Abstract
The leading cause of death in cancer patients is cancer metastasis, for which there is no effective treatment. MicroRNAs have been shown to play a significant role in cancer metastasis through regulation of gene expression. The Adenoviral type 5 E1A is associated with multiple tumor suppressing activities including the inhibition of metastasis, and E1A gene therapies have been tested in several clinical trials. However, the mechanisms involved in E1A-mediated tumor suppressing activities are not yet completely defined. Here we demonstrated that E1A down-regulated the expression of miRNA, miR-520h, which was critical for E1A-mediated cancer cell mobility and in vitro invasion activity. In addition, we identified a signal cascade, namely, E1A —| miRNA-520h —| PP2A/C —| IKK → NF-κB → Twist, in which E1A inhibited the expression of Twist through downregulation of miR-520h and the signal cascade. Our results indicated a functional link between miR-520h and tumorigenicity/invasive ability, and provided a new insight into the role of E1A-mediated miRNA regulation in tumor suppression. Therefore, the results identified a new cascade of E1A-mediated tumor suppression activity via downregulation of miRNA-520h expression.
Keywords: E1A, miR520h, breast cancer, Twist, PP2A/C
INTRODUCTION
For most cancer patients, tumor metastasis at distant sites is the main cause of death from primary solid tumors (1). Progression to metastasis involves a series of discrete biological processes. The multi-step process is commonly known as the metastatic cascade, which basically includes invasion of the adjacent tissue, intravasation (penetration through the endothelium of blood or lymphatic vessels), survival during systemic circulation, extravasation, and colonization to form a progressively growing lesion in a distant site (2). Breast cancer is the most frequently diagnosed cancer in women. Despite a recent decrease in breast cancer incidence rates in the United States (3), it remains the second leading cause of cancer deaths in women. In breast cancer, metastases may remain undetectable and latent for many years following primary tumor removal. Improving our understanding of the molecular mechanism of the metastatic process is critical for designing more effective therapies for breast cancer.
The adenoviral type 5 E1A (E1A) gene was originally identified as one of two oncogenes in the adenovirus genome. Its cooperation with other oncogenes fully transforms primary rodent cells (4) but not human cells (5). Intriguingly, an association of E1A with multiple anti-cancer activities was found in cancer cells. For instance, E1A has been shown to inhibit NF-κB and Akt (6). A series of studies have demonstrated that stable expression of the E1A gene in cancer cells can reduce their tumorigenic potential, increase contact inhibition, and promote apoptosis in vivo (7–9). Therefore, clinical trials with E1A/liposome gene therapy for breast, ovarian, and head/neck cancer have been conducted (10–12). In addition to tumor suppression activity, expression of the E1A gene in stably transfected human cancer cells can convert various human cancer cell lines from the mesenchymal phenotype into an epithelial-like phenotype and inhibit metastasis (13, 14). Thus, further understanding of the molecular mechanisms associated with E1A-mediated suppression of metastasis could improve the efficacy of E1A gene therapy in future clinical trials.
During cancer metastasis, mobility and invasiveness of cancer cells increase. To detach from neighboring cells and invade adjacent cell layers, carcinoma cells must lose cell-cell adhesion and acquire motility. The highly conserved epithelial-mesenchymal transition (EMT) program has been implicated in dissemination of carcinoma cells from primary epithelial tumors (15). Tumor progression is frequently associated with the downregulation of E-cadherin (15), which is repressed by several transcription factors including Snail, Twist and Slug (16, 17). Interestingly, E1A is known to induce expression of E-cadherin and reverses EMT (13, 14), but its molecular mechanism is not yet clear.
In the past few years, microRNAs (miRNAs) have attracted attention as candidates for therapy because they potentially act as upstream regulators of tumorigenesis (18, 19). Using post-transcriptional control mechanisms, mammalian miRNAs are involved in crucial biological processes, including development, differentiation, apoptosis, and proliferation (20). So far, the list of known miRNAs has expanded, now standing at 735 in humans, and is predicted to increase further (21). Up to 30% of human genes may be regulated by miRNAs (22) and frequent deletions and deregulation of miRNAs have been reported in many types of human tumors (23, 24). Consistent with their frequent deregulation role in cancer, more than 50% of miRNA genes are located in cancer-associated genomic regions or at fragile sites, which are preferential sites for recombination, amplification, deletion, translocation, and viral integration (25). It is becoming clear that altered expression of miRNA is a special “signature” of many human diseases, especially specific forms of cancer (19, 26). Accumulating evidence indicates that deregulation of miRNA levels might have a role not only in tumorigenesis, but also in cancer metastasis (27, 28).
A previous study indicated that E1A could reprogram transcription in tumor cells (9). E1A does not bind to the DNA directly, but instead, interacts with components of the general and specific transcriptional machinery, including the TATA-binding protein (TBP), p300 (29, 30), several TBP-associated factors (TAFs) (31, 32), and numerous transcription factors (such as ATF-2 and c-Jun; ref 51, 52). Although knowledge regarding the complex control of gene expression by miRNA has increased, our understanding of the regulation of miRNA gene transcription remains limited. In an attempt to understand E1A-mediated tumor suppression activity, we asked whether E1A might regulate the expression of miRNA.
In this study, we identified several miRNAs that are regulated by E1A in humans. One of these miRNAs, miR-520h, was further shown to be downregulated by E1A, and its downregulation was critical for E1A-mediated tumor and invasion suppression activities. We further demonstrated that E1A-mediated downregulation of miR-520h enhanced PP2A/C expression, which in turn inhibited the IKK/NF-κB pathway resulting in repression of Twist expression. Together, our results suggest that E1A-mediated Twist downregulation may contribute to E1A-mediated tumor and invasion suppression activities.
Materials and Methods
Cell Culture
The human breast cancer cell lines (MDA-MB-231, MB-453, and MB-468) and human cervical cancer cell line (HeLa) were obtained from the American Type Culture Collection (Manassas, VA). MDA-MB-231 and its E1A/vector-stable transfectants and the human ovarian cancer cell line (SKOV3-ip1) and its E1A/vector-stable transfectants have been described previously (48). MDA-MB-231/E1A and SKOV3-ip1/E1A cells were transfected with miR-520h-expressing vectors using Lipofectamine-2000 (Invitrogen, Carlsbad, CA). Forty-eight hours after transfection, cells were trypsinized and replated in Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12) with 10% fetal bovine serum (FBS) and blasticidin (8 μg/ml; Invitrogen). Blasticidin-resistant clones (231/E1A-miR-520h, ip1/E1A-miR-520h) were selected. These clones were selected and expanded for further studies. All cell lines were maintained in DMEM/F12 (1:1) supplemented with 10% FBS (Hyclone, Logan, UT), 2 mM L-glutamine (Gibco, Grand Island, NY), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Antibodies
Antibodies to the following were used: E1A (554155; BD Pharmingen, San Diego, CA), E-cadherin (610181; BD Transduction Laboratories, Lexington, KY), HA (11666606001; Roche Diagnostics, Indianapolis, IN), IKKβ (2684; Cell Signaling Technology, Beverley, MA), pIKKα/β (2681; Cell Signaling Technology), Akt (9272; Cell Signaling Technology), pAkt (9271 or 9275; Cell Signaling Technology), PP2A/C (2259; Cell Signaling Technology, or SC-6110; Santa Cruz Biotechnology), Twist (SC-15393; Santa Cruz Biotechnology, Santa Cruz, CA), Slug (SC-15391; Santa Cruz Biotechnology), Snai1 (SC-28199; Santa Cruz Biotechnology) and α-tubulin (T-5168; Sigma, St. Louis, MO).
microRNA microarray hybridization
Three micrograms of total RNA obtained from MDA-MB-231 (stably expressing control vector or E1A vector) were labeled and hybridized on miRNA microarrays (using the Ncode Multi-Species miRNA microarray V1 kit by Invitrogen). The arrays were designed to detect the 376 miRNA transcripts listed in the Sanger miRBase Release 7.0. Raw signal intensities representing hybridization to probes were mean-normalized across cell lines. The significant of miRNAs up-regulation or down-regulation were identified by Ncode Profiler software (Invitrogen).
RNA isolation, reverse transcription, and PCR
Total RNA was extracted from cultured cells using the mirVana miRNA isolation kit (Ambion, Austin, TX), according to the manufacturer’s instructions. Reverse transcription reactions contained 1 μg of RNA, 50 nM stem-loop RT primer, 0.25 mM each dNTP, 50 units of MMLV reverse transcriptase (Invitrogen), 1X reverse transcription buffer, 10 mM DTT, and 4 units of RNase inhibitor. The stem-loop RT primers were designed according to the report by Chen C et al (50). The sequences of mature miRNAs were obtained from the Sanger Center miRNA Registry (http://microrna.sanger.ac.uk/sequences/) as follows: miR-22 RT primer (5′-GTTGGCTCGGTGCAGGGCCTGAGGTATTCGCACCAGAGCCAACACAGTTC-3′); miR-27a RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGCGGAA-3′); miR-377 RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACAAAAG-3′); miR-515-3pRT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACGCTC-3′); miR-195RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGCCAAT-3′); miR-197RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGCTGGGT-3′); Let-7b RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACCACA-3′); miR-25RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCAGACC-3′); miR-92RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACAGGCC-3′); miR-107RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTGATAGC-3′); miR-15bRT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTGTAAAC-3′); miR-520h RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACTCTAA-3′); miR-18a RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTATCT-3′); miR-182 RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAGTGTTG-3′); miR-93 RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTACCT-3′); RNU6B RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAAAATAT-3′); U47 RT primer (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCTCAG-3′).
The reactions were incubated at 37°C for 50 minutes, 16°C for 30 minutes, followed by pulsed RT of 60 cycles at 20°C for 30 seconds, 42°C for 30 seconds and 50°C for 1 second. Reactions were terminated by incubating at 85°C for 5 minutes to inactivate the reverse transcriptase. In addition, 231/VC and 231/E1A as well as 231/E1A-miR520h cells were used to isolate total RNA by TRIZOL. An aliquot of 1 μg of total RNA was subjected to RT-PCR using reverse transcriptase kit (Invitrogen). Equal amount of cDNA (2 μl) was used to perform PCR. The sequence of pp2a and snail along with internal reference β-actin primers are as follows: pp2a forward: 5′-GCCTACAAGAAGTTCCCCATGAG-3′, reverse: 5′-GCACCAGTTATACCCTCCATCAC-3′; snail forward: 5′-AATGCTCATCTGGGACTCTGTCC-3′, reverse: 5′-GCCTCCA AGGAAGAGACTGAAGTAG-3′; β-actin forward: 5′-GCTCGTCGTCGACAACGGCTC-3′, reverse: 5′-CAAACATGATCTGGGTCATCTTCTC-3′. After initial denaturation at 94 °C for 4.5 min, cycling parameters were as follows: pp2a: denaturation (94 °C, 1 min), annealing (55 °C, 1 min), and extension (72 °C, 1min). The reaction included amplification 25 cycles; snail: denaturation (94 °C, 1 min), annealing (58 °C, 1 min), and extension (72 °C, 1min). The reaction included amplification 30 cycles; β-actin: denaturation (94 °C, 1 min), annealing (55 °C, 1 min), and extension (72 °C, 1min). The reaction included amplification 25 cycles.
Real-time PCR quantification
Real-time PCR was performed using a Roche LightCycler 480 Real-Time PCR system. PCR reactions contained 0.5 μM of each forward and reverse primer, 0.1 μM Universal ProbeLibrary Probe #21 (Roche), 1X LightCycler TaqMan Master, and 2 μl of cDNA. Amplification curves were generated with an initial denaturing step at 95°C for 10 minutes, followed by 50 cycles of 95°C for 5 seconds, 60°C for 10 seconds, and 72°C for 1 second. The U6 and U47 small nuclear RNAs were used as an internal control. The sequence of forward primers were shown as following: miR-2 2 (5 ′-CGGCGGAAGCTGCCAGTTGAA-3′), miR-27a (5′CGGCGGTTCACAGTGGCTAAG-3′), miR-520h (5′-TCGCGACAAAGTGCTTCCCT-3′), miR-18a (5′-CGGCGGTAAGGTGCATCTAGTGC-3′), miR-182 (5′-CGGCGGTTTGGCAATGGTAGAACT-3′), miR-93 (5′-TCGCGCAAAGTGCTGTTCGTGC-3′), miR-197 (5′-CGGCGGTTCACCACCTTCTCC-3′), Let-7b (5′-CGGCGGTGAGGTAGTAGGTTG-3′), miR-195 (5′-CGGCGGTAGCAGCACAGAAAT-3′), miR-107 (5′-CGGCGGAGCAGCATTGTACAGG-3′), miR-25 (5′-CGGCGGCATTGCACTTGTCTC-3′), miR-92 (5′-CGGCGGTATTGCACTTGTCCC-3′), miR-15b (5′-CGGCGGTAGCAGCACATCATG-3′), miR-377 (5′-CGGCGGATCACACAAAGGCAA-3′), miR-515-3p (5′-CGGCGGGAGTGCCTTCTTTTG-3′), RNU6B (5′-TTCCTCCGCAAGGATGACACGC-3′), and U47 (5′-CGGCGGTAATGATTCTGCCAAA-3′). The reverse primer for all above sets of genes was 5′-GTGCAGGGTCCGAGGT-3′.
miRNA inhibition and overexpression
The mature miRNA sequences were obtained from the Sanger Center miRNA Registry (http://microrna.sanger.ac.uk/sequences/) and the human miRNA gene was designed according to the manufacturer’s instructions. The sequences of the top strands (5′ primer) and bottom strands (3′ primer) were for miR-27a (top: 5′-TGCTGGCGGAACTTAGCCACTGTGAAGTTTTGGCCACTGACTGACTTCACAGTCTAAGTTCCGC-3′; bottom: 5′-CCTGGCGGAACTTAGACTGTGAAGTCAGTCAGTGGCCAAAACTTCACAGTGGCTAAGTTCCGCC-3′), miR-520h (top: 5′-TGCTGACTCTAAAGGGAAGCACTTTGTGTTTTGGCCACTGACTGACACAAAGTGTCCCTTTAGAGT-3′; bottom: 5′-CCTGACTCTAAAGGGACACTTTGTGTCAGTCAGTGGCCAAAACACAAAGTGCTTCCCTTTAGAGTC-3′), and control (top: 5′-TGCTGGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′; bottom: 5′-CCTGGAAATGTACTGCGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCAGTACATTTC-3′). These miRNA gene double-strands were ligated with the Block-iT™ Pol II miR RNAi Expression Vector, pcDNA6.2-GW/EmGFP-miR (Invitrogen). Subsequently, the miRNA expression vector or control vector were transfected into the MDA-MB-231/E1A cells using Lipofectamine-2000 (Invitrogen) according to the manufacturer’s instructions. The GFP+ cells accounted for over 90% of transfected cells, as determined by fluorescence-activated cell sorting (FACS) analysis were used for migration or invasion studies.
Anti-miR microRNA inhibitors (Ambion) are single-stranded chemically modified oligonucleotides designed to inhibit endogenous miRNAs. For in vitro miRNA-inhibition studies, MDA-MB-231 cells were transfected with anti-miR inhibitors (150 nM) based on the manufacturer’s instructions. Forty-eight hours after transfection, cells were used for migration assays, invasion assays, luciferase reporter assays, and immunoblot assays.
Luciferase assay
The full-length 3′UTR of the miR-520h-regulated human PP2A/C genes (GenBank Accession No. NM_002715) were amplified from MCF7 cDNA and cloned into the SacI/SpeI site of pMIR-REPORT Luciferase vector (Ambion). The primers for PP2A/C were forward (5′-GCGACATTGTTGGTCAAGAAACC-3′) and reverse (5′-GCAGGAAGAACCCACAAAGTG C-3′). The segment (base pairs 2226–2231) of the PP2A/C 3′UTR containing the mutated miR-520h target sequence (ACTTTG to TGAAAC) was also cloned into the pMIR-REPORT Luciferase vector (Ambion) using a QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The primers for PP2A/Cmt were forward (5 ′-CACATTGTTGGTGTGCTGAAACTGGGTTCTTCCTGCATATTAAC-3′) and reverse (5′-GTTAATATGCAGGAAGAACCCAGTTTCAGCACACCAACAATGTG-3′). Cells (50% confluent in 24-well plates) were transfected with indicated plasmids by Lipofetamine 2000 (Invitrogen). Firefly luciferase reporter gene construct (1 μg) and 0.1 μg of the pTK-Renilla luciferase construct (for normalization) were cotransfected per well. Cell extracts were prepared 48 h after transfection, and the luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
Trans-well migration and invasion assays
For transwell migration assays, 1 × 105 cells were plated in the top chamber onto the non-coated membrane (24-well insert; pore size, 8 μm; Corning Costar, Corning, NY). For invasion assay, 1 × 105 cells were plated in the top chamber onto the Matrigel-coated membrane. Each well was coated freshly with Matrigel (60 μg; BD Bioscience) before the invasion assay. In both assays, cells were plated in medium without serum or growth factors, and medium supplemented with serum was used as a chemoattractant in the lower chamber. The cells were incubated for 24 hours (migration assay) or 48 hours (invasion assay) and cells that did not migrate or invade through the pores were removed by a cotton swab. Cells on the lower surface of the membrane were fixed with methanol and stained with crystal violet. The number of cells migrating through or invading through the membrane were counted by light microscope (40X, three random fields per well).
Animal studies
All animal work was done in accordance with a protocol approved by the CMUH Institutional Animal Care and Use Committee. Female SCID mice age-matched, 4–6 weeks old, were used in assays for tumor growth in an orthotopic xenograft model and lung colonization metastasis in an experimental metastasis model. For primary tumor growth assay, viable cells (3 × 106; re-suspended in a 1:1 mixture of PBS and growth-factor-reduced Matrigel [BD Biosciences] in a total volume of 50 μl) were injected orthotopically into the mammary gland. Primary tumor growth rates were analyzed by measuring tumor length (L) and width (W), and calculating volume through the formula LW2/2. For experimental metastasis assays, 1 × 106 viable cells were re-suspended in 0.1 ml of PBS and introduced into the circulation via tail-vein injection. Lung metastasis was quantified after 28 days post injection.
RESULTS
E1A-mediated Regulation of miRNA Expression in Human Cancer Cells
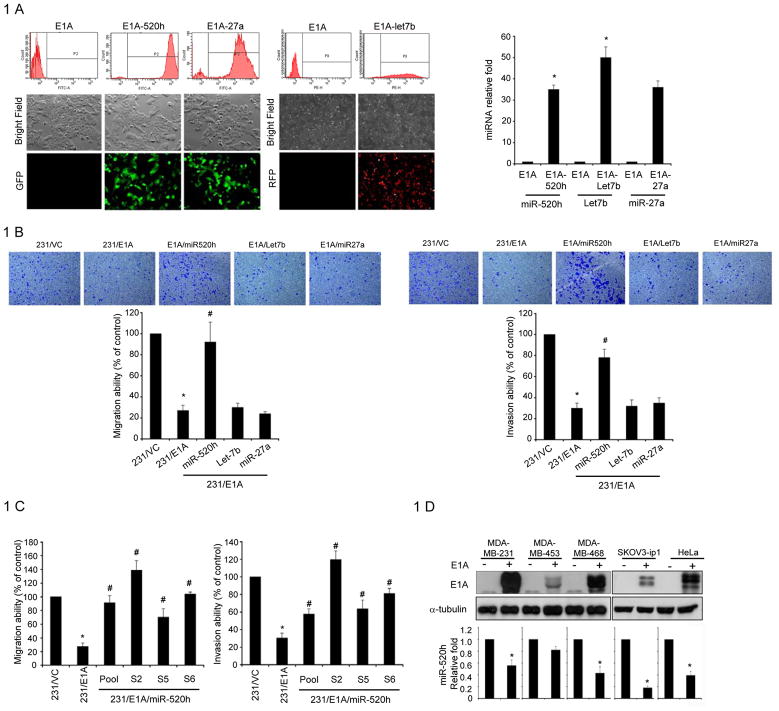
To investigate whether the Adenovirus 5 E1A gene could regulate miRNA expression, we analyzed global miRNA expression in MDA-MB-231 human breast cancer cells stably transfected with control vector (231/VC) or E1A expression vector (231/E1A) (14) using the miRNA microarray (NCode, Invitrogen). Differential expression of these down-regulated or up-regulated candidate miRNAs in 231/VC or 231/E1A cells were further validated through quantitative stem-loop polymerase chain reaction (qRT-PCR) (Table 1). Among the four potential down-regulated genes identified by the miRNA microarray, miRNA520h, miRNA-27a, and Let-7b were further confirmed by qRT-PCR to be downregulated (Table 1). To investigate whether these three miRNAs have an effect on the E1A–mediated suppression of cell invasion and migration activities, we generated stable pool transfectants by introducing these three miRNAs respectively into 231/E1A cells which were validated by green (E1A-520h and E1A-27a) or red fluorescence (E1A-let 7b) (Fig. 1A left panel) and by qRT-PCR (Figure 1A right panel). As expected, comparison between 231/E1A and 231/VC cells showed that ectopic expression of E1A reduced mobility and in vitro invasion activity. However, restoration of miR-520h, but not Let7b and miRNA-27a, in 231/E1A cells reversed these reductions (Fig. 1B). Moreover, in three different single stable miR-520h transfectants of 231/E1A cells (231/E1A/miR-520h cells), restoring the expression of miR-520h increased cell mobility and invasion activity (Fig. 1C). MiR-520h had no effect on 231/E1A cell proliferation (Supplementary Fig. S1A). Furthermore, examination of E1A’s effect on miR-520h expression in various types of cancer cells showed significantly decreased expression of miR-520h in E1A-transiently transfected cells (Fig. 1D). We also found significantly decreased cell migration and invasion abilities in various types of E1A-transiently transfected breast cancer cells (Supplementary Fig. S1B). These data suggest that E1A-mediated miR-520h downregulation may be involved in the E1A-mediated suppression of cell mobility and in vitro invasion activity.
Table 1.
Differentially expressions of miRNAs 231/E1A cells versus 231/VC cells were analyzed by miRNA microarray analysis and confirmed by qRT-PCR.
| Upregulated miRNAs by E1A | ||||||
|---|---|---|---|---|---|---|
| miRNA ID | microarray | qRT-PCR# | P-value | Function | Validated target | Refs. |
| has-mir-377 | 78.6 | 3.48±1.39 | 0.0199* | unknown | unknown | |
| has-mir-515-3p | 12.67 | 13.87±7.66 | 0.0102* | unknown | unknown | |
| has-mir-92 | 7.7 | 3.13±0.41 | 0.0450* | unknown | unknown | |
| has-mir-18a | 6.27 | 6.93±1.05 | 0.0121* | inhibition cell proliferation | K-Ras | (53) |
| has-mir-15b | 6.24 | 5.11±1.04 | 0.0051** | cell proliferation, apoptosis, cell cycle progression | cyclin E1 | (54), (55) |
| has-mir-195 | 4.71 | 3.24±1.49 | 0.0583 | block G1/S transition | cyclin D1, CDK6, E2F3 | (56) |
| has-mir-182 | 4.18 | 3.05±0.91 | 0.0548 | transformation or maintenance of an oncogenic state | FOXO1 | (57) |
| has-mir-107 | 4.06 | 4.26±1.5 | 0.1035 | cell cycle arrest | unknown | (58) |
| has-mir-93 | 3.16 | 3.77±0.34 | 0.0806 | unknown | FUS1 | (59) |
| has-mir-25 | 3.11 | 4.74±1.48 | 0.0878 | unknown | Krüppel-like factor 4 (KLF4) | (60) |
| Downregulated miRNAs by E1A | ||||||
| miRNA ID | microarray | qRT-PCR | P-value | |||
| has-mir-520h | 0.01 | 0.02±0.02 | 0.0001** | stem cell differentiation | ABCG2 | (61), (62) |
| has-mir-197 | 0.02 | 0.78±0.03 | 0.0178* | unknown | FUS1 | (63) |
| Let-7b | 0.23 | 0.30±0.17 | 0.0365* | Induces apoptosis and inhibits tumorigenesis | RAS, MYC and HMGA2 | (64) |
| has-mir-27a | 0.31 | 0.33±0.08 | 0.0352* | cancer cell growth, | Prohibitin, FOXO1 | (65) |
Each assay was performed in at least three separate experiments. The data were normalized to the level of U47 RNA in each sample.
The asterisk indicates a significant difference between 231/VC and 231/E1A cells.
, P < 0.05.
P < 0.01.
Figure 1. E1A-mediated Regulation of miRNA in Human Cancer Cells.
A, left, the expression of miR-520h, miR-27a (GFP+ cells) and let-7b (RFP+ cells) accounted for over 90% of transfected cells, as determined by fluorescence-activated cell sorting (FACS) analysis, and were used for further migration or invasion study. right: After miRNA transfection, RNA was isolated from MDA-MB-231/E1A transfected cells and the expression of miRNAs was confirmed by qRT-PCR. The data were normalized to the level of U47 RNA in each sample. Error bars indicate SD. The asterisk indicates a significant difference (P<0.05) between 231/E1A and miRNA transfected 231/E1A cells. B, transwell migration assay and matrigel invasion assay of MDA-MB-231/E1A cells transfected with different miRNA expression vectors, miR-520h, let-7b and miR-27a. Bars indicate the migration or invasion activity compared to that of 231/VC± SD. Each assay was performed in three independent experiments. The asterisk indicates a significant difference (P<0.05) between 231/VC and 231/E1A cells. E1A-dependent migration and invasion ability suppression was overturned by miR-520h to a significant degree, as indicated by the # symbol (P<0.05). C, transwell migration assay (left) and matrigel invasion assay (right) of MDA-MB-231/E1A cells with different stable miR-520 expressing transfectants (231/E1A/miR-520h). Bars indicate the migration or invasion activity compared to 231/VC ± SD. Each assay was performed in three independent experiments. The asterisk indicates a significant difference (P<0.05) between 231/VC and 231/E1A cells. E1A-dependent migration and invasion suppression was overturned by expression of miR-520h, as indicated by the # symbol (P<0.05). D, different cancer cells were transfected with E1A. Upper, 48 hours after transfection, E1A expression was confirmed by western-blotting. Bottom, RNA was isolated from transfectants to determine the expression level of miR-520h by qRT-PCR. The data were normalized to the level of U47 RNA in each sample. Error bars indicate SD. The asterisk indicates a significant difference (P<0.05) between E1A transfected and control cells; P-values based on a one-tailed Student’s t-test.
miR-520h Promotes Invasion and Metastasis of Cancer Cells
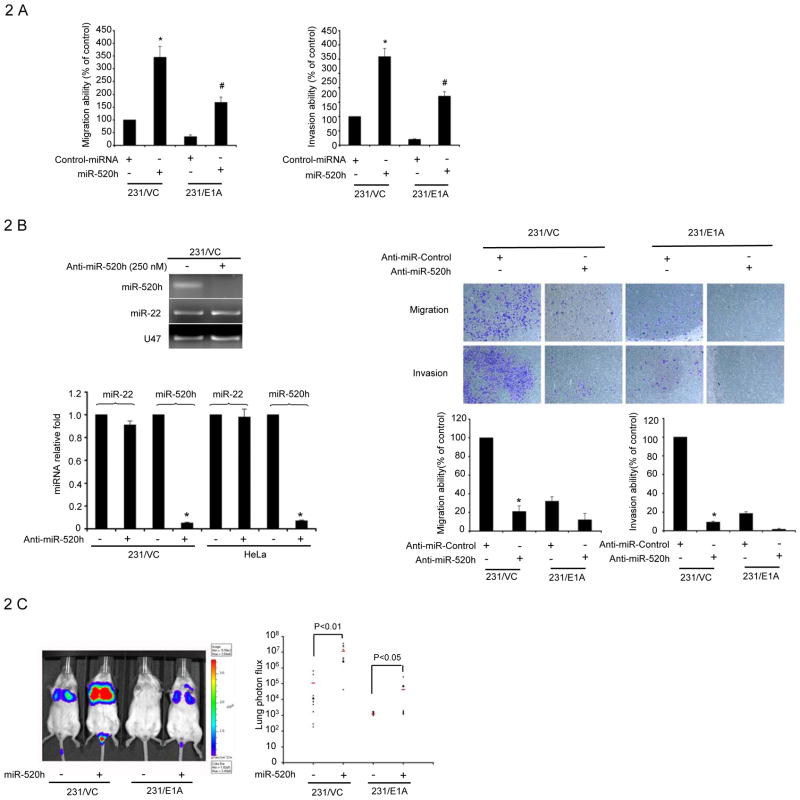
Consistently, enforced expression of miR-520h, but not a non-target sequence control miRNA, in 231/VC and 231/E1A cells enhanced cell mobility and invasion activity (Fig. 2A). Similar results were also observed in an ovarian cancer cell line (Supplementary Fig. S2). To further assess the contribution of miR-520h to cell migration, we performed in vitro loss-of-function analyses by silencing the endogenous miRNA-520h using a modified antisense miR-520h inhibitor (Ambion). Administration of miR-520h inhibitor decreased the endogenous level of miR-520h, but not another miRNA, miR-22, as detected by qRT-PCR (Fig. 2B left). At 48 hr after transfection of the miR-520h inhibitor, but not a control miRNA inhibitor, the capacity of 231/VC cells to migrate and invade was significantly reduced by more than 70% (Fig. 2B right). This reduction was not due to loss of cell viability (Supplementary Fig. S3B). Next, we screened other cancer cell lines for the effect of miR-520h inhibitor on cell mobility and found migration and invasion ability were inhibited in all cell lines by transfection of miR-520h inhibitor (Supplementary Fig. S3A). To determine whether the expression of miR-520h would promote tumor invasion in vivo, we further investigated the effects of miR-520h on E1A-mediated anti-metastatic lung colony formation in an experimental metastasis/lung colonization assay. Luciferase-tagged 231/VC, 231/VC-miR520h, 231/E1A and 231/E1A-miR520h cells were transplanted into SCID mice via tail-vein injection. As shown previously (14), no lung colonization was found in all 10 mice that had been transplanted with 231/E1A cells (Fig. 2C, 2D). However, 5 out of 10 mice that received the miR-520h-overexpressing 231/E1A cells exhibited numerous lung colonizations (Fig. 2D). Similarly, miR-520h expression enhanced metastasis from 6 out of 10 mice to 9 out of 10 mice compared with 231/VC cells. Restoring the expression of miR-520h in 231/VC and 231/E1A cells increased the lung colonization activity of these cells by more than one hundred-fold by bioluminescence imaging (Fig. 2C, lane 2 versus lane 1 and lane 4 versus lane 3). We also orthotopically implanted miR-520h overexpressed- or control vector overexpressed-231/VC and 231/E1A cells into the mammary fat pads of SCID mice and measured the growth of the resulting primary tumors. At four weeks post implantation, the growth rate of miR-520h–overexpressing tumors was higher than control tumors, indicating miR-520h may enhance tumor growth in target microenvironments (Fig. S4). Taken together, these observations suggest that miR-520h may be involved in in vitro migration/invasion, in vivo tumorigenicity and lung colonization activity in the experimental metastasis model.
Figure 2. miR-520h Is Required for Invasive Phenotype.
A, transwell migration assay and matrigel invasion assay of MDA-MB-231/VC and 231/E1A transfected with miR-520h or control miRNA. Bars indicate the migration or invasion activity compared to that of 231/VC ± SD. Each assay was performed in three separate experiments. The asterisk indicates a significant difference (P<0.05) between miR-520h transfected and control miRNA transfected 231/VC cells. The significant difference (P<0.05) between miR-520h transfected and control miRNA transfected 231/E1A cells is indicated by the # symbol (P<0.05). B, left, the expression of miR-520h and miR-22 in MDA-MB-231/VC cells and HeLa cells after miR-520h inhibitor treatment for 72 hours was determined by agarose elephoresis (upper) and qRT-PCR (lower). Right, transwell migration assay and matrigel invasion assay of MDA-MB-231/VC and 231/E1A cells transfected with miR-520h inhibitor (Anti-miR-520h) or control inhibitor (Anti-miR-Control). Bars indicate the migration or invasion activity compared to that of 231/VC ± SD. Each assay was performed in three separate experiments. The asterisk indicates a significant difference (P<0.05) between miR-520h inhibitor transfected and control inhibitor transfected 231/VC cells. C, 231/VC, 231/VC-miR520h, 231/E1A, and 231/E1A-miR520h cells stably expressing luciferase were injected intravenously into immunodeficient SCID mice. left, bioluminescence imaging of lung colonizations were measured four weeks after transplantation. Bioluminescence was used to quantify lung colonization. Right, data shown are representative mice corresponding to marked data points. n=10; horizontal line represents median signal for each cohort. P-values were obtained by one-tailed Student’s t-test. D, incidence of lung colonization in mice that received tail-vein injection of miR-520h-overexpression or control 231/VC and 231/E1A cells.
Twist is Involved in miR-520h-mediated Metastasis
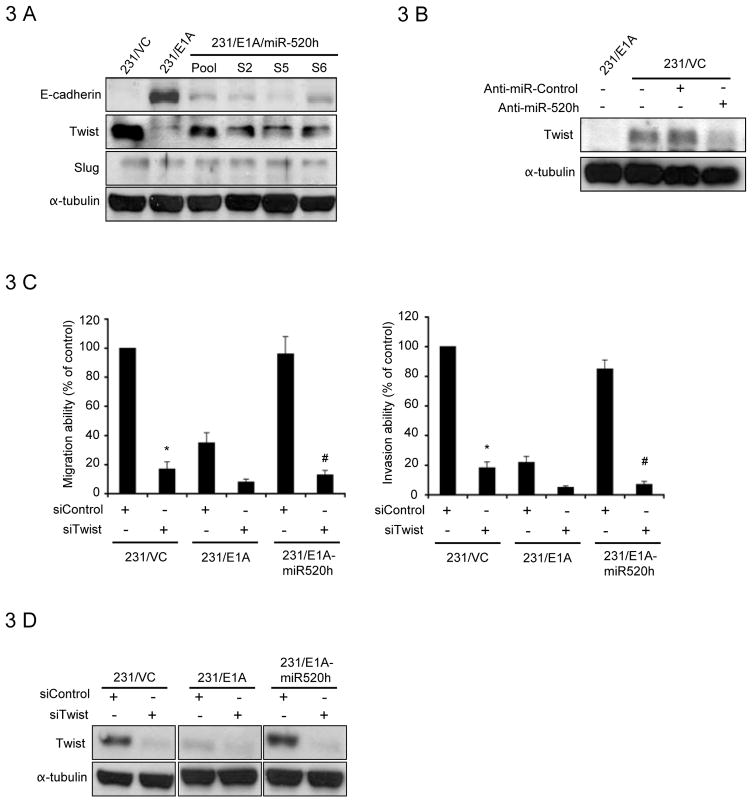
As E1A is known to induce expression of E-cadherin and reverse EMT, we examined whether miR-520h participates in EMT to enhance metastasis. E-cadherin expression was examined in 231/VC, 231/E1A, and different clones that overexpressed miR-520h using Western-blotting. The endogenous level of E-cadherin expression was reduced in miR-520h-overexpressing 231/E1A cells (Fig. 3A) and SKOV3-ip1/E1A cells (Supplementary Fig. S5A). Furthermore, we found that the expression of Vimentin, a mesenchymal marker, was downregulated by E1A and this inhibitory effect was overcome by miRNA-520h (Supplementary Fig. S5B). To further explore the mechanism of the downregulation of E-cadherin by miR-520h, we examined the expressions of three transcription factors involved in repression of E-cadherin, Twist, Slug and Snail. The results showed that Twist expression was consistently induced in miR-520h expressing cells (Fig. 3A) and inhibited in 231/VC cells transfected with miR-520h inhibitor (Fig. 3B), suggesting that miR-520h downregulates E-cadherin expression through increasing the expression of Twist. On the other hand, due to its short half-life (44), we did not clearly detect the change of Snai1 protein expression in different miR-520h-expressing cells (data not shown). This will need further investigation in the future. Since E1A downregulated miR-520h expression in cancer cells, Twist expression was also shown to be downregulated in E1A-expressing HeLa (data not shown) and SKOV3-ip1 cells (Supplementary Fig S5C). We also found that overexpression of Twist in 231/E1A cells was able to rescue E1A-mediated suppression of cell migration and invasion abilities (Supplementary Fig. S5D). We next studied whether miR-520h-induced Twist expression might contribute to miR-520h-mediated metastasis. The results showed that cells transfected with Twist siRNA strongly suppressed miR-520h-induced cell migration (by 80%) and invasion (by 85%; Fig. 3C and 3D) in 231/E1A cells. Taken together, these results suggest that E1A downregulated Twist is mediated through mir-520h, which is involved in E1A-mediated suppression of metastasis.
Figure 3. Induction of Twist is Critical for miR-520h Metastasis Promoting Function.
A, immunoblotting for E-cadherin, Twist, Slug in 231/VC, 231/E1A, and different stable clones of 231/E1A/miR-520h. B, whole cell extracts of MDA-MB-231 cells transfected with the miR-520h inhibitor (anti-miR-520h) or control inhibitor (anti-miR-control) were prepared 48 hours after transfection and analyzed by immunoblotting with Twist and tubulin antibodies. C, transwell migration assay (left) and matrigel invasion assay (right) of 231/VC, 231/E1A, and 231/E1A-miR520h overexpression cells stably transduced with Twist siRNA. Bars indicate the migration or invasion activity compared to that of 231/VC ± SD. The asterisk indicate a significant difference (P<0.05) between siTwist transduced and siControl tranduced 231/VC cells. The significant difference (P<0.05) between Twist knockdown and control knockdown 231/E1A-miR520h is indicated by the # symbol. D, 231/VC, 231/E1A, and 231/E1A-miR520h overexpression cells stably transduced with Twist siRNA and analyzed by immunoblotting with Twist and tubulin antibodies.
PP2A/C Is a Direct and Functional Target of miR-520h
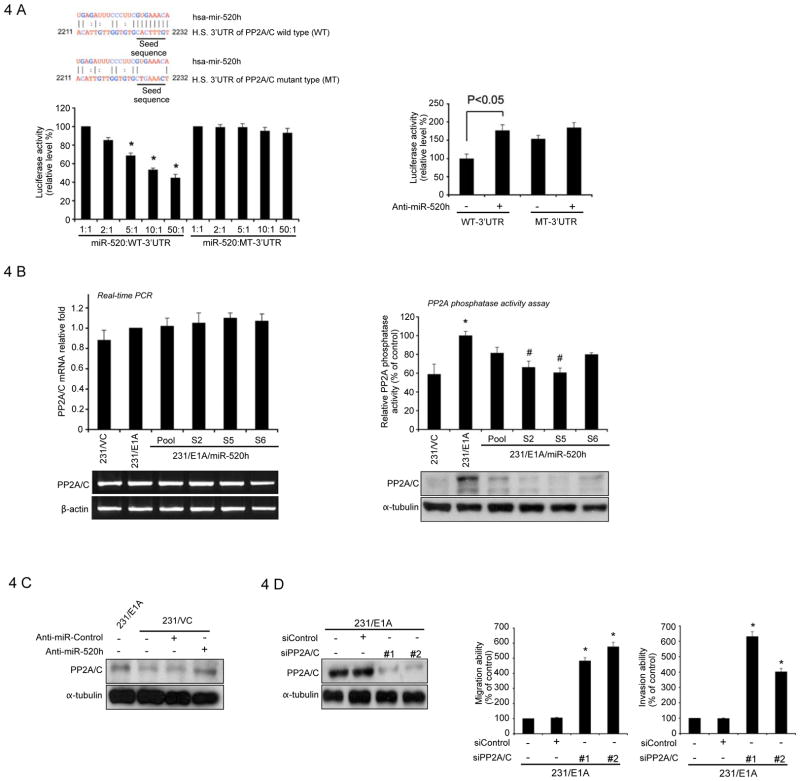
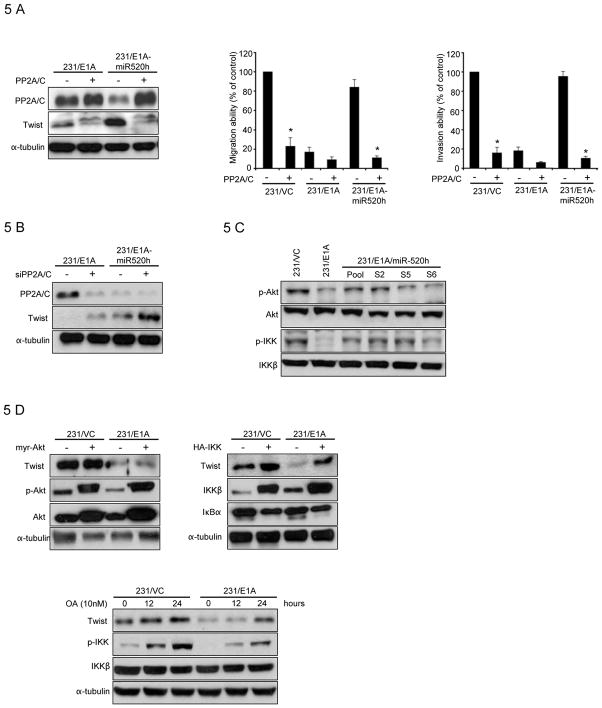
Next, we investigated the target molecules of the miR-520h that might contribute to biological activities. To this end, several computational prediction methods were used to identify miR-520h target genes in humans. Among the targets predicted by both the TargetScan Human 5.1 (33) and MIRAND Version 3.0 (21) search programs (Fig. S6), one target of miR-520h, the protein phosphatase 2A catalytic subunit (PP2A/C) gene, attracted our attention as PP2A/C was shown to be upregulated in E1A-expressing cells and required for E1A-mediated sensitization to anticancer drugs (34). Therefore, we asked whether miR-520h might downregulate PP2A/C by directly binding to sites within the 3′UTR of its message RNA, thereby promoting tumor progression. To this end, we constructed luciferase reporter vectors encoding the complete wild-type 3′UTR of the PP2A/C mRNA (WT-3′UTR) as well as parallel control vectors containing mismatches in the predicted miR-520h binding site (MT-3′UTR) (Fig. 4A , left, upper), and transfected these vectors into HEK293T cells at different miR-520h vector-to-reporter ratios. Transfection of the WT-3′UTR plasmid, but not the MT-3′UTR plasmid, resulted in decreased luciferase activity in a dose-dependent manner. The results supported a negative effect of the endogenous miR-520h molecule on PP2A/C 3′UTR expression (Fig. 4A left, bottom). Blockage of endogenous miR-520h expression using an antisense inhibitor (Ambion) resulted in a significant increase in PP2A/C WT-3′UTR luciferase expression in 231/VC cells (Fig. 4A right). Mutations in the predicted miR-520h binding site reduced sensitivity to miR-520h inhibitor (PP2A/C MT versus PP2A/C WT; Fig. 4A right). These experiments validated the regulatory potential of miR-520h via binding within the PP2A/C 3′UTR and confirmed miR-520h-mediated modulation of endogenous PP2A/C in cancer cells. Overexpression of miR-520h did not inhibit the expression of PP2A/C in message RNA level (Fig. 4B, left), but the level of endogenous PP2A/C protein was decreased in various 231/E1A clones stably expressing miR-520h, which was also supported by the reduced phosphatase activities in different miR-520h expressing 231/E1A cells (Fig. 4B, right). Consistently, introduction of miR-520 inhibitor led to increased PP2A/C expression in 231/VC cells (Fig. 4C). To determine whether reduction of PP2A/C levels might account for the induction of cell mobility and invasiveness observed following miR-520h overexpression, 231/E1A cells were transfected with siRNA against PP2A/C, which caused a >90% reduction in the level of the PP2A/C protein, and subsequently, increased cell motility and invasiveness (Fig. 4D). Together, our results indicated that PP2A/C is one target of the miR-520h, and loss of PP2A/C promotes cancer cells mobility and invasive activity. Since miR-520h inhibits PP2A/C and also activates Twist, we asked whether PP2A/C might also downregulate Twist. To test this, 231/E1A-miR520h and 231/VC cells, in which PP2A/C was downregulated by miR520h, were transfected with a construct constitutively expressing PP2A/C mRNA without the 3′UTR (an mRNA that is resistant to miR-520h-mediated inhibition of translation). Strikingly, constitutive expression of PP2A/C not only inhibited the expression of Twist but also abrogated miR-520h-induced cell motility and invasiveness (Fig. 5A). On the other hand, transfection of 231/E1A cells with PP2A/C siRNA induced the expression of Twist (Fig. 5B). Together, these results suggest that PP2A/C downregulates Twist expression. Next, we asked how PP2A/C might down-regulate Twist. PP2A/C has been shown to repress Akt and NF-κB activation through de-phosphorylation (35, 36). Both PI3K/Akt and IKK/NF-κB signaling pathways have been shown to enhance Twist expression (47, 49). Thus, we investigated whether miR-520h through down-regulation of PP2A may activate Akt and IKK by increasing their phosphorylation. Indeed, miR-520h expression enhanced Akt and IKK phosphorylation as predicted (Fig. 5C). To determine which PP2A/C-mediated pathways are involved in the regulation of Twist, we assessed Twist expression in 231/E1A cells transfected with or without Akt or IKK constitutive expression vectors (myr-Akt and HA-IKKβ). Twist expression was induced after transfection with IKKβ but not after transfection with myr-Akt (Fig. 5D), suggesting that IKK but not Akt is involved in upregulation of Twist in our experimental conditions. In addition to increased Twist expression, we found that cell invasion and migration abilities were also induced by transfection with IKKβ (Supplementary Fig. S7A). To support this notion, we showed that the addition of BAY117082, an inhibitor of IKK also suppressed Twist expression (Supplementary Fig. S7B). In contrast, LY294002, a PI3K inhibitor did not significantly affect Twist expression (data not shown). Treatment of 231/E1A cells with the PP2A/C phosphatase inhibitor (okadaic acid; OA) restored the level of IKK phosphorylation and increased Twist expression as well (Fig. 5E). Together, in the current study, we identified a signal cascade, namely, E1A —| miRNA520h —| PP2A/C —| IKK → NF-κB → Twist, which demonstrates that E1A, through downregulation of miR-520h, results in inhibition of Twist expression, which may contribute to the E1A associated tumor/metastasis suppression activity.
Figure 4. Direct Downregulation of PP2A/C by miR-520h.
A, left, upper, a schematic diagram represents the predicted miR-520h binding sequences or mutated versions. Lower panel, luciferase activity of DNA constructs carrying the wild-type PP2A/C 3′UTR (WT-3′UTR) or mutated-type PP2A/C 3′UTR (MT-3′UTR) downstream of the luciferase reporter. 293T cells were cotransfected with WT-3′UTR or mutant MT-3′UTR reporter (carrying the luciferase-PP2A/C-3′UTR chimera) and a plasmid expressing miR-520h at different ratios. miR-520h reduced luciferase activity in transfectant with wild-type (WT-3′UTR), but not with mutant (MT-3′UTR), reporter in a dose-dependent manner. Luciferase activity was normalized for transfection efficiency as indicated in the experimental procedures. Data are shown as separate assays at five different luciferase/miRNA ratios as indicated. Bars indicate Firefly/Renilla luciferase activity ratios that are normalized to the activity ratio (1:1) of 293T transfected with WT-3′UTR reporter or MT-3′UTR and miR-520h. Data are shown as three independent assays, and each sample was assayed in triplicate. Error bars represent SD. The asterisk indicates a significant difference (P<0.05) between miR-520h transfected different ratio (5:1, 10:1, 50:1) and control ratio (1:1) cells. Right, cotransfection with the inhibitor for miR-520h (anti-mir-520h) increased PP2A/C WT-3′UTR luciferase expression but not PP2A/C MT-3′UTR luciferase expression in 231/VC cells. Bars indicate Firefly/Renilla luciferase activity ratios normalized to that of 231/VC transfected with WT-3′UTR reporter. Data are shown as three independent assays, and each sample was assayed in triplicate. Error bars represent SD. P-value based on one-tailed Student\s t-test. B, left, upper, expression of PP2A/C mRNA in stable clones of 231/VC, 231/E1A, and different 231/E1A/miR-520h by qRT-PCR. Data were normalized to the level of GAPDH mRNA. Bottom, RT-PCR results of PP2A/C mRNA expression in 231/VC, 231/E1A and different stable clones of 231/E1A-miR-520h. β-actin served as the internal control. Right, upper, assay of PP2A/C phosphatase activities in stable clones of 231/VC, 231/E1A, and different 231/E1A/miR-520h. Bars indicate the phosphatase activity normalized to that of 231/E1A ± SD. The asterisk indicates a significant difference (P<0.05) between 231/VC and 231/E1A cells. E1A-dependent PP2A/C activity induction was overturned by miR-520h to a significant degree, as indicated by the # symbol (P<0.05). Bottom, immunoblotting of PP2A/C in stable clones of MDA-MB-231/VC, 231/E1A, and different 231/E1A/miR-520h. C, whole cell extracts of MDA-MB-231 cells transfected with the miR-520h inhibitor (anti-miR-520h) or non-target sequence control inhibitor (anti-miR-control) were prepared 48 hours after transfection and analyzed by immunoblotting with PP2A/C and tubulin antibodies. D, left, 231/E1A cells stably transduced with different sequences of siRNA that specifically target to PP2A/C (siPP2A/C) and analyzed by immunoblotting with PP2A/C and tubulin antibodies. Middle and right, transwell migration assay (middle) and matrigel invasion assay (right) of 231/E1A cells stably transduced with different sequences of siRNA that specifically target to PP2A/C (siPP2A/C). Bars indicate the migration or invasion activity in compare with 231/E1A ± SD. The asterisk indicates a significant difference (P<0.05) between siPP2A/C- and siControl-transduced 231/E1A cells.
Figure 5. Twist is Regulated by PP2A/C-mediated Signal Transduction Cascades.
A, left, immunoblotting of PP2A/C and Twist in 231/E1A and 231/E1A-miR-520h cells transiently transfected with PP2A/C expressing vector lacking the endogenous 3′UTR. Transwell migration assay (middle) and Matrigel invasion assay (right) of 231/VC, 231/E1A, and 231/E1A-miR-520h transiently transfected with PP2A/C expressing vector lacking the endogenous 3′UTR. Bars indicate the migration or invasion activity compared to that of 231/VC ± SD. The asterisk indicates a significant difference (P<0.05) between PP2A/C transfected and control vector transfected cells. B, immunoblotting of PP2A/C and Twist in 231/E1A and 231/E1A-miR-520h cells transiently transfected with PP2A/C siRNA. C, immunoblots showing the phosphorylation status of Akt and IKK. D, immunoblotting of Twist in 231/VC and 231/E1A cells transfected with a constitutive expression vector for myr-Akt (left) or IKK (right). E, immunoblotting of Twist and p-IKK in 231/VC and 231/E1A cells after the treatment of OA (10nM), PP2A phosphatase inhibitor at indicated time points.
Discussion
In this present work, we demonstrated that miR-520h can promote cell migration/invasion in vitro and tumorigenicity and metastasis potential in vivo through regulation of PP2A/IKK/NF-κB/Twist pathway. However, the oncogenic role of miR-520h can be downregulated and suppressed by E1A. Specifically, we showed that E1A downregulates miR-520h and blocks the synthesis of the PP2A/C protein, and E1A-induced PP2A/C expression inhibits expression of Twist through the IKK/NF-κB pathway. In addition, miR-520c (which is located within the same gene cluster as the miR-520h gene) has recently been reported to target the cell surface receptor, CD44 (27), which acts as a metastasis suppressor in cancers of the breast, prostate, and colon (37), indicating that miR-520c likely functions similar to miR-520h as a metastasis promoter in breast cancer. In addition to suppressing metastasis, E1A gene therapy has been shown to increase sensitivity to different categories of anticancer drug in multiple clinical trials. Recent studies have implicated miR-27a in drug-resistance in cancer cells (38). We also show that miR-27a was downregulated by E1A. Whether miR-27a and other miRNA family members are involved in E1A-mediated chemosensitization have yet to be investigated. On the other hand, E1A regulates the expression of several miRNAs (Table 1), but its involvement in the transcriptional control of miRNA expression is not clear. Since E1A regulates multiple miRNAs expression, it would be of interest to determine whether these E1A-regulated miRNAs may contribute to functionality of E1A protein.
Increasing data suggest that miRNAs might affect metastasis through regulation of EMT, which is a genetic developmental program regulated by a number of transcription factors (39, 40). Twist plays a key role in EMT-mediated intravasation of tumor cells (i.e., entry into the circulation to seed metastases) (41). Furthermore, Twist is required to bypass oncogene-induced cellular senescence (42). Recent research shows that tumor cells undergoing EMT resemble cancer stem cells (43). While investigating E1A-mediated mesenchymal-epithelial transition (MET), we discovered that Twist was markedly reduced by ectopic expression of E1A in various cancer cells. Overexpression of miR-520h in 231/E1A and ip1/E1A cells led to restoration of Twist expression. On the other hand, administration of miR-520h inhibitor lowered Twist level in 231/VC cells. In addition, transfection of Twist siRNA in cells overexpressing miR-520h inhibited cell mobility. Therefore, our data suggest that Twist is a downstream target of E1A. The results raise an interesting question as whether E1A gene therapy (through its ability to downregulate Twist) might suppress growth of cancer stem cells.
Protein phosphatase 2A (PP2A) is a ubiquitously expressed serine/threonine phosphatase that suppresses tumor growth. The report that PP2A might be a tumor suppressor was based on the observation that okadaic acid, a strong inhibitor of PP2A, was a potent tumor promoter (45). Mutation of PP2A was found in human cancers including breast, colon, and lung cancers and melanoma (46). In this study, we identified PP2A/C as a target of miR-520h, which inhibited PP2A/C protein translation by binding to PP2A/C 3′UTR and suggests that E1A induces PP2A/C expression through an miRNA regulatory mechanism. In addition, we found that overexpression of PP2A/C in various cancer cells inhibited their motility, suggesting a putative role for PP2A/C in the suppression of metastasis. Furthermore, Twist has been identified as the conserved target of NF-κB (47), which is known to be regulated by IKK. The current study suggests that E1A-mediated suppression of IKK/NF-κB signaling may participate in the regulation of Twist.
We have identified Twist and PP2A/C as downstream targets of E1A, suggesting that E1A may act through different mechanisms to inhibit the expression of putative oncogenic miRNAs (miR-520h) and oncogenes. Moreover, we also provided a new insight into the regulation of miRNA expression by E1A and identified a signal cascade involving PP2A/C, IKK, NF-κB and Twist that are regulated by the E1A-mediated miR520h downregulation. Based on our results shown here, it is likely that this signaling cascade plays a role in the E1A associated tumor/metastasis suppression activities.
Supplementary Material
Acknowledgments
We would like to thank Drs. Jennifer Hsu and Stephanie Miller and the Office of Scientific Publications for their careful editing. We thank RNAi Core (Academia Sinica, Taiwan) for providing specific siRNAs. This work was partially supported by grants Breast SPORE CA116199, Ovarian SPORE CA83639, DOD COE W81WXH-06-2-0033, National Breast Cancer Foundation, Inc, NSC-96-3111-B National Science Council, Taiwan, and by China Medical University and Hospital-M. D. Anderson Cancer Center Sister Institution Fund to M.-C.H.; by National Science Council grant NSC 96-2320-B-004-MY2, NSC 97-2320-B-039-039-MY3, NSC 98-2815-C-039-082-B; a Taiwan Merit Scholarship TMS-094-2-B-023 from National Science Council of Taiwan; by National Health Research Institutes grant from Taiwan (NHRI-EX97-9712BC, NHRI-EX97-9712BC); by Department of Health, Executive Yuan grant from Taiwan (DOH97-TD-G111-024); by grants from China Medical University (CMU96-220, CMU96-189 and CMU97-277) and an Odyssey Scholarship from M. D. Anderson Cancer Center to J.-L.S. In memoriam, Mr. Tiong Loi Ang for his courageous fight against cancer
References
- 1.Nguyen D, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nature medicine. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Ravdin PM, Cronin KA, Howlader N, Berg CD, Chlebowski RT, Feuer EJ, Edwards BK, Berry DA. The decrease in breast-cancer incidence in 2003 in the United States. The New England journal of medicine. 2007 Apr 19;356(16):1670–4. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 4.Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–6. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 5.Byrd P, Grand R, Gallimore P. Differential transformation of primary human embryo retinal cells by adenovirus E1 regions and combinations of E1A + ras. Oncogene. 1988;3:477–84. [PubMed] [Google Scholar]
- 6.Yan DH, Shao R, Hung MC. E1A cancer gene therapy. In: Panner C, editor. Gene therapy of cancer. 2. Academic Press; San Diego, CA: 2001. pp. 465–77. [Google Scholar]
- 7.Yu D, Scorsone K, Hung M-C. The adenovirus-5 E1A gene products act as a transformation suppressor of neu oncogene. Mol Cell Biol. 1991;11:1745–50. doi: 10.1128/mcb.11.3.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueno NT, Yu D, Hung MC. E1A: tumor suppressor or oncogene? Preclinical and clinical investigations of E1A gene therapy. Breast Cancer. 2001;8:285–93. doi: 10.1007/BF02967526. [DOI] [PubMed] [Google Scholar]
- 9.Frisch SM, Mymryk JS. ADENOVIRUS-5 E1A: PARADOX AND PARADIGM. Nat Rev Mol Cell Biol. 2002;3:441–52. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 10.Madhusudan S, Tamir A, Bates N, Flanagan E, Gore ME, Barton DP, Harper P, Seckl M, Thomas H, Lemoine NR, Charnock M, Habib NA, Lechler R, Nicholls J, Pignatelli M, Ganesan TS. A multicenter Phase I gene therapy clinical trial involving intraperitoneal administration of E1A/lipid complex in patients with recurrent epithelial ovarian cancer overexpressing HER-2/neu oncogene. Clin Cancer Res. 2004;10:2986–96. doi: 10.1158/1078-0432.ccr-03-0291. [DOI] [PubMed] [Google Scholar]
- 11.Yoo GH, Hung M-C, Lopez-Berestein G, LaFollete S, Ensley JF, Carey M, Baston E, Reynolds TC, Murray JL. Phase I of intratumoral liposome E1A gene therapy in patients with recurrent breast and head and neck cancer. Clinc Cancer Res. 2001;7:1237–45. [PubMed] [Google Scholar]
- 12.Hung MC, Hortobagyi GN, Ueno NT. Development of clinical trial of E1A gene therapy targeting HER-2/neuoverexpressing breast and ovarian cancer. Adv Exp Med Biol. 2000;465:171–80. doi: 10.1007/0-306-46817-4_16. [DOI] [PubMed] [Google Scholar]
- 13.Frisch S, Reich R, Collier IE, Genrich LT, Maring G, Goldberg GI. Adenovirus E1A represses protease gene expression and inhibits metastasis of human cancer cells. Oncogene. 1990;5:75–83. [PubMed] [Google Scholar]
- 14.Yu D, Hamada J, Zhang H, Nicolson GL, Hung MC. Mechanisms of c-erbB2/neu oncogene-induced metastasis and repression of metastatic properties by adenovirus 5 E1A gene products. Oncogene. 1992;7:2263–70. [PubMed] [Google Scholar]
- 15.Thiery JP. Epithelial-mesenchymal transitions in tumor progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 16.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–66. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 17.Yang Ja W, RA Epithelial-mesenchymal transtion: at the crossroads of development and tumor metastasis. Developmental Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 19.Slack AE-KaFJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 20.Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–5. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosine, indicating that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Caldas CB, JD Sizing up miRNAs as cancer genes. Nature Med. 2005;11:712–4. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 24.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri SMS, Rattan S, Bullrich F, Negrini M, Croce aCM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 27.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 28.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008:451. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song CZ, Loewenstein PM, Toth K, Green M. Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc Natl Acad Sci USA. 1995;92:10330–3. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Hung MC. Involvement of co-activator p300 in the transcriptional regulation of the HER-2/neu gene. J Biol Chem. 1997;272:6101–4. doi: 10.1074/jbc.272.10.6101. [DOI] [PubMed] [Google Scholar]
- 31.Geisberg JV, Chen JL, Ricciardi RP. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol Cell Biol. 1995;15:6283–90. doi: 10.1128/mcb.15.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazzarelli JM, Atkins GB, Geisberg JV, Ricciardi RP. The viral oncoproteins Ad5 E1A, HPV16 E7 and SV40 TAg bind a common region of the TBPassociated factor-110. Oncogene. 1995;11:1859–64. [PubMed] [Google Scholar]
- 33.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 34.Liao Y, Hung MC. A new role of protein phosphatase 2A in adenoviral E1A protein-mediated sensitization to anticancer drug-induced apoptosis in human breast cancer cells. Cancer Res. 2004;64:5938–5942. doi: 10.1158/0008-5472.CAN-04-1533. [DOI] [PubMed] [Google Scholar]
- 35.Palkowitsch L, Leidner J, Ghosh S, Marienfeld RB. Phosphorylation of serine 68 in the IkappaB kinase (IKK)-binding domain of NEMO interferes with the structure of the IKK complex and tumor necrosis factor-alpha-induced NF-kappaB activity. J Biol Chem. 2008;283:76–86. doi: 10.1074/jbc.M708856200. [DOI] [PubMed] [Google Scholar]
- 36.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–92. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 37.Lopez JI, Camenisch TD, Stevens MV, Sands BJ, McDonald J, Schroeder JA. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005;65:6755–63. doi: 10.1158/0008-5472.CAN-05-0863. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of Micro RNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–8. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 45.Cohen PaC, PTW Protien phosphatases come of age. J Biol Chem. 1989;264:21435–8. [PubMed] [Google Scholar]
- 46.Schonthal AH. Role of serine/threonine protein phosphatase 2A in cancer. Cancer Lett. 2001;170:1–13. doi: 10.1016/s0304-3835(01)00561-4. [DOI] [PubMed] [Google Scholar]
- 47.Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, Franzoso G. Upregulation of Twist-1 by NF-kappaB blocks cytotoxicity induced by chemotherapeutic drugs. Mol Cell Biol. 2007;27:3920–35. doi: 10.1128/MCB.01219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu DWJ, Scanlon M, Price JE, Hung MC. Enhanced c-erbB-2/neu expression in human ovarian cancer cells correlates with more severe malignancy that can be suppressed by E1A. Cancer Res. 1993;53:891–8. [PubMed] [Google Scholar]
- 49.Yang MH, Wu KJ. TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle. 2008;7(14):2090–6. doi: 10.4161/cc.7.14.6324. [DOI] [PubMed] [Google Scholar]
- 50.Chen CRD, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.