed with NSCLC, were previously treated with chemotherapy, had good performance status (0-1), stage IIIB (with pleural effusion) or stage IV, and nonsquamous histology. Additional details regarding patient demographics were described previously [9]. The previously developed algorithm was applied in a fully blinded manner to the patients' serum samples. Serum aliquots were diluted 1:20 in a saturated sinapinic acid solution (35 mg/ml sinapinic acid (Sigma, St. Louis, MO); 50% acetonitrile (Burdick & Jackson, Muskegon, MI); and 0.1% trifluoroacetic acid (Sigma, St. Louis, MO)) and randomly spotted in triplicate on gold, 100-well, sample plates. Mass spectra for all samples were generated in a linear mode and in an automated manner using the Voyager-DE STR™ workstation. Results from 500-525 independent spectrum acquisitions for each sample were averaged to generate each spectrum. Raw spectra were coded and sent electronically to Biodesix (Steamboat Springs, CO). Spectral pre-processing was performed, which included background (BG) and noise estimation, BG subtraction, normalization to partial ion current and alignment [8]. The classification algorithm (VeriStrat®) was based on eight distinct m/z features (5843, 11446, 11530, 11685, 11759, 11903, 12452 and 12580 Da) [8]. The identities of these features and their underlying biological significance are currently under investigation. The integrated intensities of these eight peaks were used as input for the fixed kNN classifier (k = 7), which then returned a label, either “good” or “poor”. The procedure was identical to the one described in Taguchi et al [8]. The entire procedure was performed in a fully blinded manner, i.e. all clinical data were kept blinded until the classification into good and poor groups had been obtained. The log-rank test was used to determine whether progression free survival and overall survival of the two groups (“good” and “poor”) were statistically different. All statistical calculations and graphs were generated using PRISM 5 (GraphPad Software, La Jolla, CA)
Results
From the available 35 samples with associated with clinical data, we generated 276 spectra, with 5-9 replicates per sample. A concordant classification among the replicates was obtained, except in 1 patient. In this sample, of the 7 spectra obtained, 3 were classified as “good” and 4 were classified as “poor”, likely due to tiny variations in the replicates in this borderline case. This patient was classified as “undefined” and excluded from the survival analyses.
Representative baseline corrected spectra for one “good”- (upper panel) and one “poor”- patient (lower panel) are shown in Figure 1A. Figure 1B displays the mean peak intensities of the eight mass spectral features used in the classifier from all 35 patients. Higher mean peak intensities are observed in the “poor” group when compared with the group that was classified as “good”. While this demonstrates that each feature is individually associated with the classification, classifiers based on only single features (peaks) lose substantial predictive power and robustness, as previously reported [8].
Figure 1.
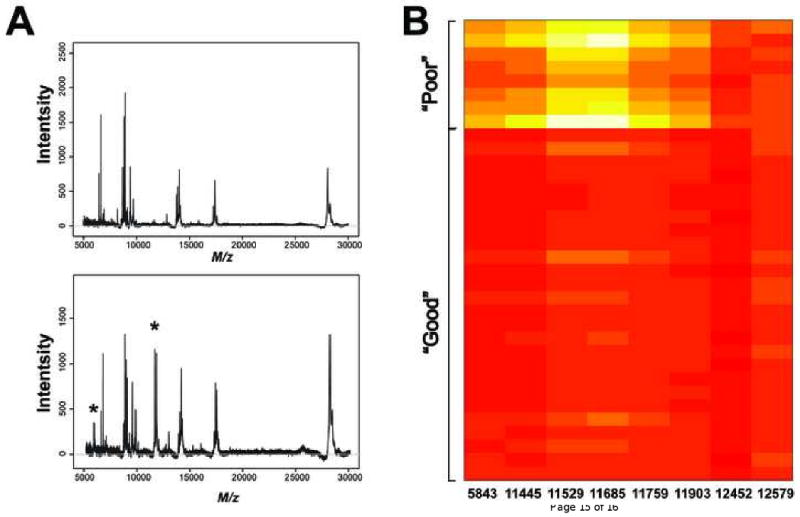
(A) Representative spectra from a patient classified as “good” (upper panel) versus another patient who was classified as “poor”. A peak around 6000 Da and another cluster of peaks in the 11000 to 12000 Da range are observed in the spectrum from a patient who was classified as “poor”. (B) A heatmap is used to visually summarize peak intensities of the eight peaks in the VeriStrat algorithm from the cohort treated with bevacizumab and erlotinib. Differences can be observed between the patients who were classified as “poor” and those who were classified as “good”.
The group that was classified as “poor” had statistically significant lower overall survival and progression-free survival (log rank p=0.007 and p=0.0003, respectively) than the group that was classified as “good” (Figure 2). For the “good” outcome group, the median OS time was 61 weeks compared to the 24 weeks for the group that classified as “poor” (hazard ratio (HR)=0.14, 95% confidence interval (CI)=0.03 to 0.58). Median PFS was 36 weeks for the “good” outcome group versus 8 weeks for the “poor” outcome group (HR=0.045, 95% CI=0.008 to 0.237).
Figure 2.
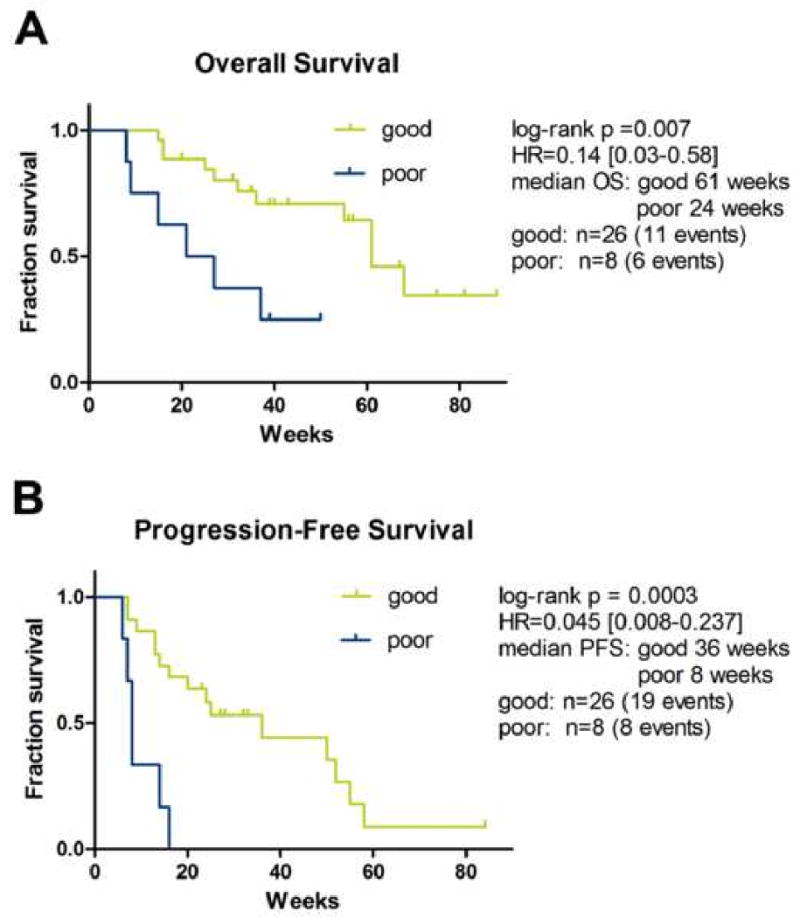
Kaplan-Meier analysis is used to plot survival curves of patients treated with both bevacizumab and erlotinib according to “good” or “poor” classification. There was a statistically significant difference in progression-free survival and overall survival as determined by the log-rank test.
Discussion
Identification of biomarkers is important to select the patients most likely to benefit from specific targeted therapies. It is also crucial for these biomarkers to be validated and be reproducible between independent studies and patient cohorts. We have shown in our current study that VeriStrat®, which was developed on spectra from patients treated with single agent gefitinib, can accurately classify this new cohort of patients, who received both erlotinib and bevacizumab, into good and poor survival groups. We have previously shown that this classifier was not predictive of clinical outcome in two cohorts of patients treated with chemotherapy alone and another cohort treated with surgery alone, suggesting that VeriStrat's predictive power is specific to treatment with EGFR TKIs. It is thus interesting that survival is accurately classified with the combination of erlotinib and bevacizumab, which yields a significantly improved response rate and progression-free survival compared to erlotinib alone. This observation may reflect the fact that the improved clinical outcome of the combination results from a synergy of these agents through a common mechanism (i.e. enhancement of erlotinib activity). This is supported by the fact that no clinical activity has been observed in lung cancer with bevacizumab alone. In our current study, the observed hazard ratio between “good” and “poor” groups for OS is 0.14, which is substantially smaller than the hazard ratio reported for EGFR TKI monotherapy [8], though the current sample size (n = 35) is small and the confidence intervals for the two predictions have some overlap.
Confidence in our conclusions is increased by another study using the same samples in which an academic statistical group derived a classifier containing many of the same spectral features [10]. The present study, however, has significant immediate clinical implications, as VeriStrat is now performed in a CLIA-certified laboratory with strict quality control and is commercially available for clinical use.
Conclusions
We have demonstrated that an established proteomic classifier based on MALDI MS of pretreatment serum samples can accurately classify patients into two groups differing in their survival benefit from treatment with the clinically active combination of erlotinib and bevacizumab. The classifier has demonstrated no such ability on pretreatment samples in multiple cohorts treated with chemotherapy or surgery, suggesting that this classifier may be predictive and not prognostic. The biology underlying this classifier is unclear, but is under investigation and may reflect the activity of EGFR ligand-releasing proteases on abundant serum proteins. The study involves only a limited number of patients, and yet finds highly statistically significant differences. Further validation with larger cohorts and randomized prospective trials is needed, but our findings suggest that this mass spectrometry-based serum classifier may have clinical utility for selecting patients most likely to have improved survival after treatment with this costly and occasionally toxic but potentially effective, regimen.
Table 1.
| Characteristics | Bevacizumab/Erlotinib (n 35*) |
Good | Poor |
|---|---|---|---|
| Sex (%) | |||
| Male | 14 (40) | 11 (42) | 3 (37.5) |
| Female | 21 (60) | 15 (58) | 5 (62.5) |
| Age, y | |||
| Median | 59 | 58 | 63 |
| Range | 36-72 | 36-72 | 49-72 |
| Stage | |||
| IIIB | 6 | 5 | 0 |
| VI | 27 | 19 | 8 |
| Recurrent | |||
| RECIST (%) | |||
| Partial Response | 8 | 8 | 0 |
| Stable Disease | 21 | 14 | 6 |
| < 16 weeks | 11 | 8 | 5 |
| > 16 weeks | 10 | 6 | 1 |
| Progressive Disease | 6 | 4 | 2 |
Stage was not available for 2 of the patients classified as “Good”
Acknowledgments
This study was funded by the Vanderbilt Lung Cancer SPORE (P50 CA-90949) and SPECS (UO1 CA114771) grants.
Footnotes
Conflict of interest statement: David Carbone is an unpaid advisor to BioDesix, but holds no patents or ownership interest with them.
Heinrich Roder, Joanna Roder, and Maxim Tsypin are Biodesix employees who performed the proteomic classification.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P, Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Onn A, Sandler A. Angiogenesis and lung cancer: prognostic and therapeutic implications. J Clin Oncol. 2005;23:3243–3256. doi: 10.1200/JCO.2005.18.853. [DOI] [PubMed] [Google Scholar]
- 3.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, O'Neill VJ, Fehrenbacher L, Belani CP, Bonomi PD, Hart L, Melnyk O, Ramies D, Lin M, Sandler A. Phase II study of efficacy and safety of bevacizumab in combination with chemotherapy or erlotinib compared with chemotherapy alone for treatment of recurrent or refractory non small-cell lung cancer. J Clin Oncol. 2007;25:4743–4750. doi: 10.1200/JCO.2007.12.3026. [DOI] [PubMed] [Google Scholar]
- 5.Douillard JY, Laporte S, Fossella F, Georgoulias V, Pujol JL, Kubota K, Monnier A, Kudoh S, Rubio JE, Cucherat M. Comparison of docetaxel- and vinca alkaloid-based chemotherapy in the first-line treatment of advanced non-small cell lung cancer: a meta-analysis of seven randomized clinical trials. J Thorac Oncol. 2007;2:939–946. doi: 10.1097/JTO.0b013e318153fa2b. [DOI] [PubMed] [Google Scholar]
- 6.Stinchcombe TE, Socinski MA. Bevacizumab in the treatment of non-small-cell lung cancer. Oncogene. 2007;26:3691–3698. doi: 10.1038/sj.onc.1210366. [DOI] [PubMed] [Google Scholar]
- 7.Dowlati A, Gray R, Sandler AB, Schiller JH, Johnson DH. Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non-small cell lung cancer treated with chemotherapy with or without bevacizumab--an Eastern Cooperative Oncology Group Study. Clin Cancer Res. 2008;14:1407–1412. doi: 10.1158/1078-0432.CCR-07-1154. [DOI] [PubMed] [Google Scholar]
- 8.Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, Nishio M, Brahmer J, Spreafico A, Ludovini V, Massion PP, Dziadziuszko R, Schiller J, Grigorieva J, Tsypin M, Hunsucker SW, Caprioli R, Duncan MW, Hirsch FR, Bunn PA, Jr, Carbone DP. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 9.Herbst RS, Johnson DH, Mininberg E, Carbone DP, Henderson T, Kim ES, Blumenschein G, Jr, Lee JJ, Liu DD, Truong MT, Hong WK, Tran H, Tsao A, Xie D, Ramies DA, Mass R, Seshagiri S, Eberhard DA, Kelley SK, Sandler A. Phase I/II trial evaluating the anti-vascular endothelial growth factor monoclonal antibody bevacizumab in combination with the HER-1/epidermal growth factor receptor tyrosine kinase inhibitor erlotinib for patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:2544–2555. doi: 10.1200/JCO.2005.02.477. [DOI] [PubMed] [Google Scholar]
- 10.Salmon S, Chen H, Chen S, Herbst R, Tsao A, Tran H, Sandler A, Billheimer D, Shyr Y, Lee JW, Massion P, Brahmer J, Schiller J, Carbone D, Dang TP. Classification by mass spectrometry can accurately and reliably predict outcome in patients with non-small cell lung cancer treated with erlotinib-containing regimen. J Thorac Oncol. 2009;4:689–696. doi: 10.1097/JTO.0b013e3181a526b3. [DOI] [PMC free article] [PubMed] [Google Scholar]


