Abstract
Fibroblasts are one of the most abundant cell types in connective tissues. These cells are responsible for tissue homeostasis under normal physiological conditions. When tissues are injured, fibroblasts become activated and differentiate into myofibroblasts, which generate large contractions and actively produce extracellular matrix (ECM) proteins to facilitate wound closure. Both fibroblasts and myofibroblasts play a critical role in wound healing by generating traction and contractile forces, respectively, to enhance wound contraction. This review focuses on the mechanisms of force generation in fibroblasts and myofibroblasts and techniques for measuring such cellular forces. Such a topic was chosen specifically because of the dual effects that fibroblasts/myofibroblasts have in wound healing process— a suitable amount of force generation and matrix deposition is beneficial for wound healing; excessive force and matrix production, however, result in tissue scarring and even malfunction of repaired tissues. Therefore, understanding how forces are generated in these cells and knowing exactly how much force they produce may guide the development of optimal protocols for more effective treatment of tissue wounds in clinical settings.
Keywords: Fibroblasts, myofibroblasts, wounds, contraction, measurement
1. Introduction
Typical wound healing consists of three phases: inflammation, proliferation, and remodeling. Contraction occurs during the proliferative phase, during which a cell-rich granulation tissue is formed. Contraction is an important part of wound healing, as it enables wound closure. However, wound contraction in humans has both positive and negative effects. It is beneficial to wound healing by narrowing the wound margins, which leads to wound closure; however, excessive contraction causes formation of undesirable contracture and scarring, leading to cosmetic and functional problems.
Widely present in the majority of tissues, fibroblasts are a type of cell which have mesenchymal origin and can exhibit either non-contractile or highly contractile phenotype. Under normal conditions, fibroblasts function to maintain tissue homeostasis by regulating the turnover of extracellular matrix (ECM). When tissues are injured, however, fibroblasts around the injured region differentiate into myofibroblasts, a type of highly contractile cells that produce abundant ECM proteins. While the mechanisms of wound healing are not completed understood, it has become clear that both fibroblasts and myofibroblasts play a critical role in the wound healing process. Specifically, the traction forces of fibroblasts and coordinated contraction of myofibroblasts are believed to be responsible for wound contraction and closure (1). However, excessive myofibroblast activity, accompanied by elevated levels of mechanical stress in the healing region, often causes scar tissue formation, and in the worst case, contracture of tissues (e.g. Dupuytren's contracture), leading to local immobilization and loss of function (2). Therefore, it is important to understand how mechanical forces are generated in fibroblasts and myofibroblasts as well as the ways in which forces are modulated. Moreover, precise measurement of cellular forces is essential to quantitative study of cellular forces, which may aid in designing optimal regimens for wound healing.
In this article, we briefly review the characteristics of fibroblasts and myofibroblasts, their force generation and transmission, and major techniques for measuring mechanical forces generated by these cells. Finally, our perspectives on this research area are provided.
2. Fibroblasts and myofibroblasts in wound healing
Fibroblasts are spindle-shaped cells and are widely distributed in most types of tissues, particularly connective tissues. These cells are of mesenchymal origin and express vimentin, but not desmin or α-smooth muscle actin (α-SMA). Fibroblasts play a critical role in regulating the turnover of ECM under normal conditions. In injured tissues, fibroblasts are activated and differentiate into myofibroblasts, which contract and participate in healing by reducing the size of wound and secreting ECM proteins. The differentiation of fibroblasts to myofibroblasts is a key event in connective tissue wound healing (Fig. 1).
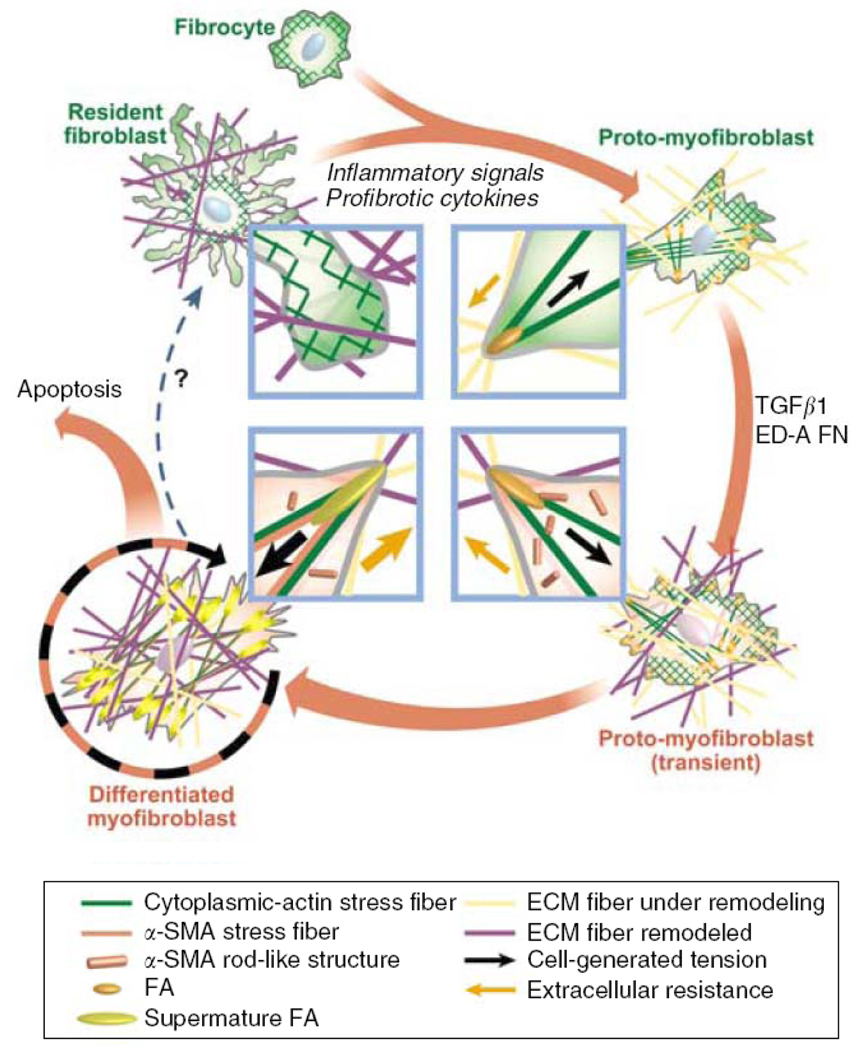
Figure 1.
Schematic illustration of the mechanical feedback loop in myofibroblast development. Fibroblasts in intact tissue are stress-shielded by ECM and do not develop contractile features and cell–matrix adhesions. Upon injury, inflammatory signals activate fibroblasts to spread into the provisional wound matrix and become proto-myofibroblasts. TGF-β1 stimulates proto-myofibroblasts to express α-SMA and become differentiated myofibroblasts. Myofibroblasts may exit this cycle when ECM is reconstituted and takes over the mechanical load; stress-released myofibroblasts eventually undergo apoptosis. (Reproduced with permission from Fig. 3 in Hinz, J Invest Dermatol 127: 526, 2007).
Phenotypically, myofibroblasts are an intermediate cell type between fibroblasts and smooth muscle cells (SMCs). Myofibroblasts start to appear in the early phase of granulation tissue formation, become most abundant in the proliferation phase of wound healing, and progressively disappear in the later stage of healing, possibly by an apoptotic mechanism. Myofibroblasts have ultra-structures distinctive from those of fibroblasts, including extensive cell-matrix adhesions, abundant intercellular adherens and gap junctions, and bundles of contractile cytoplasmic microfilaments (or stress fibers), which are associated with contractile proteins such as non-muscle myosin and are composed of the cell’s contractile apparatus (1, 3). Similar to SMCs, myofibroblasts express α-SMA, an early differentiation marker of vascular SMCs. However, myofibroblasts have only relatively low expression of smooth muscle myosin heavy chain; in addition, they do not express smoothelin, a later marker of SMC differentiation (4). Along with vimentin, desmin, and myosin, α-SMA is used to further classify myofibroblasts into various subcategories (5).
Multiple sources of myofibroblasts exist in order to meet the temporarily high demand for contractile cells in wound repair. Myofibroblasts in skin wounds are generally believed to originate from local recruitment of fibroblasts in the dermis and subcutaneous tissues surrounding the wound (6). However, pericytes and vascular SMCs are also potential sources of myofibroblasts, which are especially important in vascular wound healing (4, 7). There has been evidence that myofibroblasts may be derived from tubular epithelial cells through epithelial-mesenchymal transition (EMT) (4, 8). In addition, myofibroblasts may come from peripheral blood fibrocytes, the circulating precursor cells which are a sub-population of bone marrow-derived leukocytes with fibroblast characteristics (4, 7, 9, 10). As many as 30–50% of myofibroblasts in the wound may potentially be derived from such fibrocyte progenitors (4). Additionally, another possible origin of myofibroblasts is tissue specific stem cells, which, upon exposure to certain stimuli including mechanical loading, may differentiate into myofibroblasts (11, 12). Nonetheless, it remains unclear whether myofibroblasts of different origins exhibit similar or different characteristics and functions in wound healing.
A major function of myofibroblasts is to synthesize ECM proteins, notably collagen types I–VI and XVIII, glycoproteins, and proteoglycans for normal growth, differentiation, and wound repair (5). For example, the depth of subepithelial collagen is highly correlated with the number of myofibroblasts (13). In addition, myofibroblasts also secrete many other matrix molecules including laminin and thrombospondin, glycosaminoglycans (GAGs), hyaluronic acid (HA), and heparan sulfuate (HS), as well as matrix-modifying proteins such as matrix metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases (TIMPs) (5).
In addition to producing matrix components, myofibroblasts also generate contractile forces which bring together the edges of an open wound and therefore facilitate wound closure. However, excessive myofibroblast activities, including excessive contraction and over-production of ECM, are also the major cause of tissue fibrosis and scar formation, e.g., Dupuytren’s contracture. In fact, under normal healing conditions myofibroblasts begin to undergo apoptosis once the wound proceeds to full epithelialization. On the other hand, persistence of myofibroblasts in granulation tissues may lead to the formation of scar tissues, including hypertrophic and keloidal scars.
3. Force generation by fibroblasts and myofibroblasts
Like many other types of non-muscle cells, fibroblasts and myofibroblasts generate intracellular tension by ATP-powered sliding of actin–myosin filaments (14–16). This tension is transmitted to the ECM via focal adhesions that are located at both ends of the stress fiber (17) and as a result, cell traction forces (CTFs) are produced. Besides the actin-myosin interaction, another source of traction forces is actin polymerization, which drives forward protrusion of the leading edge of a migrating cell (18). Compared with the Ca2+-regulated rapid and reversible contraction of smooth muscle cells, actin-myosin interaction-mediated cell traction is relatively slow, sustained, and non-reversible (7, 19–21). CTFs are regulated by a number of molecules that are associated with actin and myosin. These include myosin light chain kinase (MLCK), MLC phosphatase (22–24), Rho, Rac, and Cdc42 (25). In addition, neo-expressed contractile proteins, such as α-SMA, are also involved in force generation by myofibroblasts (26). For example, α-SMA expression in fibroblasts treated with TGF-β1 increases cell contraction (27). Similarly, compared to wild-type 3T3 fibroblasts or fibroblasts transfected with α-cardiac and β- or γ-cytoplasmic actin, 3T3 fibroblasts transiently or permanently transfected with α-SMA cDNA exhibit significantly higher contractile forces, as indicated by a significantly greater collagen lattice contraction (27). However, α-SMA is not required for the induction of CTFs. Rather, expression of this protein enhances CTFs through modification of stress fibers (28).
One important function of CTF is to enable fibroblasts and myofibroblasts to migrate in the ECM (29, 30). In addition to enabling cell migration (31, 32), CTFs deform the ECM network and result in stress and strain in the network, which in turn modulate cellular functions (30, 33) and maintain cellular tensional homeostasis (34–36). Fibroblasts generate traction forces that are far greater than needed for cell migration (14, 30). As a result, large traction forces dramatically deform the ECM. This morphogenetic process of ECM rearrangement is believed to be the primary function of fibroblast traction forces (1, 30). During tissue development and repair, locomotive fibroblasts alone can generate sufficient traction forces, leading to contraction and re-organization of collagen matrix (37–41). Ehrlich et al. even argued that the traction of fibroblasts serves as individual units to contract the wound, and that the presence of myofibroblasts may signify termination of the contractile force generation process (39, 40). It has recently become clear, however, that both traction forces of fibroblasts and contraction of myofibroblasts are essential for wound healing (1), sequentially playing major roles. To address this issue, Tomasek et al. proposed a model describing a three-phase healing process in a three-dimensional collagen lattice (Fig. 2) (1). In the first phase, when fibroblasts attach to collagen fibrils, spread, and migrate, they exert traction forces to the matrix and cause slow compaction of the collagen lattice. As a result, mechanical stresses start to develop within the lattice, which induce differentiation of fibroblasts into pro-myofibroblasts. Meanwhile, stress fibers are formed within the cells and fibronexus adhesion complexes are formed between the cells and matrix (phase II). In this phase, the pro-myofibroblasts become further differentiated into myofibroblasts in the presence of TGF-β1. Both types of cells exert contractile forces on the ECM. In phase I, traction force increases almost linearly as a result of fibroblasts exerting forces on the matrix. As mechanical stress continues to develop, the cells generate contractile forces and enter phase II, eventually maintaining the forces at a constant level until remodeling is complete (19). If this process continues and myofibroblasts do not progressively disappear after repair, healing activity becomes excessive and phase III appears, during which detrimental contracture occurs.
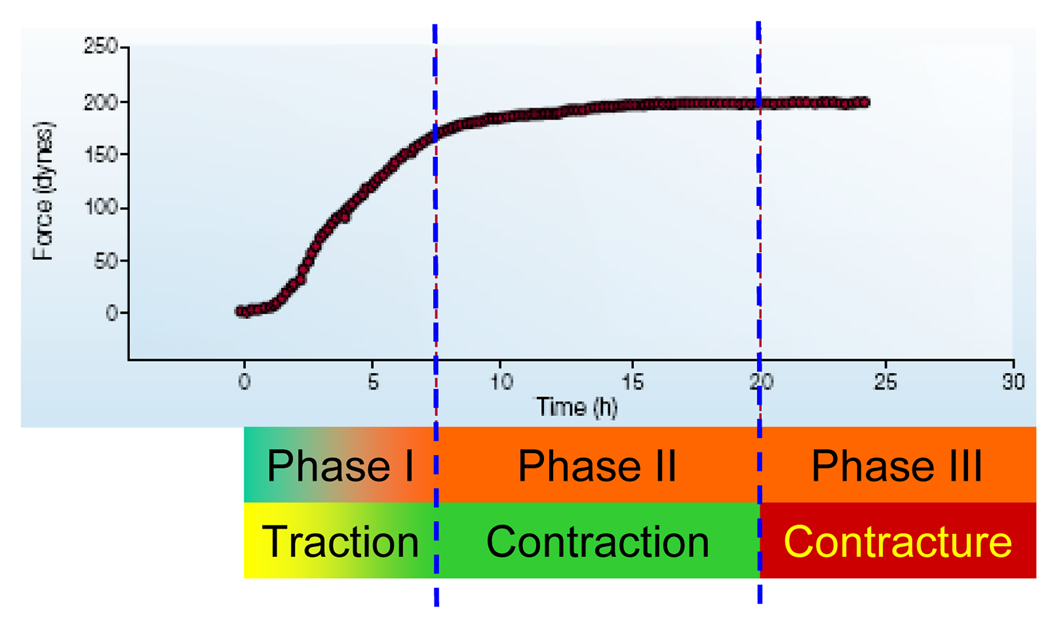
Figure 2.
A three-phase healing process in a three-dimensional collagen lattice model. In phase I, fibroblasts exert traction forces to the matrix and cause slow compaction of the collagen lattice and as a result, mechanical stresses start to develop within the lattice, inducing transformation of fibroblasts into pro-myofibroblasts. Traction force increases almost linearly as a result of fibroblasts exerting forces on the matrix. In phase II, pro-myofibroblasts become further differentiated into myofibroblasts in the presence of TGF-β1. Cells generate contractile forces and eventually keep the forces at a constant level until remodeling is completed. If this process continues and myofibroblasts do not progressively disappear, healing activity becomes excessive and enters phase III, during which detrimental contracture occurs. (Adapted with permission from Fig. 3 in Tomasek et al., Nat Rev Mol Cell Biol 3: 349, 2002).
4. Measurement of cellular mechanical forces
Measurement of the forces generated by fibroblasts and myofibroblasts can be performed on cell populations or single cells. To measure forces of a cell population, many studies used a fibroblast-populated collagen lattice (FPCL) model, in which fibroblasts are embedded in a collagen gel lattice and shrink the lattice as a result of cellular traction and contraction. The contractile forces of cells are then indirectly measured by changes in gel volume or area, or directly quantified by a so-called “culture force monitor” (CFM) technique (38, 42, 43). On the other hand, the traction forces of individual fibroblasts and myofibroblasts can be determined by a variety of single cell-based techniques, which provide either qualitative or quantitative force measurements. Such techniques include wrinkleable silicone membrane, micro-machined cantilever beam array, micropost force sensor array (MFSA), and cell traction force microscopy (CTFM). Note that while the following description of cellular force measurement methods is intentionally focused on fibroblasts and myofibroblasts, these methods are applicable to all types of adherent cells.
4.1. Cell population-based techniques
Cell population-based techniques have been developed based essentially on a FPCL model or its derivatives. The FPCL was originally developed by Bell et al. as a dermal equivalent, an engineered skin graft substitute for burn patients (44, 45). Although it did not gain success in this capacity, the system has since evolved into a widely used in vitro model for wound healing-related studies (46), largely because this model provides a measurement of the contractility of resident cells.
In general, there are three types of FPCL models. The first is a free-floating FPCL (FF-FPCL), in which the lattice floats in medium without any constraints. Isotonic contraction is created in this model, leading to decrease in lattice diameter. The second model is tethered FPCL (T-FPCL), in which the lattice is tightly attached to a substrate and so cannot move or relax. This results in isometric contraction of the lattice, leading to a decrease in the height of the tethered lattice but not its diameter. The third one is tethered-delayed-released FPCL (TDR-FPCL). In this model, a cell-populated lattice is first attached to a substrate for a certain period of time to allow tension to be developed within the lattice. The lattice is then released and starts to contract isotonically as a result of loss of substrate constraint (47, 48). Using FPCL models, initial cell spreading and elongation, traction force-related cell migration, and cell contraction have been shown to be responsible for lattice contraction by resident cells (47). However, FPCL contraction resulting from fibroblast spreading and elongation is much smaller compared to those generated by cell migration and contraction.
a. Free-floating fibroblast-populated collagen lattice
The use of FF-FPCLs represents a classical approach to measure cellular contraction semi-quantitatively. This approach is based on optical measurement of collagen lattice shrinkage (40, 49). During culture, FF-FPCL progressively reduces its size to balance the contraction generated by the cells embedded inside (Fig. 3A). Therefore, measuring the reduction in the geometric features of the collagen lattice provides indirect quantification of the contractility of these cells. Usually, changes in lattice volume, diameter of sphere-shaped lattices, area, or length of rectangular lattices are measured (48, 50, 51).
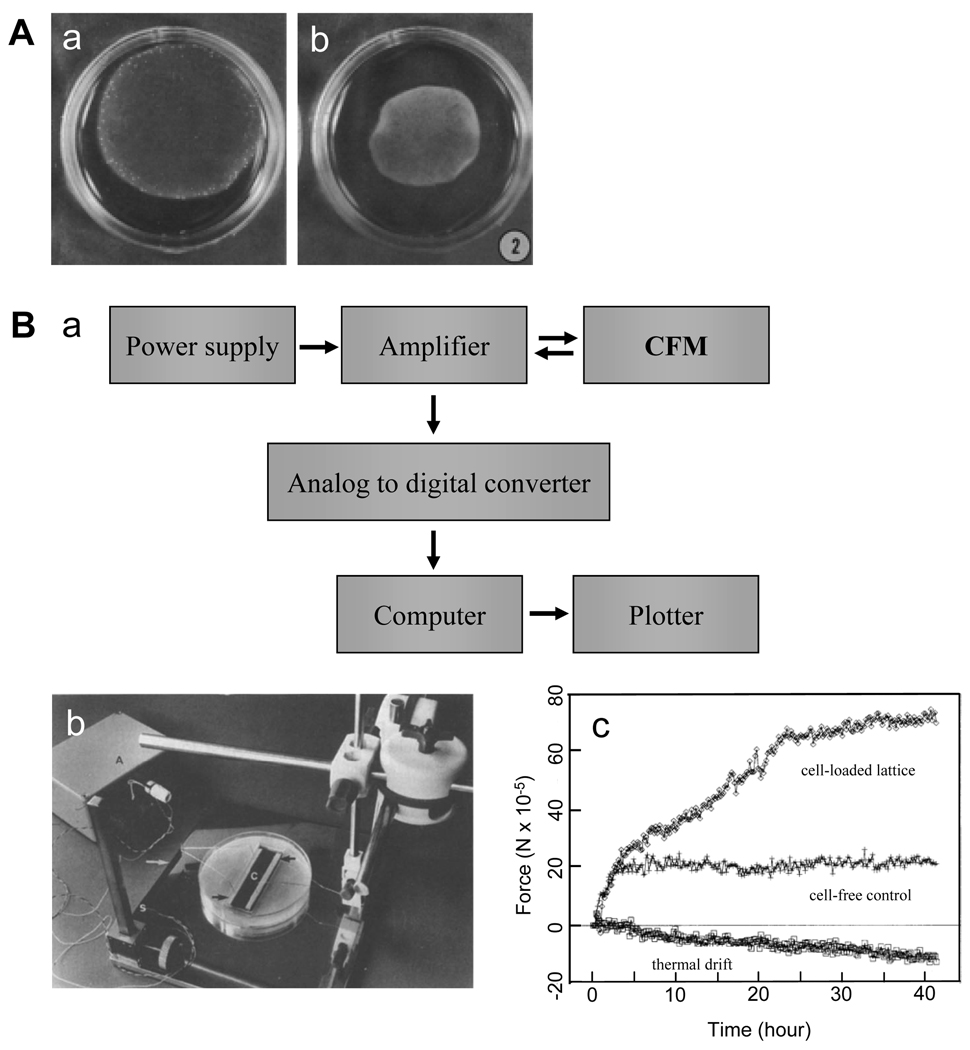
Figure 3.
Fibroblast-populated collagen lattice (FPCL) based cell contraction measurements. (A). A free-floating fibroblast-populated collagen lattice (FF-FPCL) for estimating fibroblast contraction. a and b show the lattice upon cell loading and after 48 hours in culture, respectively. Measuring the reduction in the geometric features of collagen lattice provides indirect quantification of the contractility of these cells. (Adapted with permission from Fig. 2 in Ehrlich and Rajaratnam, Tissue Cell 22: 407, 1990). (B). a, flow chart of a culture force monitor (CFM). b, a picture of a CFM setup. c. a typical CFM measurement. Three curves are shown, including thermal drift from the system, cell-free collagen gel response, and gross force produced by 5.5 × 106 cells in a lattice. (Adapted with permission from Fig. 1–3 in Eastwood et al., Biochim Biophys Acta 1201: 186, 1994).
Using FF-FPCL models, the kinetics of FPCL contraction caused by fibroblasts cultured in vitro can be monitored simply by tracking the collagen lattice changes over time. The lattice contracting process has been shown to consist of three distinct phases — a lag phase before the initiation of contraction, followed by a rapid contraction phase, and finally by a slow contraction phase (49). Various factors affect the contraction profile of FPCL, including cell density, culture temperature, serum concentration, collagen concentration, and drug intervention. A significant advantage of the FPCL approach is that measuring collagen lattice contraction is relatively simple. However, because of large variation and instability of the geometry of the collagen lattice during culture, this geometry-dependent measurement method provides only a rough estimate of the contractility of cells in the lattice. In addition, whether the remodeling of FPCL is due to cell migration or cell contraction is not clear (40).
Recently, a study used a collagen-GAG foam-like gel construct to measure the contractile force of cells embedded inside (52). In addition to calculating the average contractile force of a cell population from gross gel deformation, the open-cell structure of this construct also enabled quantification of contraction by individual cells using conventional column buckling relationships. When cells are grown in the scaffold, they deform their surrounding struts. By determining the deformation of struts the cell-mediated contractile force (Fc) can then be calculated from Euler’s buckling relation and the hydrostatic compression end restraint. This approach extends previous methods for analyzing cell buckling of 2-D substrates to 3-D constructs and can therefore estimate the contractile forces of individual cells in 3-D condition. For example, the mean contractile force generated by individual dermal fibroblasts within such a collagen-GAG scaffold was calculated to be 11~41 nN.
b. Culture force monitor
One disadvantage of the FPCL model is its inability to quantify the real contractile forces of fibroblasts/myofibroblasts. Quantification of cellular contractile forces is important when comparing the contractility of cells subjected to different treatments as well as correlating in vitro data to in vivo findings. Another disadvantage of the FPCL model is that it lacks sufficient sensitivity toward relatively small contractile forces. Therefore, direct measurement of the stress within FPCL with reasonably high sensitivity represents a more favorable approach. This is achieved by attaching strain or stress gauges to the FPCL to continuously track the changes in the strain or stress of the collagen lattice. Such a technique is termed “culture force monitor” (CFM). Depending on the sensitivity of the strain or force gauges, the CTM technique can measure cellular contractile forces in a very low force range when collagen lattice changes are hardly visible.
In an early CFM system (Fig. 3B), two attachment bars in a collagen lattice were connected to a central measuring beam (a thin alloy slab). Strain gauges were attached to the beams in a full bridge network to give the maximum sensitivity. The output signal from strain gauge was enhanced with an amplifier, which was further channeled into a digital voltameter and then into an analog-to-digital converter in a computer. Data were collected at a rate of 1 Hz, enabling fast and continuous monitoring of force changes within the collagen lattice (43). The sensitivity of the CFM method is markedly higher compared to the volumetric gel contraction method. For example, the device of Eastwood et al. detected a displacement of 0.5 mm or 3.3% of total collagen lattice length, which is barely measurable using FF-FPCL method (43). Taking advantage of the high sensitivity of CFM, one may determine fibroblast-mediated force generation during the initial stages of contraction, which is otherwise too small to be detected (43). Using such a CFM system, Eastwood et al. also identified three distinct phases (initial contraction, linear increase, and equilibrium) of contraction of FPCLs within the first 24 hours. This finding was consistent with the results from a previous study using an FF-FPCL model, but with considerably higher resolution (49). Moreover, they found that force generation was parallel to cell attachment and therefore concluded that most of the forces resulted from fibroblast attachment and migration within the lattice, which agrees with the findings from other groups (40). Finally, by comparing the contraction profiles of cells, different populations of fibroblasts could be distinguished on the basis of their specific contraction patterns. Thus, such a technique may serve as a useful biophysical cell profiling tool to identify cells (53). For example, using CFM measurement, ocular fibroblasts were shown to exhibit a marked difference in their contraction profiles— corneal fibroblasts generated the strongest contraction while scleral fibroblasts produced the weakest (54).
In CFM systems, the collagen lattice can either float in medium or be tethered to an underlying substrate. In the setup developed by Delvoye et al., a floating collagen lattice was restrained at both ends by curing collagen on immobilized glass rods, one of which was connected to a strain gauge (50). In tethered collagen lattices, CFM measures the isometric contraction generated within the lattice with minimal changes in its dimensions (55, 56).
To improve the efficiency of measurement, a multi-station CFM system consisting of four vertical cantilever beams with semiconductor strain gages was developed. Such a CFM system is able to test multiple samples simultaneously and therefore can facilitate statistical design and analysis of experiments (57, 58). In addition, a dynamic CFM (D-CFM) system has also been developed. By using computer-controlled linear actuators in the system to independently apply precise motion waveforms to multiple FPCLs, this system was able to detect the differences in force patterns resulting from various motion waveforms imparted to the lattice. Thus, such a system may facilitate the study of the effect of dynamic mechanical loads on cells, ECM remodeling, and cell-matrix interactions (59). A combination of CFM and time lapse reflection microscopy, a simultaneous imaging and micro-culture force monitor (SIM-CFM) system was developed to measure the mechanical strain generated during matrix contraction whilst simultaneously recording cell and matrix behavior (19).
4.2. Single cell-based techniques
Cells are heterogeneous and the forces they generate are often widely variable. Cell population-based techniques only provide an estimation of the “averaged” forces of a group of cells. Therefore, knowing the level and pattern of forces generated by individual cells is important in terms of correlating cell behaviors with the mechanical characteristics of cells. To this end, a number of single cell-based techniques for measuring cellular forces have been developed.
Using optical or microelectronic approaches, the traction forces of individual cells can be determined qualitatively and quantitatively. An early study using thin silicone membrane demonstrated that individual fibroblasts transmitted traction forces to their substrate because cells created wrinkles on the membrane (Fig. 4A) (30, 60, 61). Such a technique was further improved by estimating traction forces by applying a flexible micro-needle of known stiffness to reverse the wrinkles generated by the cell (62). However, there is currently no mathematical solution available for accurately predicting the wrinkles caused by a complex, non-isotropic traction force field. As a result, absolute values of traction forces cannot be determined through this approach. Alternatively, techniques using micro-beads or fluorescent dot embedded non-wrinkleable elastic membranes were developed in an attempt to track the displacements and thereby determine CTFs at certain locations. However, these methods generally lacked sufficient resolution in CTF measurement (17, 32, 63, 64). Later on, instead of using continuum substrate, a technique based on a micro-machined array of cantilever beams with calibrated stiffness was developed (Fig. 4B) (65). Cells were plated on pads underlined with cantilever beams. When a cell generated a traction force and bent a cantilever beam, the extent of bending was recorded and the traction force was then determined accordingly. Such a technique can reliably determine the traction forces of individual cells. Its limitations, however, are obvious because first, it can only determine the forces in one direction and cannot determine the traction force field within whole cell area; second, fabrication of the device is complicated and again, its resolution is not satisfactory.
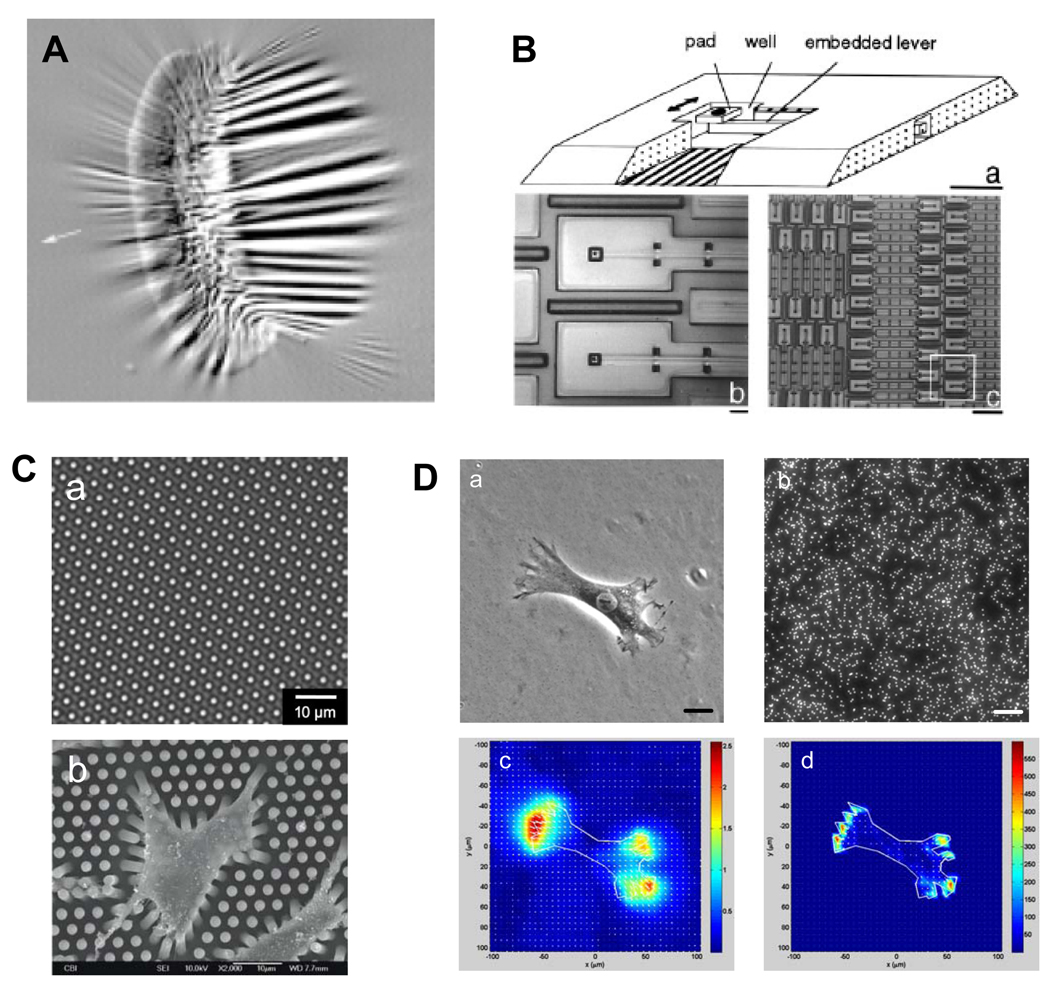
Figure 4.
Techniques for measuring traction forces of single cells. (A). A cell produces wrinkles on a silicone membrane. (Reproduced with permission from Fig. 1 in Beningo and Wang, Trends Cell Biol 12: 79, 2002). (B). Different magnifications of a micromachined device for cell traction force measurement. (Adapted with permission from Fig. 1 in Galbraith and Sheetz, Proc Natl Acad Sci USA 94: 9114, 1997). (C). A micropost force sensor array. a, phase contrast microscopy image of MFSA. b, SEM image of rat aorta SMCs adhering to the top of MFSA. (Adapted with permission from Fig. 5 and 6 in Li et al., Cell Motil Cytoskeleton 64: 509, 2007). (D). Cell traction force microscopy (CTFM). a, a human patellar tendon fibroblast (HPTF) on a polyacrylamide gel; b, fluorescence image of fluorescent beads embedded in gel; c, substrate displacement field; and d, cell traction force field. Scale bars, 20 µm. (Adapted with permission from Fig. 1 in Chen et al., Cell Motility & Cytoskeleton 64: 248, 2007).
a. Micropost force sensor array
To overcome the aforementioned limitations, techniques based on micropost force sensor array (MFSA) have been developed to measure CTFs (66–69). In a MFSA, each micropost functions as an individual force-sensing unit and independently senses the traction forces locally applied by the cell. Specifically, when a cell adheres to the microposts, it exerts tensile forces and bends the microposts (Fig. 4C) (67). The traction forces can then be determined according to the deflection of microposts using beam theory (70). Compared to micro-cantilevers which can only determine traction forces in a single direction (65), microposts in an array can detect traction forces in all directions. Therefore, MFSA technology is especially useful in mapping the forces during cell migration (66). Moreover, such microposts could be used to apply localized mechanical forces to a cell by embedding magnetic nano-wires inside them (71). Similarly, taking advantage of the periodicity of the micropost array, an optical Moire-based traction forces mapping technique was explored by acquiring the diffracted Moire fringe pattern of the array instead of tracking the displacements of individual microposts (69). This method may potentially determine CTFs at higher resolution because of the magnification effect of Moire fringe pattern as well as having no need to track or visualize individual microposts.
b. Cell traction force microscopy
One shortcoming of the MFSA technique is that it can only measure CTFs at predetermined discreet points. A further advancement in traction force measurement, cell traction force microscopy (CTFM) determines traction forces in the entire cell spreading area (72–74). In CTFM (Fig. 4D), fluorescent microbead-embedded elastic hydrogels such as polyacrylamide gel (PAG) or gelatin gel are used as a cell culture substrate. With appropriate ECM protein coating (e.g., type I collagen), cells adhere to and spread out on the gel. They generate traction forces, deform the gel, and cause the embedded beads to be displaced. A pair of images, referred to as "force-loaded" and "null-force" images, are taken during CTFM measurement. The “force-loaded” image is taken while the adherent cells remain on the gels, whereas the "null-force" image is taken after the cells have been removed. Two steps are involved in deriving CTFs from this pair of images. The first step is to solve an image registration problem in which beads from two images are matched and thus gel displacement field is derived. The second step is to solve an inverse problem in which CTFs are computed that will give a best match to the corresponding displacement field. By dividing each image into a number of overlapped regions, cross correlation functions provide a convenient way of matching each region from one image to the other. By further assuming that the gel is thick enough (e.g. > 100 µm) to behave like an elastic half space, the application of the analytical Boussinesq solution obtains the discrete loads as an estimate of the traction forces. This can be achieved either by applying an inverse Fourier transform (72), or through solving a general regularized inverse problem (73). The latter approach is more involved, but allows for the incorporation of a priori information such as levels of errors. Recently, a new CTFM solution has been developed, in which effective pattern recognition algorithms and finite element method (FEM) were introduced (75). In this approach, a feature registration scheme was devised to match individual beads from the image pair. Furthermore, the gel is modeled in FEM as a 3D object with its actual thickness using brick elements. By applying static condensation, FEM obtains traction forces in the form of forces on the surface nodes that lie within the boundary of the cells. This approach ensures reliable displacement calculation and enables simultaneous determination of CTFs of multiple cells.
5. Application of micropatterning techniques in cell traction force measurement
Cells are heterogeneous in size and morphology, which leads to large variations in their behaviors. For example, cell size and shape alone have been shown to affect cell motility, proliferation, differentiation, apoptosis, as well as lineage commitment of stem cells (76–79). Accordingly, cells also display large variations in the magnitude and distribution of their CTFs. One way to reduce such variation and to obtain higher statistical confidence in CTF measurement, therefore, is to keep the size of cells constant. Based on lithography or soft lithography microfabrication techniques, cell micropatterning has become a powerful way to precisely control cell size and morphology, as well as cell-cell and cell-matrix interactions (80). While different in technical approaches, the majority of cell micropatterning techniques depends on fabricating regions of cell-adhesive ECM proteins or cell-repellent substances of specific geometry on cell culture substrate. When cells are cultured on such a substrate, they selectively attach to cell-adhesive regions or avoid cell-repellent regions, and thus eventually adopt specific shapes prescribed by substrate fabrication. By micropatterning cells in various sizes, it has been shown that CTF increases almost linearly with cell spreading area (81).
On the other hand, and more importantly, understanding the relationship between cell shape and cellular function is critical for the study of cell biology and specifically for regulation of cell phenotype. Strategic integration of cell micropatterning and CTF monitoring may add new insights to fundamental understanding of certain biological phenomena or help to establish useful models for disease studies. Here, a few examples are given to demonstrate how cell micropatterning can be a useful tool in conjunction with CTF measurement to address specific biological questions.
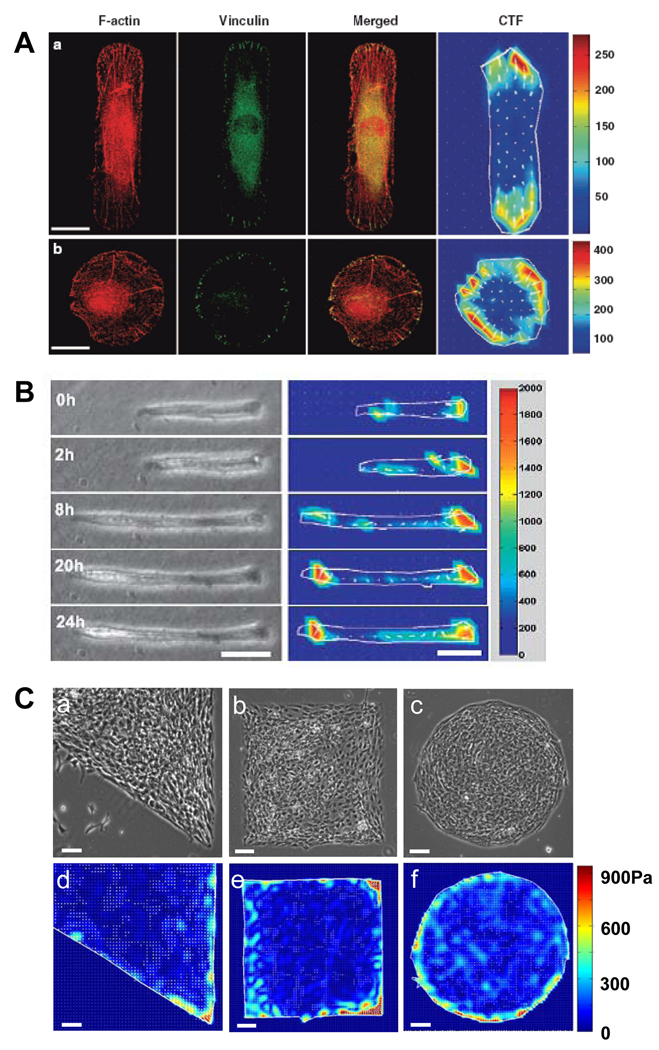
The first example shows how cell shape affects its internal structure and protein expression by micropatterning human patellar tendon fibroblasts (HPTFs) into different shapes with the same area (82). Not surprisingly, cells exhibited distinctive cytoskeletal structure, spatial arrangement of focal adhesions, and spatial distribution and magnitude of CTFs, all of which depend on cell shape (Fig. 5A). In rectangular cells, long and parallel actin filament bundles were formed along the longitudinal direction; in circular cells, however, filament bundles existed along the circumference of cells. Focal adhesions were concentrated at the two ends of rectangular cells but uniformly distributed along the edge of circular cells. These regions were also where the largest traction forces were located in both shapes of cells. As a result, rectangular cells have considerably higher expression of type I collagen compared with circular cells. Hence, the shape of cells may directly determine cell phenotype via changes in gene and protein expression.
Figure 5.
CTFM for micropatterned cells. (A). Application of CTFM to determine traction forces of micropatterned cells. Immunofluorescence microscopy images of F-actin, vinculin, and their overlay, as well as CTFs in micropatterned HPTFs. a, rectangular HPTF; b, circular HTF. Development of focal adhesions and stress fibers is seen to be closely related to cell traction force distribution. Scale bars, 20 µm. (Adapted with permission from Fig. 6 in Li et al., Cell Motil Cytoskeleton 65: 332, 2008). (B). Traction force development of micropatterned C2C12 cells during differentiation. Phase contrast images (left panel) and corresponding traction force maps (right panel), respectively. The color bar represents traction force level. Scale bars, 50 µm. (Reproduced with permission from Fig. 5 in Li et al., J Biomech 41: 3349, 2008). (C). Mechanical stress distribution of patterned cell aggregates. a–c, micropatterned cell islands of distinctive shapes. d–f. CTF distribution of cell islands determined by CTFM. Scale bars, 100 µm. (Adapted with permission from Fig. 2 in Li et al., J Biomech 42: 1622, 2009).
In another example, an in vitro model was developed to investigate the contractility of micropatterned C2C12 skeletal muscle cells using CTFM (83). In this model, C2C12 cells were micropatterned such that they assumed a rectangular shape similar to a typical myotube. During differentiation, these cells gradually fused into a myotube; meanwhile, their contractile forces, represented by CTFs, continually increased until the myotube reached maturation (Fig. 5B). Therefore, CTFM can directly quantify the contractile forces of a myotube, a precursor of myofibers which are the fundamental structural unit of muscle tissues and is responsible for muscle contraction. Such a model can potentially be used as a fast screening approach for therapy of Duchenne muscular dystrophy (DMD).
In addition to measuring traction forces of individual cells, CTFM can also be applied to cell aggregates. In a recent study, fibroblasts were micropatterned to form cell islands of specific shapes (Fig. 5C). Using CTFM, it was found that cells produced larger mechanical stresses around the perimeter of the cell island compared to the inner region. The stress pattern closely resembled cell proliferation and differentiation patterns. However, when overall mechanical stress levels of cell islands were elevated by application of mechanical stretching, neither cell proliferation nor differentiation patterns followed the new mechanical stress pattern although both proliferation and differentiation quantitatively increased. Thus, these results indicated that mechanical stress magnitude, rather than the stress gradient as previously proposed, determines the formation of specific cell proliferation and differentiation patterns, leading to local tissue pattern formation during tissue morphogenesis (84).
6. Concluding remarks
Based on research in the past few decades, it has become increasingly clear that both fibroblasts and myofibroblasts play an important role in wound healing. Despite our current understanding of the characteristics of these cells, as well as the importance of the mechanical forces both applied to the cells and generated by the cells in the repair process, many fundamental questions remain to be answered. First, the origins of myofibroblasts remain to be determined. It also remains to be determined whether myofibroblasts of various origins exhibit different characteristics and functions during wound healing. The independent and interactive roles of cytokines, ECM, and mechanical forces in regulating the structure and function of fibroblasts and myofibroblasts must be investigated further. Second, while appropriate in vitro models are valuable, in vivo associations should be established to guide in vitro studies. Techniques for directly measuring force generation by these cells are to be developed to enable correlation between in vitro and in vivo findings. Third, factors that affect intrinsic cellular forces and dynamic activities remain to be defined. Identification of such factors may allow us to predict the ability of tissues to contract and form scar tissues. Finally, the fate of fibroblasts and myofibroblasts upon completion of wound healing must be determined. While the current prevailing opinion is apoptosis of myofibroblasts, it is equally possible that phenotypic reversion (i.e. myofibroblasts dedifferentiate into fibroblasts) or further phenotypic modulation occurs. This is especially fundamental information for exploring solutions for preventing scar tissue formation. Such investigations are in many ways complementary and show promise in providing fundamental answers to many lingering questions regarding regulation of tissue repair and the roles of myofibroblast in the reparative process.
Abbreviations
- α-SMA
α-smooth muscle actin
- CFM
Culture force monitor
- CTF
Cell traction force
- CTFM
Cell traction force microscopy
- D-CFM
Dynamic culture force monitor
- DMD
Duchenne muscular dystrophy
- ECM
Extracellular matrix
- EMT
Epithelial-mesenchymal transition
- FEM
Finite element method
- FF-FPCL
Free-floating fibroblast-populated collagen lattice
- FPCL
Fibroblast-populated collagen lattice
- GAG
Glycosaminoglycan
- HA
Hyaluronic acid
- HPTF
Human patellar tendon fibroblast
- HS
Heparan sulfuate
- MFSA
Micropost force sensor array
- MLC
Myosin light chain
- MLCK
Myosin light chain kinase
- MMP
Matrix metalloproteinase
- PAG
Polyacrylamide gel
- SIM-CFM
Simultaneous imaging and micro-culture force monitor
- SMC
Smooth muscle cell
- TDR-FPCL
Tethered-delayed-released fibroblast-populated collagen lattice
- T-FPCL
Tethered fibroblast-populated collagen lattice
- TGF-β
Transforming growth factor-β
- TIMP
Tissue inhibitor of metalloproteinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 2.Kozma EM, Olczyk K, Bobinski R, Kasperczyk M, Szpyra K. Chir Narzadow Ruchu Ortop Pol. 2002;67:73–79. [PubMed] [Google Scholar]
- 3.Hinz B. J Invest Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- 4.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Am J Physiol. 1999;277:C1–C9. doi: 10.1152/ajpcell.1999.277.1.C1. [DOI] [PubMed] [Google Scholar]
- 6.Ross R, Everett NB, Tyler R. J Cell Biol. 1970;44:645–654. doi: 10.1083/jcb.44.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmouliere A, Chaponnier C, Gabbiani G. Wound Repair Regen. 2005;13:7–12. doi: 10.1111/j.1067-1927.2005.130102.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Liu Y. Am J Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe R, Donnelly SC, Peng T, Bucala R, Metz CN. J Immunol. 2001;166:7556–7562. doi: 10.4049/jimmunol.166.12.7556. [DOI] [PubMed] [Google Scholar]
- 10.Chesney J, Bucala R. Curr Rheumatol Rep. 2000;2:501–505. doi: 10.1007/s11926-000-0027-5. [DOI] [PubMed] [Google Scholar]
- 11.Szczodry M, Zhang J, Lim C, Davitt HL, Yeager T, Fu FH, Wang JH. J Orthop Res. 2009;27:1373–1378. doi: 10.1002/jor.20878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Wang JH. J Orthop Res. 2009 [Google Scholar]
- 13.Brewster CE, Howarth PH, Djukanovic R, Wilson J, Holgate ST, Roche WR. Am J Respir Cell Mol Biol. 1990;3:507–511. doi: 10.1165/ajrcmb/3.5.507. [DOI] [PubMed] [Google Scholar]
- 14.Adelstein RS, Pato MD, Sellers JR, de Lanerolle P, Conti MA. Soc Gen Physiol Ser. 1982;37:273–281. [PubMed] [Google Scholar]
- 15.Kolega J, Janson LW, Taylor DL. J Cell Biol. 1991;114:993–1003. doi: 10.1083/jcb.114.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger JW, Sanger JM, Jockusch BM. J Cell Biol. 1983;96:961–969. doi: 10.1083/jcb.96.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 18.Bereiter-Hahn J. Med Eng Phys. 2005;27:743–753. doi: 10.1016/j.medengphy.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Bogatkevich GS, Tourkina E, Abrams CS, Harley RA, Silver RM, Ludwicka-Bradley A. Am J Physiol Lung Cell Mol Physiol. 2003;285:L334–L343. doi: 10.1152/ajplung.00417.2002. [DOI] [PubMed] [Google Scholar]
- 20.Hinz B, Gabbiani G. Curr Opin Biotechnol. 2003;14:538–546. doi: 10.1016/j.copbio.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Katoh K, Kano Y, Amano M, Onishi H, Kaibuchi K, Fujiwara K. J Cell Biol. 2001;153:569–584. doi: 10.1083/jcb.153.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chrzanowska-Wodnicka M, Burridge K. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartshorne DJ, Ito M, Erdodi F. J Muscle Res Cell Motil. 1998;19:325–341. doi: 10.1023/a:1005385302064. [DOI] [PubMed] [Google Scholar]
- 24.Somlyo AV, Bradshaw D, Ramos S, Murphy C, Myers CE, Somlyo AP. Biochem Biophys Res Commun. 2000;269:652–659. doi: 10.1006/bbrc.2000.2343. [DOI] [PubMed] [Google Scholar]
- 25.Ridley AJ. J Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 26.Hinz B, Dugina V, Ballestrem C, Wehrle-Haller B, Chaponnier C. Mol Biol Cell. 2003;14:2508–2519. doi: 10.1091/mbc.E02-11-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Mol Biol Cell. 2001;12:2730–2741. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Li H, SundarRaj N, Wang JH. Cell Motil Cytoskeleton. 2007;64:248–257. doi: 10.1002/cm.20178. [DOI] [PubMed] [Google Scholar]
- 29.Burridge K, Chrzanowska-Wodnicka M. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 30.Harris AK, Stopak D, Wild P. Nature. 1981;290:249–251. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- 31.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. J Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. J Cell Biol. 1994;127:1957–1964. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tranquillo RT, Durrani MA, Moon AG. Cytotechnology. 1992;10:225–250. doi: 10.1007/BF00146673. [DOI] [PubMed] [Google Scholar]
- 34.Sawhney RK, Howard J. Cell Motil Cytoskeleton. 2004;58:175–185. doi: 10.1002/cm.20004. [DOI] [PubMed] [Google Scholar]
- 35.Eckes B, Krieg T. Clin Exp Rheumatol. 2004;22:S73–S76. [PubMed] [Google Scholar]
- 36.Harris AK. J Cell Sci Suppl. 1987;8:121–140. doi: 10.1242/jcs.1987.supplement_8.7. [DOI] [PubMed] [Google Scholar]
- 37.Andujar MB, Melin M, Guerret S, Grimaud JA. J Submicrosc Cytol Pathol. 1992;24:145–154. [PubMed] [Google Scholar]
- 38.Brown RA, Talas G, Porter RA, McGrouther DA, Eastwood M. J Cell Physiol. 1996;169:439–447. doi: 10.1002/(SICI)1097-4652(199612)169:3<439::AID-JCP4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich HP. Eye. 1988;2(Pt 2):149–157. doi: 10.1038/eye.1988.28. [DOI] [PubMed] [Google Scholar]
- 40.Ehrlich HP, Rajaratnam JB. Tissue Cell. 1990;22:407–417. doi: 10.1016/0040-8166(90)90070-p. [DOI] [PubMed] [Google Scholar]
- 41.Kanekar S, Borg TK, Terracio L, Carver W. Cell Adhes Commun. 2000;7:513–523. doi: 10.3109/15419060009040308. [DOI] [PubMed] [Google Scholar]
- 42.Freyman TM, Yannas IV, Yokoo R, Gibson LJ. Exp Cell Res. 2002;272:153–162. doi: 10.1006/excr.2001.5408. [DOI] [PubMed] [Google Scholar]
- 43.Eastwood M, McGrouther DA, Brown RA. Biochim Biophys Acta. 1994;1201:186–192. doi: 10.1016/0304-4165(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 44.Bell E, Ehrlich HP, Buttle DJ, Nakatsuji T. Science. 1981;211:1052–1054. doi: 10.1126/science.7008197. [DOI] [PubMed] [Google Scholar]
- 45.Bell E, Ivarsson B, Merrill C. Proc Natl Acad Sci U S A. 1979;76:1274–1278. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson MA, Longaker MT. Wound Repair Regen. 2004;12:134–147. doi: 10.1111/j.1067-1927.2004.012208.x. [DOI] [PubMed] [Google Scholar]
- 47.Dallon JC, Ehrlich HP. Wound Repair Regen. 2008;16:472–479. doi: 10.1111/j.1524-475X.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- 48.Grinnell F. J Cell Biol. 1994;124:401–404. doi: 10.1083/jcb.124.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishiyama T, Tominaga N, Nakajima K, Hayashi T. Coll Relat Res. 1988;8:259–273. doi: 10.1016/s0174-173x(88)80045-1. [DOI] [PubMed] [Google Scholar]
- 50.Delvoye P, Wiliquet P, Leveque JL, Nusgens BV, Lapiere CM. J Invest Dermatol. 1991;97:898–902. doi: 10.1111/1523-1747.ep12491651. [DOI] [PubMed] [Google Scholar]
- 51.Qi J, Chi L, Wang J, Sumanasinghe R, Wall M, Tsuzaki M, Banes AJ. Exp Cell Res. 2009;315:1990–2000. doi: 10.1016/j.yexcr.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Harley BA, Freyman TM, Wong MQ, Gibson LJ. Biophys J. 2007;93:2911–2922. doi: 10.1529/biophysj.106.095471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eastwood M, Porter R, Khan U, McGrouther G, Brown R. J Cell Physiol. 1996;166:33–42. doi: 10.1002/(SICI)1097-4652(199601)166:1<33::AID-JCP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 54.Dahlmann-Noor AH, Martin-Martin B, Eastwood M, Khaw PT, Bailly M. Exp Cell Res. 2007;313:4158–4169. doi: 10.1016/j.yexcr.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasugai S, Suzuki S, Shibata S, Yasui S, Amano H, Ogura H. Arch Oral Biol. 1990;35:597–601. doi: 10.1016/0003-9969(90)90025-6. [DOI] [PubMed] [Google Scholar]
- 56.Kolodney MS, Wysolmerski RB. J Cell Biol. 1992;117:73–82. doi: 10.1083/jcb.117.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell BH, Agarwal C, Wang JH. Biomech Model Mechanobiol. 2004;2:239–245. doi: 10.1007/s10237-004-0041-z. [DOI] [PubMed] [Google Scholar]
- 58.Campbell BH, Clark WW, Wang JH. J Biomech. 2003;36:137–140. doi: 10.1016/s0021-9290(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 59.Peperzak KA, Gilbert TW, Wang JH. Med Eng Phys. 2004;26:355–358. doi: 10.1016/j.medengphy.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Beningo KA, Wang YL. Trends Cell Biol. 2002;12:79–84. doi: 10.1016/s0962-8924(01)02205-x. [DOI] [PubMed] [Google Scholar]
- 61.Harris AK, Wild P, Stopak D. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 62.Burton K, Taylor DL. Nature. 1997;385:450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 63.Das T, Maiti TK, Chakraborty S. Lab Chip. 2008;8:1308–1318. doi: 10.1039/b803925a. [DOI] [PubMed] [Google Scholar]
- 64.Iwadate Y, Yumura S. Biotechniques. 2008;44:739–750. doi: 10.2144/000112752. [DOI] [PubMed] [Google Scholar]
- 65.Galbraith CG, Sheetz MP. Proc Natl Acad Sci U S A. 1997;94:9114–9118. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proc Natl Acad Sci U S A. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li B, Xie L, Starr ZC, Yang Z, Lin JS, Wang JH. Cell Motil Cytoskeleton. 2007;64:509–518. doi: 10.1002/cm.20200. [DOI] [PubMed] [Google Scholar]
- 68.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng XY, Zhang X. Journal of Micromechanics and Microengineering. 2008;18:125006. [Google Scholar]
- 70.Timoshenko S, Woinowskey-Kreiger S. Theory of plates and shells. New York: McGraw-Hill; 1959. [Google Scholar]
- 71.Sniadecki NJ, Lamb CM, Liu Y, Chen CS, Reich DH. Rev Sci Instrum. 2008;79:044302. doi: 10.1063/1.2906228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Am J Physiol Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 73.Dembo M, Oliver T, Ishihara A, Jacobson K. Biophys J. 1996;70:2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang JH, Lin JS. Biomech Model Mechanobiol. 2007;6:361–371. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 75.Yang Z, Lin JS, Chen J, Wang JH. J Theor Biol. 2006;242:607–616. doi: 10.1016/j.jtbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 77.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 78.Bailly M, Yan L, Whitesides GM, Condeelis JS, Segall JE. Exp Cell Res. 1998;241:285–299. doi: 10.1006/excr.1998.4031. [DOI] [PubMed] [Google Scholar]
- 79.Dike LE, Chen CS, Mrksich M, Tien J, Whitesides GM, Ingber DE. In Vitro Cell Dev Biol Anim. 1999;35:441–448. doi: 10.1007/s11626-999-0050-4. [DOI] [PubMed] [Google Scholar]
- 80.Folch A, Toner M. Annu Rev Biomed Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 81.Wang N, Ostuni E, Whitesides GM, Ingber DE. Cell Motil Cytoskeleton. 2002;52:97–106. doi: 10.1002/cm.10037. [DOI] [PubMed] [Google Scholar]
- 82.Li F, Li B, Wang QM, Wang JH. Cell Motil Cytoskeleton. 2008;65:332–341. doi: 10.1002/cm.20263. [DOI] [PubMed] [Google Scholar]
- 83.Li B, Lin M, Tang Y, Wang B, Wang JH. J Biomech. 2008;41:3349–3353. doi: 10.1016/j.jbiomech.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li B, Li F, Puskar KM, Wang JH. J Biomech. 2009;42:1622–1627. doi: 10.1016/j.jbiomech.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]