Abstract
Inadequate protein intake initiates adverse changes in skeletal muscle function and structure (i.e., an accommodative response). mRNA level changes due to short-term inadequate dietary protein might be an early indication of subsequent accommodation. The aims of this study were to assess the effects of dietary protein and the diet-by-age interaction on the skeletal muscle transcriptome. Twelve younger (21-43 y) and 10 older (63-79 y) men completed three controlled feeding trials with protein intakes of 0.50, (LPro: lower protein), 0.75 (MPro: medium protein), and 1.00 g protein•kg body weight−1•day−1 (HPro: higher protein). A fasting state biopsy was taken on day 12 of each trial. Global changes in transcript levels were assessed with Affymetrix genechips and expression patterns determined using self-organizing maps. 958 transcripts were differentially expressed (P<0.05) by diet and 853 had a diet-by-age interaction (P<0.05). The results for diet alone revealed that LPro was associated with up-regulation of transcripts related to ubiquitin-dependent protein catabolism and muscle contraction and LPro and MPro resulted in up-regulation of transcripts related to apoptosis and down-regulation of transcripts related to cell differentiation; muscle and organ development; extracellular space; and responses to stimuli and stress. The diet-by-age effect on protein modification transcripts was consistent with the older males being less responsive to anabolic stimuli (lower protein synthesis at HPro) and more responsive to a catabolic state (protein breakdown at LPro). Changes in skeletal muscle mRNA levels in younger and older males to protein intake near or below the RDA are indicative of an early accommodative response.
Keywords: young and elderly, dietary protein, adaptation and accommodation, vastus lateralis, gene expression
INTRODUCTION
Dietary protein provides essential amino acids which are necessary for the synthesis of proteins and other biomolecules [1]. During an extended period of inadequate protein intake, structural and functional skeletal muscle proteins are catabolized [2] to obtain essential amino acids and a new steady state is developed with (i.e. accommodation [3]) or without (i.e. adaptation [4]) reduced physiological function. Accommodative responses to inadequate protein intake observed in older adults include decreased lean body and muscle masses, decreased muscle strength and function, and a decreased immune response [5-7]. These adverse physiological consequences may be especially problematic for older adults who already experience the involuntary decrease in skeletal muscle mass and strength that accompanies advancing age (sarcopenia). The Food and Nutrition Board of the Institute of Medicine has highlighted the need for more research to understand the events which precede the accommodative response to inadequate protein intake in skeletal muscle [8].
While microarray analysis does not determine mechanisms, it gives an unbiased global view of the molecular events that occur during various physiologic states and diseases. For example, skeletal muscle transcript levels respond to changes in energy [9-11] and nutrient availability [12-14] and mRNA level changes occur in human skeletal muscle [14] and rat liver [15] in response to inadequate protein intake. We have previously reported the global changes in skeletal muscle transcript levels that occur in older adults (67 ± 7 y) following one week of inadequate protein intake (63% of the recommended dietary allowance (RDA) for protein [14]). The changes in the transcript profile included up-regulation of transcripts related to contraction, movement and development; extracellular connective tissue; and immune, inflammatory, and stress responses, and down-regulation of transcripts related to energy metabolism, protein synthesis, and proliferation. However, that study could not determine if these were short-term responses to a decrease in protein intake or whether they were the first indication that the inadequate protein diet was stimulating an accommodative response in these older subjects. In addition, the study did not permit us to determine whether these changes are unique to older adults or could be generalized to younger adults. The present study was designed to assess these issues of age, dietary protein level, and diet-by-age interactions on skeletal muscle transcript profiles. We hypothesize that the dietary protein-related transcript profile will reflect an accommodative response to inadequate protein intake and that age will blunt the beneficial effects of added protein intake on the muscle transcriptome.
METHODS
Subjects and Preliminary Testing
Twenty-five healthy, relatively sedentary, free-living men were recruited to participate in this study. Twenty-two men (12 aged 29 ± 7 (younger) and 10 aged 72 ± 6 y (older)) successfully completed the study. Prior to starting the study, each subject completed an evaluation that included a resting-state electrocardiogram, routine clinical blood and urine chemistries, and a written medical history. All subjects had clinically normal heart, liver, and kidney functions and did not have diabetes mellitus. Each subject received written and oral descriptions of the protocol, signed a written informed consent agreement, and received a monetary stipend for participating. The study protocol and consent form were approved by the Institutional Review Board at Purdue University, West Lafayette, IN.
Experimental Design and Dietary Control
A controlled diet was provided to each subject for three 18-d periods in a randomized, cross-over design consisting of three levels of dietary protein intake: LPro (lower protein, 0.50 g•kg−1•d−1, 63% of the RDA for protein), MPro (medium protein, 0.75 g•kg−1•d−1, 94% of the RDA for protein), and HPro (higher protein, 1.00 g•kg−1•d−1, 125% of the RDA for protein). Each subject was provided customized meals that contained sufficient energy for body weight maintenance as described previously [16, 17]. Briefly, estimated energy requirements were predicted to equal the subjects resting energy expenditure (using sex-specific Harris-Benedict equation [18]) times 1.7 for most of the men (range 1.7-2.6) to account for physical activity related energy expenditure. The non-protein energy came from 65% carbohydrate and 35% fat. As described previously, these diets provided weight maintenance energy and fat content [17]. The energy, macronutrient and micronutrient components of the diet were calculated using Nutritionist Pro computer software (version 1.5; First Databank, Inc, San Bruno, CA). Dietary protein sources; meal consumption instructions and setting; the protocols for alcohol, water, and multi-vitamin ingestion; and the protocol for the washout periods have been described previously [19]. Fasting-state nude body weight was determined on each weekday throughout the study using a digital platform scale (model ES200L; Ohaus Corporation, Pine Brook, NJ). Subjects were relatively sedentary but asked to maintain their regular physical activity levels throughout the duration of the study. The energy expenditure (kcal/day) and the summary index (total units) for physical activity were determined using the Yale Physical Activity Survey as previously described [20]. There were no differences between the younger and older men (data not shown) or across the trials for recorded energy expenditure of physical activity (mean ± SD; LPro 1113.4 ± 730.8 (n=22); MPro 1090.1 (n=19) ± 684.6; and HPro 1470.7 ± 1572.3 (n=19) kcal/d, P>0.05) or the summary index (LPro 46.1 ± 19.5 (n=22); MPro 52.3 ± 18.1 (n=19); and HPro 47.0 ± 17.7 (n=19) total units, P>0.05), suggesting physical activity levels were relatively similar for all trials.
Urine and Blood Collections and Analyses
During participant screening prior to the study and on days 7-10 of each trial, 24-hour urine collections were obtained, put into containers, and refrigerated at 4 °C. Aliquotted urine samples were frozen at −20 °C until later testing. Total urinary nitrogen concentrations were measured using a nitrogen analyzer (model FP-528, Leco, St Joseph, MI).
A fasting state blood sample was obtained by catheter from an antecubital vein of every subject on day 12 of each trial. The blood samples were placed into tubes without anticoagulants, maintained at room temperature for ~30 min, and centrifuged at 3000 × g for 10 min at 4 °C. Serum was aliquotted and frozen at −80 °C for later testing. Blood urea nitrogen (BUN) concentration, albumin concentration, and aspartate aminotransferase were measured in the serum samples at the Laboratory Corporation of America, Burlington, NC.
Muscle Collection and RNA Isolation
On day 12 of each trial, a fasting state vastus lateralis muscle sample was obtained using a percutaneous muscle biopsy technique [21]. Following the baseline blood draw, a small portion of the mid-thigh of each subject's dominant leg was anaesthetized, the muscle sample was extracted using a 6-mm Bergstrom biopsy needle (Microsurgical Instruments, Lake Forest, IL), and the tissue was frozen in liquid nitrogen until further processed.
Total RNA was isolated from the frozen muscle sample using Tri-Reagent following the manufacturer's instructions (Molecular Research Center, Cincinnati, OH). This RNA was further purified using the RNeasy procedure (QIAGEN Inc., Valencia, CA) as we have described previously [14]. Ultraviolet light spectrophotometry and gel electrophoresis (1.5% agarose) were used to determine RNA quantity and integrity, respectively.
Microarray Analysis
The transcript profile of each of the 66 available skeletal muscle RNA samples was determined using the Affymetrix U133 Plus 2.0 GeneChip® and standard Affymetrix protocols (Affymetrix, Santa Clara, CA) at the Indiana University Center for Medical Genomics (Indianapolis, IN).
Real-Time Polymerase Chain Reaction Validation
RT-PCR was used to cross-validate the diet transcript profiling results with three of the transcripts that had the most statistically significant changes from the microarray analysis. For the diet comparison, equal amounts of RNA from the 12 younger males were combined into three pools of RNA based on the protein intake level (LPro, MPro, and HPro). The pooled RNA was reverse transcribed to cDNA as previously described [22]. Transcript levels were examined by real-time PCR using the BioRad My IQ real-time PCR system and the BioRad SYBR Green supermix following the manufacturer's protocol (BioRad, Hercules, CA). The primer information and cycle conditions were as follows: 1) ERGIC and golgi 2 (GenBank accession number: NM_016570), forward, 5′-GGTGACAGTTACTGAGGAGCA-3′, reverse 5′-CCAAGTCTGAAACGACAGCA-3′, 40 cycles, annealing temperature (Ta) = 51.0 °C; 2) lysophosphatidic acid acyltransferase (NM_032717), forward, 5′-ACCTGCTTCGAATGATGACC-3′, reverse 5′-CCCTGTTAGCAAACTGGACTG-3′, Ta = 51.0 °C; and 3) forkhead box K1 (NM_001037165), forward, 5′-GACCCAGAACGGAAAGCAT-3′, reverse 5′-GTCGCAAGGAGCTGCAAAG-3′, Ta = 51.0 °C. The expression of each of the transcripts was determined using the threshold cycle (Ct) value procedure described by Livak et al. [22].
Statistical Analysis
Clinical parameters
All data are presented as mean ± SD. For the diet, urine, and blood parameters, a two-factor, repeated measures ANOVA was used to assess age (younger and older; between subject effects), dietary protein intake (LPro, MPro, and HPro; within subject effect), and the interaction of diet and age on the dependent variables. Post hoc analyses were conducted using the Tukey adjustment. SAS software (version 9.1, SAS Institute, Cary, NC) was used for the statistical analyses of all clinical and microarray parameters. Significance was determined at P<0.05.
Microarray analysis
A detailed description of the microarray processing, statistics, clustering, and functional annotation is provided as supplemental data. The following is a brief overview of the procedures used. Prior to beginning the study, a sample size estimate of eight subjects was determined as appropriate to detect fold changes in transcript levels of greater than 1.8-fold due to dietary protein intake based on the variance estimates from our previous skeletal muscle microarray analysis and using a very conservative significance level of α=7.3 × 10−6 and β = 0.8.
Initially, raw Affymetrix readings for the 66 individual arrays were evaluated for quality using the relative log expression (RLE) test in the Bioconductor package (http://bioconductor.org [23]). Ten arrays were identified as having processing artifacts and removed from future analysis. The expressions in the remaining arrays were normalized with the robust multi-array (RMA [24]) probe level model using R software (http://r-project.org [25]) and the Bioconductor package. The MAS 5.0 present/absent call was used to filter the normalized data; only the 15,737 transcripts deemed “present” or “marginal” on >80% of the chips for one of the age groups during at least one of the three protein trials were included in subsequent analyses. A two-sample Z-test was used to assess differences in transcript levels due to age, diet, and diet × age. A nonparametric smoothing function was fit to describe the relationship between variance and total intensity of each transcript [26]. The estimated variance was used to test the effect of age, diet, and the diet-by-age interaction at the transcript level. The false discovery rate (FDR) correction was used to balance the risk of type I and type II errors and determine differential expression of transcripts [27].
Determining Expression Patterns and Functional Groups
Self-organizing maps
The GeneCluster 2.0 [28, 29] software program was used for unsupervised classification of the diet- and the diet-by-age-related differentially expressed transcripts. The mean transcript level was determined for each dietary trial (younger and older males combined) as well as for each of the six diet-by-age groups. Mean values were clustered using self organizing maps (SOM) after the transcript levels were normalized to have a mean of zero and a variance of one. Default settings were used for all other parameters.
GenMAPP
GenMAPP 2.0 (www.GenMapp.org) was used to functionally characterize the differentially regulated transcripts that fell into each SOM cluster for the diet and diet-by-age analyses. This program identifies over-represented Gene Ontology (GO) groups using a z score [30, 31]. The lists were filtered to include only GO terms that had a z score ≥ 2.0 and had >5 and <100 genes measured in the experiment (i.e., this reduces terms that are too narrow or too broad, respectively).
Functional Gene Networks and Pathways Analysis
Transcripts which had a diet-by-age interaction were analyzed through the use of Ingenuity Pathways Analysis (IPA) 5.0 (Ingenuity Systems Inc., www.ingenuity.com) to determine networks of interacting genes (i.e. nodes) based on their connectivity. Networks were generated first using the fold-change (LPro to HPro) values for the older men and then compared with the fold changes values for the younger men (LPro to HPro). The Functional Analysis of a network identified the biological functions and/or diseases that were most significant to the genes in the network.
Availability of the Microarray Dataset
The Affymetrix GeneChip data have been submitted to the GEO database (http://www.ncbi.nlm.hih.gov/geo/) for use by the scientific community. The sample series accession number is GSE9419. Detailed versions of the lists which include the fold changes and cluster information for the differentially expressed transcripts will be made available upon request.
RESULTS
Subject Characteristics and Dietary Intervention
The subject characteristics, dietary intakes, and blood parameters of these subjects during the controlled diet interventions were previously published [19]. Briefly, there were no differences in height, weight, and BMI between younger and older males. Albumin concentration, blood leukocyte content, and serum aspartate aminotrasferase concentration were within clinically normal ranges and were not different among the trials and between the younger and older males.
Protein intake (on a g•kg−1•d−1 basis) increased from LPro to MPro to HPro (P<0.0001) in both the younger and older males and was not different between the two groups (0.51 ± 0.01, 0.76 ± 0.02, and 1.01 ± 0.02 g•kg−1•d−1, respectively). Energy intake on a MJ/d basis was not different among the trials but was higher for the younger vs. older males (for the younger males, LPro, 13.5 ± 2.7; MPro, 13.2 ± 2.4; and HPro, 13.3 ± 2.5 MJ/d and for the older males, LPro, 11.3 ± 1.8; MPro, 11.5 ± 1.6; and HPro, 11.8 ± 1.9 MJ/d, P<0.05). The dietary intakes of energy (kJ•kg−1•d−1), carbohydrate, and fat were higher in younger vs. older subjects independent of trial (P<0.01).
During the study trials, the older males had a higher BUN concentration compared to the younger adults (P<0.0001), independent of dietary protein intake. Habitual intake of dietary protein was not assessed prior to starting the study; however, there was no difference in BUN concentration between the younger and older men prior to starting the study (5.2 ± 0.9 and 7.2 ± 0.9 mmol/L, P>0.05, respectively). Changes in dietary protein intake were confirmed by changes in BUN and urinary total nitrogen excretion; in both younger and older males, as dietary protein intake increased, BUN concentration increased (younger: LPro, 2.8 ± 1.1; MPro, 3.5 ± 0.8; and HPro, 4.0 ± 1.0 mmol/L and older: LPro, 4.0 ± 1.4; MPro, 4.8 ± 1.3; and HPro, 5.9 ± 1.1 mmol/L, P<0.0001) and urinary total nitrogen excretion increased (younger, LPro, 5.0 ± 1.0; MPro, 6.6 ± 1.8; and HPro, 8.2 ± 2.2 g/d and older, LPro, 5.8 ± 0.9; MPro, 7.4 ± 1.7; and HPro, 9.1 ± 2.3 g/d, P<0.0001). Serum BUN concentration, collected prior to starting the study, was higher in both the younger and older adults compared to all three study trials (P<0.05) which crudely suggests that the habitual protein intakes of the subjects were greater than 1.0 g protein•kg−1•d−1.
Number of Differentially Expressed Transcripts
Of the 15,737 transcripts deemed “present”, 32.1% (5052) were differentially expressed by age (P<0.05) and 16.2% (2556) met the FDR criterion (2155 up-regulated and 401 down-regulated, Supplemental Table 1). Approximately 90% of the 5052 age-related transcript level differences were unique to the main effect of age. Forty-four functional categories were established for age-related changes in skeletal muscle mRNA using NIH DAVID (http://david.abcc.ncifcrf.gov/; Supplemental Table 1). The age-related changes in the skeletal muscle profile will be addressed with greater detail in a future paper.
958 (6.1%) “present” transcripts were differentially expressed by diet (P<0.05, Table 1), but there were no dietary protein-related differences in transcript levels that met the FDR criterion. 21 transcripts met a significance cut-off of P<0.001 for diet. Three of these transcripts (ERGIC and golgi 2, lysophosphatidic acid acyltransferase, forkhead box K1) were examined by RT-PCR and the transcript level fold changes (HPro trial/LPro trial) were directionally comparable between techniques; for RT-PCR: 1.26 (UP), 1.13 (DOWN), and 1.70 (DOWN), respectively and for microarray analysis: 1.10 (UP), 1.20 (DOWN), and 1.07 (DOWN), respectively. There were 853 (5.4%) “present” transcripts influenced by a diet-by-age interaction (Table 1, P<0.05), but none of these transcripts met the FDR criterion. Ten transcripts had a P-value < 0.001 for diet-by-age. Ninety-seven of the 958 differentially expressed transcripts for diet (10% of total number of transcripts for diet) and the 853 transcripts for the diet-by-age interaction (11% of total number of transcripts for diet-by-age) were common between the two datasets. The fold changes were generally modest for both diet and the diet-by-age interaction; there were no fold changes greater than 2 (Supplemental data).
TABLE 1.
The number of transcripts at P-value cut-offs and the number of transcripts that met the FDR criterion for age, diet, and diet-by age
| Variable |
|||
|---|---|---|---|
| Criterion | Age | Diet | Diet-by-age |
| 1P<0.05 (Total) | 5052 | 958 | 853 |
|
| |||
| P<0.05 to P=0.01 | 2280 | 744 | 717 |
| P<0.01 to P=0.001 | 1628 | 193 | 126 |
| P<0.001 to P=0.0001 | 755 | 16 | 10 |
| P<0.0001 | 389 | 5 | 0 |
|
| |||
| 2FDR | 2556 | 0 | 0 |
Differentially expressed transcripts based on a two-sample Z-test
False discovery rate (FDR) for age P=0.0081
Self-organizing Map (SOM) and GenMAPP Analysis for Dietary Protein
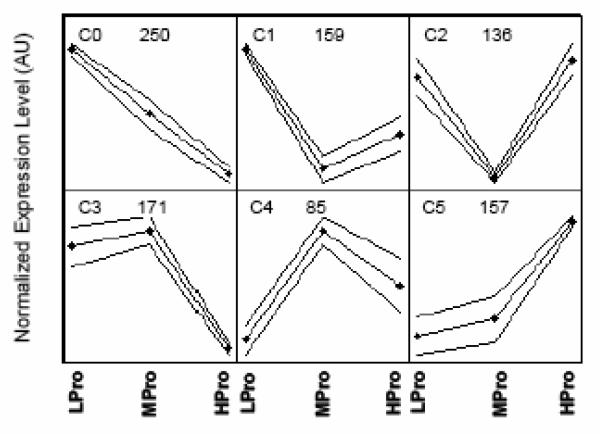
A 2 × 3 SOM was used to visualize expression patterns and cluster the 958 transcripts that were differentially expressed for diet (P<0.05, Figure 1). After the clusters were established, the GenMAPP program was used to functionally annotate the transcripts within each of the six clusters (Supplemental Table 2). Transcripts involved with muscle contraction and ion transport were over-represented in C0, a pattern where transcript levels were down-regulated with increasing dietary protein intake. In C1 the LPro diet up-regulated transcript levels related to “ubiquitin-dependent protein catabolism”, “chromatin assembly/disassembly”, and the ubiquitin cycle. Of the seven differentially expressed transcripts that fell under the GO term “ubiquitin-dependent protein catabolism” five (71%) were in C1. In C2, transcripts were up-regulated when dietary protein intake was below or above the RDA, these transcripts were for the “response to DNA damage”, nuclear transport, regulation of metabolism, cellular morphogenesis, “intracellular signaling cascade”, and “organelle organization and biogenesis”. Transcripts for the function “cytoskeletal protein binding” were over-represented in C3, a cluster where diet HPro down-regulated transcript levels. Cluster C4 includes transcripts for the “G-protein coupled receptor protein signaling pathway”, ion transport, cell differentiation, phosphorylation, and transferase activity that were down-regulated when dietary protein intake was below the RDA. Lastly, the expression pattern in cluster C5 showed that increasing protein intake up-regulated transcripts for functions such as “negative regulation of apoptosis”, “stress and stimuli responses”, “muscle and organ development”, and cell differentiation. Two of the largest % changed and z scores were for the GO terms “negative regulation of apoptosis” and “response to external stimulus”: four (67%) of the six differentially expressed transcripts for these GO terms clustered to pattern in C5 (z score 3.7).
FIGURE 1.
Self-organizing Map showing the patterns of significantly differentially expressed transcripts resulting from changes in dietary protein (958 transcripts, p<0.05). Each point within a cluster is equal to the mean transcript level for younger and older males combined at the dietary protein trial indicated at the bottom of the graph: LPro (Lower protein, 0.50 g•kg−1•d−1), MPro (Medium protein, 0.75 g•kg−1•d−1), and HPro (Higher protein, 1.00 g•kg−1•d−1). Numbers in the top center within each cluster are the number of transcripts that had that expression pattern.
Self-organizing Map and GenMAPP Analysis for Diet-by-Age Interactions
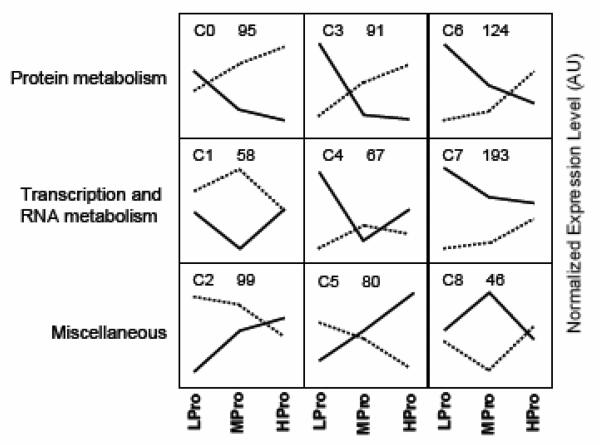
A 3×3 SOM was used to determine common expression patterns and cluster the 853 transcripts with a diet-by-age interaction (P<0.05, Figure 2). After the nine SOM clusters were established, GenMAPP was used to functionally annotate the transcripts within each cluster (Supplemental Table 3). There was a common functional theme of protein metabolism for the three clusters where expression rose in younger subjects but fell in older subjects, i.e. clusters C0, C3, and C6. In cluster C0 expression of transcripts for protein synthesis and “response to chemical stimulus” were similar on the low protein diet but diverged between the age groups as protein intake increased (older males = down-regulated; younger males = up-regulated). In cluster C3, transcripts related to protein modification, the “structural constituent of ribosome”, amino acid metabolism, and “unfolded protein binding” were elevated in older males but this pattern was reversed as diet protein level increased. The transcripts in C6 were for “ubiquitin-dependent protein catabolism” and they were up-regulated in the older compared to the younger males during the LPro diet period.
FIGURE 2.
Self-organizing Map showing the patterns of significantly differentially expressed transcripts resulting from the diet-by-age interaction (853 transcripts, P<0.05). Each point within a cluster is equal to the mean transcript level for the younger (dashed line) or older (solid line) males at the dietary protein trial indicated at the bottom of the graph; LPro (Lower protein, 0.50 g•kg−1•d−1), MPro (Medium protein, 0.75 g•kg−1•d−1), and HPro (Higher protein, 1.00 g•kg−1•d−1). Numbers in the top center within each cluster are the number of transcripts that had that expression pattern.
There was a common functional theme of transcription and RNA metabolism for the clusters where the expression pattern converged for younger and older subjects as protein intake increased, i.e. C1, C4, and C7. In cluster C1, transcripts for “transport and regulation of transcription” were lower in the older subjects on the LPro and MPro diet but converged with the younger group on the HPro diet. In clusters C4 and C7 transcript levels were higher in the older males during the LPro diet (C4 and C7) and MPro diet (C7 only) and converged at the highest protein intake level. There were no over-riding functional themes in common for the remaining clusters: C2, C5, and C8. Transcripts for transport, cell-to-cell signaling, and neuro-physiological processes were down-regulated with age during the LPro diet in C2. In cluster C5 there was a progressive increase in transcript levels in the older males and decrease in transcript levels in the younger males with increasing protein intake; this contained transcripts for the functions oxygen metabolism and apoptosis.
Ingenuity Pathways Analysis (IPA) for diet-by-age interactions
Using IPA we identified a significant network specific to post translational modification, protein degradation, and protein synthesis for the transcripts with a diet-by-age interaction (26 out of 35 possible nodes were differentially expressed; P = 1 × 10−35; Table 2 and Supplemental Figure 1). Of the 26 differentially expressed transcripts on this network, only two changed in the same direction in younger and older men when diet was changed from LPro to HPro (THOC4 and FMA98A, Table 2).
TABLE 2.
Twenty-six differentially expressed and interactive transcripts which form the network identified using Ingenuity Pathway Analysis
| 1Symbol | Name | Affymetrix |
2FC Young |
FC Old |
|---|---|---|---|---|
| CETN2 | Centrin, EF-hand protein, 2 | 209194_at | −1.04 | 1.04 |
| CFLAR | CASP8 and FADD-like apoptosis regulator |
210564_x_at | −1.15 | 1.16 |
| FAM98A | Family with sequence similarity 98, member A | 239487_at | 1.01 | 1.20 |
| HLA-C | Major histocompatibility complex, class I, C |
211911_x_at | −1.08 | 1.08 |
| KIAA1553 | KIAA1553 | 227920_at | 1.07 | −1.04 |
| LCP2 | Lymphocyte cytosolic protein 2 | 205269_at | −1.00 | 1.08 |
| LPXN | Leupaxin | 242778_at | −1.05 | 1.03 |
| MAGOH | Mago-nashi homolog, proliferation- associated |
210093_s_at | −1.06 | 1.13 |
| NOS1 | Nitric oxide synthase 1 | 239132_at | 1.16 | −1.30 |
| NSFL1C | NSFL1 (p97) cofactor (p47) | 217831_s_at | −1.05 | 1.12 |
| NUFIP2 | Nuclear fragile X mental retardation protein interacting protein 2 |
224958_at | 1.02 | −1.01 |
| PAIP2 | Poly(A) binding protein interacting protein 2 |
222983_s_at | −1.12 | 1.08 |
| PDIA3 | Protein disulfide isomerase family A, member 3 |
208612_at | −1.08 | 1.13 |
| PDPN | Podoplanin | 221898_at | 1.05 | −1.05 |
| RAD23B | RAD23 homolog B | 201222_s_at | 1.11 | −1.27 |
| RARRES3 | Retinoic acid receptor responder 3 | 204070_at | −1.12 | 1.04 |
| RFFL | Ring finger and FYVE-like domain containing 1 |
228980_at | −1.14 | 1.35 |
| STK4 | Serine/threonine kinase 4 | 1569791_at | 1.04 | −1.07 |
| SUMO3 | SMT3 suppressor of mif two 3 homolog 3 |
200739_s_at | −1.04 | 1.07 |
| TGFBR3 | Transforming growth factor, beta receptor III |
204731_at | −1.05 | 1.22 |
| THOC4 | THO complex 4 | 226319_s_at | 1.02 | 1.12 |
| UBA1 | Ubiquitin-like modifier activating enzyme 1 |
200964_at | −1.08 | 1.09 |
| UBC | Ubiquitin C | 242214_at | −1.11 | 1.05 |
| UBE2D3 | Ubiquitin-conjugating enzyme E2D 3 | 200669_s_at | −1.15 | 1.13 |
| UBE2L3 | Ubiquitin-conjugating enzyme E2L 3 | 200682_s_at | −1.12 | 1.04 |
| UBE2N | Ubiquitin-conjugating enzyme E2N | 201523_x_at | −1.12 | 1.04 |
Symbol is the abbreviation for the transcript
FC = fold change; Expression value LPro/HPro
DISCUSSION
Previous studies have suggested that the current RDA for protein (0.8 g•kg−1•d−1) is inadequate to maintain protein homeostasis [5, 32-34], fat-free mass, and skeletal muscle cross-sectional area in older adults [7, 35] while protein intake above the RDA may be needed to maintain lean body mass and type I muscle fiber cross sectional area in older women [5, 6]. The results of our study reveal a molecular signature of a biological response to inadequate dietary protein intake that is consistent with this hypothesis. We believe that the observation that transcripts for “muscle and organ development” and “cell differentiation” increase when the protein intake is greater than the RDA (i.e. cluster C5 in Figure 1) and previous phenotypic data [5-7, 32-35] suggest that there are positive responses to consuming protein in quantities greater than the RDA in skeletal muscle. Thus, our array data are consistent with previous reports that suggest a higher protein intake contributes to a healthier physiological profile [5, 6, 36] and better muscle responses to resistance training [37]. The study duration was not, however, sufficient to directly link changes in the skeletal muscle transcript profile with previously reported phenotypes of adaptation and accommodation [5-7, 32-35].
The dietary protein-related changes we observed in skeletal muscle transcript levels appeared to be an early indication of an accommodative response to LPro and MPro diets in both the younger and older men. Many of these changes are consistent with phenotypes of accommodation observed in the study by Castaneda et al. where a 0.56 g protein•kg−1•d−1 diet was fed for nine-weeks [5, 6]. Transcripts related to ubiquitin-dependent protein catabolism and the ubiquitin cycle are higher after consumption of the lower protein diet and this suggests that inadequate dietary protein intake may initiate muscle protein degradation. Ubiquitin-dependent protein catabolism plays a primary role in skeletal muscle proteolysis during catabolic states [38, 39] and this pathway was previously shown to be sensitive to leucine status [40]. The IPA network developed from our data (Supplemental Figure 1) incorporates several ubiquitinconjugating enzymes (UBE2N, UBE2D3, UBE2L3), UBA1 (the protein involved in the initial steps of protein degradation) and other molecules related to proteolysis (CFLAR AND SUMO3). We speculate that this response is initiated to maintain skeletal muscle remodeling and metabolism when dietary protein intake is limiting. However, these were up-regulated with the LPro diet in the older adults while they were down-regulated in the younger adults suggesting that youth provides some protection from the consequence of inadequate protein intake. Future research on age-dependent differences in the role of ubiquitin pathways in accommodative and adaptive responses to changes in dietary protein intake is warranted.
The suggestion that inadequate protein intake quickly induces an early molecular signature of the accommodative response in skeletal muscle is thematically consistent with the interpretation from our previous research. In that report we showed that seven days of inadequate protein intake (0.5 g•kg−1•d−1, immediately after 7 d at 1.2 g•kg−1•d−1) induced alterations in the skeletal muscle transcript profile of older adults (67 ± 7 y) [14]. However, interesting, and potentially important differences exist between our current and previous studies [14]. First, the dietary protein-related changes in transcript levels were less robust in the current study (no differentially expressed transcripts met the FDR criterion and only 21 transcripts were differentially expressed at P <0.001) compared with our earlier study (85 transcripts that met the FDR criterion (P=0.0006) and 49 transcripts that were significantly altered at P<0.0001 [14]). Second, only 17 differentially expressed transcripts (at P<0.05) were common between the two studies and of these, only two were regulated in the same direction (Supplemental Table 4). The differences we observed between the transcript profile results from our two studies may be due to differences in study design. For example, the full cross-over design in the current study may have eliminated spurious findings due to inadequate controls in the pre/post analysis we used for our previous study. Additionally, the abrupt decrease in dietary protein intake (−0.7 g•kg−1•d−1) in the earlier study may have imposed a transient stress on the muscle that was reflected in the transcript profile. In contrast, the body may have had more time to recover from the shift to inadequate protein in the current study (i.e. 12 day feeding period and more modest shifts in dietary protein intake 0.25-0.5 g•kg−1•d−1). Collectively, our interpretation is that both studies are correct and that the differences reflect time-dependent changes that occur at the molecular level in the skeletal muscle in response to changes in dietary protein intake. This hypothesis must be tested directly in a longitudinal study with multiple sampling points and a longer feeding period.
Results from other microarray studies show that the muscle transcript profile changes with advancing age [41-46] and they reveal an up-regulation of transcripts involved with “RNA binding and splicing”, “hormone, growth factor, cytokine, and signaling proteins”, and “protein degradation” as well as a down-regulation of transcripts related to “energy and mitochondrial metabolism” and “stress and inflammatory responses”. While these earlier studies did not control dietary intake and physical activity, the overall results are consistent with our current study and with the phenotypes of aging skeletal muscle (e.g., mitochondrial dysfunction; increased oxidative stress, apoptosis, and RNA splicing; decreased energy metabolism). However, we also observed several age-related changes in the skeletal muscle transcript profile that are inconsistent with the physiological phenotype of sarcopenia, e.g. enrichment of transcripts for skeletal muscle and nervous system development. These transcript-level changes suggest aging muscle has initiated a compensatory mechanism to combat the age-related morphological and physiological changes in skeletal muscle.
The current study provides a unique opportunity to evaluate the interaction between age and dietary protein intake on muscle biology (Figure 2, Supplemental Figure 1). Our data are consistent with reports of a blunted anabolic response of skeletal muscle to increasing amino acid intake in older adults [40, 47, 48]. For example, the cluster analysis shows that transcripts related to protein metabolism were progressively up-regulated by increasing protein intake in the younger adults but were down-regulated in the older adults (clusters C0, C3, and C6, Figure 2). Compared to the younger adults, when more exogenous amino acids are available (i.e. HPro) the older men had lower protein synthesis related transcript levels (C0) which may result in a decreased anabolic response to the high protein load. Additionally, the older men had higher transcript levels related to ubiquitin-dependent protein catabolism (C6) on the LPro which suggests an increased catabolic state. The expression of transcripts related to protein modification (e.g. protein folding) corresponds to the effects on protein synthesis (down with HPro) and protein catabolism (up with LPro) in the older men. The important role of dietary protein in these processes is further demonstrated by identification of a network specific to post translational modification, protein degradation, and protein synthesis for the transcripts with a diet-by-age interaction.
While the present study provides preliminary data and derived hypotheses for future mechanistic and phenotypic research, these data cannot be taken as causal and are merely descriptive. The transcript-level is only one of the many regulatory steps that could be affected by changes in dietary protein intake; therefore, a longitudinal study, with sufficient time for measureable changes in the phenotype to occur and with multiple sampling points is a necessary future step to directly relate the transcript-level phenotype to biochemical (e.g. total and phosphor-proteome), structural, and functional alterations in skeletal muscle. Additional limitations of this study warrant further caution when evaluating these findings. The intensity of the expression changes we observed was modest and there are likely to be false positives in transcripts identified as differentially expressed given the P-value of 0.05 that we used. However, several factors raise our confidence that our results are valid, i.e. real-time PCR analysis confirmed the direction of change in several transcripts, when multiple probe sets were available for a given transcript they were expressed and clustered similarly, family members and isoforms for a number of differentially expressed transcripts were frequently co-regulated. Due to the limited availability of human muscle biopsy tissue we were unable to confirm whether molecular changes observed in a biological system are reflective of changes that occur in the protein or metabolic space. In addition, the RNA obtained from the biopsies was limited and we were therefore, forced to pool RNA to conduct follow-up RT-PCR validation analyses. Lastly, the differential results (discussed above), between the current and previous transcript profiling studies from our laboratory, presents a limitation to interpretation of these results. Future studies will allow us to resolve whether the differences between our two studies are due to acute, but transient changes in the skeletal muscle transcript profile or whether they are a consequence of our earlier study design.
In summary, we report transcript level changes in younger and older males following just two weeks of inadequate and marginal protein intakes that are consistent with an early disruption of muscle biology. These molecular events may precede the changes in skeletal muscle function and structure that occur during accommodation to prolonged intake of diets containing inadequate protein. In older men we also identify transcript level responses to changes in dietary protein intake that reflect a limit on anabolic responses and promotion of catabolic ones. If confirmed in future studies, this could explain why older adults are vulnerable to skeletal muscle atrophy with advancing age. In addition, our data are consistent with earlier reports that indicate physiologic benefits for older subjects that consume protein intakes that are moderately higher than the current RDA [5-7, 32-35]. The current study was not designed and cannot be used to determine the adequacy of the RDA for dietary protein intake for adults; however, if our transcript-level changes are confirmed and extended in necessary future studies, they could present a complementary assessment to other measures (e.g., nitrogen balance, isotope labeling) for addressing the adequacy of the RDA for dietary protein.
Supplementary Material
Acknowledgements
We appreciate the hard work of the research staff of WWC's Laboratory for Nutrition, Fitness, and Aging. We would also like to thank the staff at the Center for Medical Genomics at Indiana University School of Medicine for processing the microarray chips.
WWC and JCF conceived and designed the experiment. AET, JCF, BAC, and WWC participated in processing and analyzing the samples. AET, JCF, and WWC interpreted the data and wrote the manuscript.
The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant numbers 03-35200-13779 (to WWC) and 98-35200-6151 (to WWC), NIH R01 AG15750 (to WWC), and a Purdue Research Foundation Research Assistantship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors had any personal or financial conflicts of interest.
REFERENCE
- 1.Swick RW, Benevenga NJ. Labile protein reserves and protein turnover. J Dairy Sci. 1977;60:505–15. doi: 10.3168/jds.S0022-0302(77)83896-4. [DOI] [PubMed] [Google Scholar]
- 2.Hansen RD, Raja C, Allen BJ. Total body protein in chronic diseases and in aging. Ann N Y Acad Sci. 2000;904:345–52. doi: 10.1111/j.1749-6632.2000.tb06480.x. [DOI] [PubMed] [Google Scholar]
- 3.Young VR, Marchini JS. Mechanisms and nutritional significance of metabolic responses to altered intakes of protein and amino acids, with reference to nutritional adaptation in humans. Am J Clin Nutr. 1990;51:270–89. doi: 10.1093/ajcn/51.2.270. [DOI] [PubMed] [Google Scholar]
- 4.Waterlow JC. Metabolic adaptation to low intakes of energy and protein. Annu Rev Nutr. 1986;6:495–526. doi: 10.1146/annurev.nu.06.070186.002431. [DOI] [PubMed] [Google Scholar]
- 5.Castaneda C, Charnley JM, Evans WJ, Crim MC. Elderly women accommodate to a low-protein diet with losses of body cell mass, muscle function, and immune response. Am J Clin Nutr. 1995;62:30–9. doi: 10.1093/ajcn/62.1.30. [DOI] [PubMed] [Google Scholar]
- 6.Castaneda C, Gordon PL, Fielding RA, Evans WJ, Crim MC. Marginal protein intake results in reduced plasma IGF-I levels and skeletal muscle fiber atrophy in elderly women. J Nutr Health Aging. 2000;4:85–90. [PubMed] [Google Scholar]
- 7.Campbell WW, Trappe TA, Wolfe RR, Evans WJ. The recommended dietary allowance for protein may not be adequate for older people to maintain skeletal muscle. J Gerontol A Biol Sci Med Sci. 2001;56:M373–80. doi: 10.1093/gerona/56.6.m373. [DOI] [PubMed] [Google Scholar]
- 8.Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids. The Institute of Medicine of the National Academies; Washington D.C.: 2002. [DOI] [PubMed] [Google Scholar]
- 9.Kayo T, Allison DB, Weindruch R, Prolla TA. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci U S A. 2001;98:5093–8. doi: 10.1073/pnas.081061898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–3. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 11.Weindruch R, Kayo T, Lee CK, Prolla TA. Microarray profiling of gene expression in aging and its alteration by caloric restriction in mice. J Nutr. 2001;131:918S–923S. doi: 10.1093/jn/131.3.918S. [DOI] [PubMed] [Google Scholar]
- 12.Cameron-Smith D, Burke LM, Angus DJ, Tunstall RJ, Cox GR, Bonen A, Hawley JA, Hargreaves M. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. Am J Clin Nutr. 2003;77:313–8. doi: 10.1093/ajcn/77.2.313. [DOI] [PubMed] [Google Scholar]
- 13.Arkinstall MJ, Tunstall RJ, Cameron-Smith D, Hawley JA. Regulation of metabolic genes in human skeletal muscle by short-term exercise and diet manipulation. Am J Physiol Endocrinol Metab. 2004;287:E25–31. doi: 10.1152/ajpendo.00557.2003. [DOI] [PubMed] [Google Scholar]
- 14.Thalacker-Mercer AE, Fleet JC, Craig BA, Carnell NS, Campbell WW. Inadequate protein intake affects skeletal muscle transcript profiles in older humans. Am J Clin Nutr. 2007;85:1344–52. doi: 10.1093/ajcn/85.5.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endo Y, Fu Z, Abe K, Arai S, Kato H. Dietary protein quantity and quality affect rat hepatic gene expression. J Nutr. 2002;132:3632–7. doi: 10.1093/jn/132.12.3632. [DOI] [PubMed] [Google Scholar]
- 16.Thalacker-Mercer AE, Campbell WW. Dietary protein intake affects albumin fractional synthesis rate in younger and older adults equally. Nutr Rev. 2008;66:91–5. doi: 10.1111/j.1753-4887.2007.00012.x. [DOI] [PubMed] [Google Scholar]
- 17.Campbell WW, Johnson CA, McCabe GP, Carnell NS. Dietary protein requirements of younger and older adults. Am J Clin Nutr. 2008;88:1322–9. doi: 10.3945/ajcn.2008.26072. [DOI] [PubMed] [Google Scholar]
- 18.Harris JA, Benedict FG. A Biometric Study of Human Basal Metabolism. Proc Natl Acad Sci U S A. 1918;4:370–3. doi: 10.1073/pnas.4.12.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thalacker-Mercer AE, Johnson CA, Yarasheski KE, Carnell NS, Campbell WW. Nutrient ingestion, protein intake, and sex, but not age, affect the albumin synthesis rate in humans. J Nutr. 2007;137:1734–40. doi: 10.1093/jn/137.7.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruskall LJ, Campbell WW, Evans WJ. The Yale Physical Activity Survey for older adults: predictions in the energy expenditure due to physical activity. J Am Diet Assoc. 2004;104:1251–7. doi: 10.1016/j.jada.2004.05.207. [DOI] [PubMed] [Google Scholar]
- 21.Evans WJ, Phinney SD, Young VR. Suction applied to a muscle biopsy maximizes sample size. Med Sci Sports Exerc. 1982;14:101–2. [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2006. [Google Scholar]
- 26.Davison A, Hinkley D. Bootstrap methods and their applications. Cambridge University Press; Cambridge, UK: 1997. [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 28.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 29.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–12. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 32.Campbell WW, Crim MC, Dallal GE, Young VR, Evans WJ. Increased protein requirements in elderly people: new data and retrospective reassessments. Am J Clin Nutr. 1994;60:501–9. doi: 10.1093/ajcn/60.4.501. [DOI] [PubMed] [Google Scholar]
- 33.Gersovitz M, Motil K, Munro HN, Scrimshaw NS, Young VR. Human protein requirements: assessment of the adequacy of the current Recommended Dietary Allowance for dietary protein in elderly men and women. Am J Clin Nutr. 1982;35:6–14. doi: 10.1093/ajcn/35.1.6. [DOI] [PubMed] [Google Scholar]
- 34.Uauy R, Scrimshaw NS, Young VR. Human protein requirements: nitrogen balance response to graded levels of egg protein in elderly men and women. Am J Clin Nutr. 1978;31:779–85. doi: 10.1093/ajcn/31.5.779. [DOI] [PubMed] [Google Scholar]
- 35.Campbell WW, Trappe TA, Jozsi AC, Kruskall LJ, Wolfe RR, Evans WJ. Dietary protein adequacy and lower body versus whole body resistive training in older humans. J Physiol. 2002;542:631–42. doi: 10.1113/jphysiol.2002.020685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vellas BJ, Hunt WC, Romero LJ, Koehler KM, Baumgartner RN, Garry PJ. Changes in nutritional status and patterns of morbidity among free-living elderly persons: a 10-year longitudinal study. Nutrition. 1997;13:515–9. doi: 10.1016/s0899-9007(97)00029-4. [DOI] [PubMed] [Google Scholar]
- 37.Campbell WW, Barton ML, Jr., Cyr-Campbell D, Davey SL, Beard JL, Parise G, Evans WJ. Effects of an omnivorous diet compared with a lactoovovegetarian diet on resistance-training-induced changes in body composition and skeletal muscle in older men. Am J Clin Nutr. 1999;70:1032–9. doi: 10.1093/ajcn/70.6.1032. [DOI] [PubMed] [Google Scholar]
- 38.Mitch WE, Goldberg AL. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med. 1996;335:1897–905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 39.Hasselgren PO, Fischer JE. Muscle cachexia: current concepts of intracellular mechanisms and molecular regulation. Ann Surg. 2001;233:9–17. doi: 10.1097/00000658-200101000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Combaret L, Dardevet D, Rieu I, Pouch MN, Bechet D, Taillandier D, Grizard J, Attaix D. A leucine-supplemented diet restores the defective postprandial inhibition of proteasome-dependent proteolysis in aged rat skeletal muscle. J Physiol. 2005;569:489–99. doi: 10.1113/jphysiol.2005.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics. 2003;14:149–59. doi: 10.1152/physiolgenomics.00049.2003. [DOI] [PubMed] [Google Scholar]
- 42.Welle S, Brooks AI, Delehanty JM, Needler N, Bhatt K, Shah B, Thornton CA. Skeletal muscle gene expression profiles in 20-29 year old and 65-71 year old women. Exp Gerontol. 2004;39:369–77. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Aged human muscle demonstrates an altered gene expression profile consistent with an impaired response to exercise. Mech Ageing Dev. 2000;120:45–56. doi: 10.1016/s0047-6374(00)00178-0. [DOI] [PubMed] [Google Scholar]
- 44.Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Molecular characteristics of aged muscle reflect an altered ability to respond to exercise. Int J Sport Nutr Exerc Metab. 2001;11(Suppl):S9–15. doi: 10.1123/ijsnem.11.s1.s9. [DOI] [PubMed] [Google Scholar]
- 45.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol Genomics. 2005;21:253–63. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 46.Roth SM, Ferrell RE, Peters DG, Metter EJ, Hurley BF, Rogers MA. Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genomics. 2002;10:181–90. doi: 10.1152/physiolgenomics.00028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. Aging is associated with diminished accretion of muscle proteins after the ingestion of a small bolus of essential amino acids. Am J Clin Nutr. 2005;82:1065–73. doi: 10.1093/ajcn/82.5.1065. [DOI] [PubMed] [Google Scholar]
- 48.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. Am J Physiol Endocrinol Metab. 2006;291:E381–7. doi: 10.1152/ajpendo.00488.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.