Abstract
Puberty is an important developmental stage during which reproductive capacity is attained. The timing of puberty varies greatly among healthy individuals in the general population and is influenced by both genetic and environmental factors. Although genetic variation is known to influence the normal spectrum of pubertal timing, the specific genes involved remain largely unknown. Genetic analyses have identified a number of genes responsible for rare disorders of pubertal timing such as hypogonadotropic hypogonadism and Kallmann syndrome. Recently, the first loci with common variation reproducibly associated with population variation in the timing of puberty were identified at 6q21 in or near LIN28B and at 9q31.2. However, these two loci explain only a small fraction of the genetic contribution to population variation in pubertal timing, suggesting the need to continue to consider other loci and other types of variants. Here we provide an update of the genes implicated in disorders of puberty, discuss genes and pathways that may be involved in the timing of normal puberty, and suggest additional avenues of investigation to identify genetic regulators of puberty in the general population.
Keywords: puberty, pubertal timing, genetics, hypogonadotropic hypgonadism, Kallmann syndrome, genetic regulation
Introduction
The timing of puberty varies greatly in the general population and is influenced by both genetic and environmental factors [reviewed in [1–6]]. The high correlation of the onset of puberty seen within racial/ethnic groups, within families, and between monozygotic compared to dizygotic twins all provide evidence for genetic regulation of pubertal timing. Taken together, these data suggest that 50–80% of the variation in pubertal timing is determined by genetic factors [1,3–10]. Environmental and physiologic effects also influence the timing of puberty, and there is evidence supporting secular trends in the timing of puberty [1,11–13]. It is possible that gene by environment interactions play an important role in regulating the timing of puberty. However, despite changing environmental and secular influences, genetic background still plays a significant role in regulating the variation of pubertal timing within a population at any particular point in time. Thus, while acknowledging the importance of environmental factors, in this review we highlight the use of genetic methodologies to investigate the regulation of pubertal timing.
Much progress has been made in identifying genetic causes of disorders of puberty, such as hypogonadotropic hypogonadism (HH) and Kallmann syndrome (KS), but the specific genetic factors that regulate the variation in pubertal timing in the general population are just emerging. The identification of these genes has been difficult because pubertal timing is a complex genetic trait, where a direct, often one-to-one relationship between genotype and phenotype does not exist [14], likely due to multigenic influences and interactions between genetic variants and environmental exposures [15].
Insights from Single Gene Disorders
Investigation of HH and KS has led to the identification of many genes that play critical roles in the development and regulation of the hypothalamic pituitary gonadal (HPG) axis (reviewed in [4,16–20]). For example, this work has defined roles for the genes that lead to HH (GNRHR, GNRH1, GPR54, FGFR1, FGF8, PROK2, PROKR2, TAC3, TACR3, and CHD7), to X-linked (KAL1) and autosomal (FGFR1, PROK2, PROKR2, FGF8, and CHD7) forms of KS, to obesity and HH (LEP, LEPR, and PC1), and to abnormal HPG development (DAX1, SF-1, HESX-1, LHX3, and PROP-1).
Genetic causes of other disorders of pubertal development, such as precocious puberty, have been previously reviewed [18,21]. In this review, we focus on discussion of genes that are hypothesized to regulate pubertal timing at the hypothalamic level.
Normosmic hypogonadotropic hypogonadism
HH with normal olfaction has been primarily associated with mutations in the genes for the gonadotropin-releasing hormone receptor (GNRHR) and the G-protein coupled receptor 54 (GPR54), the G-protein coupled receptor for kisspeptins (the products of KISS1) [22–30]. Estimates of the frequency of GNRHR mutations in normosmic HH range from 3.5–10.4% [31,32]. Recently, mutations in GNRH1 were also identified in patients with normosmic HH [33,34]. A case of constitutional delay of growth and puberty (CDGP) was reportedly associated with a homozygous partial loss of function mutation in GNRHR [35], and pedigrees of probands with HH can include individuals with delayed but otherwise normal puberty. However, more extensive analyses suggest that genetic variation in neither GNRH nor GNRHR is a common cause of late puberty in the general population [36,37].
Research into the KISS-1/GPR54 system in both animal and human studies has identified it as a critical component of the HPG axis, necessary for pubertal onset. The first indications of the importance of the KISS-1/GPR54 signaling complex as a regulator of the HPG axis came in 2003 when two independent groups reported deletions and inactivating mutations of GPR54 in patients with HH [23,25]. Subsequently, activating mutations in this pathway were associated with precocious puberty in a report of central precocious puberty in the case of a female with an autosomal dominant mutation in GPR54 [38]. Thus, it is clear that activation of GPR54 by kisspeptins plays a pivotal role in the onset of puberty. It is not yet known, however, whether the KISS-1/GPR54 system is the initial trigger of puberty or whether it acts as a downstream effector of other yet to be identified regulatory factors [39,40]. Recently, mutations in TAC3 and TACR3 were identified in HH patients [41]. These genes encode neurokinin B and its receptor, which are highly expressed in the same neurons that express kisspeptin, further emphasizing the role of kisspeptin in the regulation of pubertal timing.
Kallmann syndrome
Several genes critical to HPG axis function and olfactory development have been identified through investigation of Kallmann syndrome (hypogonadotropic hypogonadism with anosmia/hyposmia). Mutations in Kallmann syndrome 1 (KAL-1) [42–44] and fibroblast growth factor receptor 1 (FGFR1) [45] have been implicated in the X-linked and autosomal dominant forms of the disease, respectively, but appear to account for only approximately 20% of patients with KS [46]. Recently, mutations in the prokineticin receptor-2 gene (PROKR2), a G-protein coupled receptor, and in its ligand prokinetcin-2 (PROK2) were identified in a cohort of KS patients [46], demonstrating that prokineticin signaling is important for olfactory and HPG axis development. One of the patients in this series was heterozygous for both a PROKR2 mutation and a KAL1 mutation, suggesting a possible digenic mode of inheritance [46]. Finally, mutations in the nasal embryonic luteinizing hormone releasing hormone factor (NELF), which plays a role in migration of GnRH neurons and olfactory axon outgrowth [47], have also been implicated in the pathogenesis of KS [48]. A heterozygous deletion in NELF has been reported as a component, along with FGFR1, of digenic KS, but it is not clear whether mutations in NELF alone lead to KS [49].
The distinction among the different abnormalities of pubertal development may not be absolute. For example, mutations in FGFR1 can cause both KS and HH [50], and a homozygous mutation in PROK2 has been reported to cause both KS and HH within a single kindred [51]. A more comprehensive study of PROK2 and PROKR2 in HH and KS patients found mutations in both genes distributed in both groups of patients [52]. Mutations in the FGF8 gene, which encodes a ligand for FGFR1, have been observed in HH patients accompanied by variable olfactory phenotypes [53]. Recently, mutations in CHD7, a gene responsible for CHARGE syndrome, which shares some developmental features with KS, were identified in patients with both normosmic HH and KS [54,55]. Moreover, it has been reported that loss of function mutations in FGFR1 can cause delayed puberty in members of HH pedigrees [50,56], although variation in FGFR1 does not appear to be a major cause of CDGP [37]. Cases of reversible HH have also been reported [57], further blurring the distinction between HH and CDGP.
Leptin and other genes
HH (accompanied by obesity) can also result from defects in the leptin (LEP) or the leptin receptor (LEPR) genes, highlighting the importance of nutrition in modulating the HPG axis. Leptin appears to act as a permissive factor in pubertal maturation [5]. However, recent association studies have found no substantial association between common polymorphisms in LEP and LEPR and CDGP or age at menarche in the general population [37,58].
Other causes of HH include mutations in genes that are critical to HPG development. This category includes the genes for the orphan nuclear receptors DAX1 (dosage-sensitive sex reversal adrenal hypoplasia critical (DSS-AHC) region on the X chromosome, gene 1) and steroidogenic factor-1 (SF-1). Mutations in several pituitary transcription factors, including HESX-1, LHX3, and PROP1, can lead to combined pituitary hormone deficiencies that include HH as a phenotype. Finally, the gene for prohormone convertase-1 (PC-1) has been associated with obesity and HH, possibly as a result of defective processing of neuropeptides or prohormones that are components of GnRH secretion [20,59].
Genetic Variation in Normal Puberty
Candidate gene-based association studies
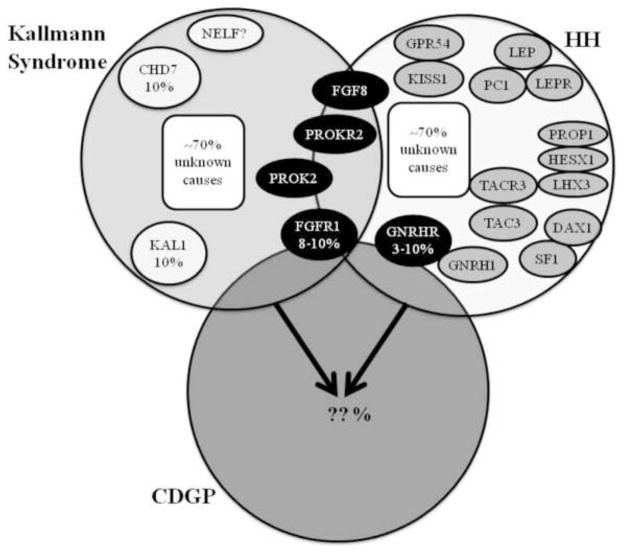
The most commonly used approach for identifying the variants that affect the timing of puberty in the general population has been candidate gene-based association studies. Such studies typically involve genotyping a panel of common variants, usually single nucleotide polymorphisms (SNPs), and determining if the frequency of any of the variants is correlated in a statistically significant manner with phenotype. Because it is clear that the information gained from the study of HH and KS is critical to our understanding of the reproductive-endocrine axis, the genes and pathways identified in these disorders provide attractive candidate genes for association studies investigating variation in pubertal timing within the general population. In many instances, SNPs in the same genes that cause monogenic forms of a phenotype or disease have been associated with the corresponding common, complex trait, and it is possible that this could be the case for puberty as well [60–64]. A substantial fraction of loci associated with traits such as obesity, height, type 2 diabetes, and lipid levels include genes that are mutated in related monogenic disorders [65]. Other disorders of pubertal timing such as CDGP, which likely represents the extreme end of normal pubertal timing, and idiopathic central precocious puberty have significant genetic components as well and may represent additional sources of candidate genes as more genes that cause these conditions are identified [66–72], [73]. This paradigm is illustrated in Figure 1, which depicts what is known and unknown about the overlap among the genetic causes of the different categories of delayed puberty, highlighting the need to study HH genes in the general population.
Figure 1. Genetic basis of delayed puberty.
A paradigm for understanding the genetics of puberty is shown. Some genes underlying the pathogenesis of Kallmann Syndrome (KS) and hypogonadotropic hypogonadism (HH) have been identified and are depicted with each diagnosis; less is known about the genetic basis of Constitutional Delay of Growth and Puberty (CDGP). There is overlap between the three clinical entities as illustrated by the overlapping circles. There is also likely overlap in their genetic bases, as has been reported for FGFR1 (in KS, HH, and CDGP), GNRHR (in HH and possibly CDGP), PROK2 (in KS and HH), PROKR2 (in KS and HH), FGF8 (in KS and HH), and CHD7 (in KS and HH). Thus far, mutations in only GNRHR and FGFR1 have been found in cases of delayed puberty who are members of families with HH or KS but who themselves have no features of the more severe disorders [35,49,50]. Whether genetic variation in these genes plays a role in modulating pubertal timing in the general population (outside of families with HH or KS) is not clear [36,37] and has yet to be proven experimentally. In both KS and HH, approximately 70% of the genetic causes are still unknown. (Please see text for a more detailed discussion of these genes and their roles in these disorders.)
Figure modified with permission from Kaminski BA and Palmert MR. Human Puberty: Physiology, Progression, and Genetic Regulat ion of Variation in Onset. In: D Pfaff, A Arnold, A Etgen, S Fahrbach, and R Rubin, editors. Hormones, Brain, and Behavior (2nd ed). Elsevier Science (USA), San Diego, CA. Copyright Elsevier 2008.
In a recent study, we used association studies to test for associations between common variants in ten HH-related genes (GNRH, GNRHR, GPR54, KISS1, LEP, LEPR, FGFR1, KAL1, PROK2, and PROKR2) and age at menarche. However, only nominally significant associations between SNPs in several of the genes and age at menarche were identified, indicating that genetic variation in these ten genes does not appear to be a substantial modulator of pubertal timing in the general population [37]. Other work has also shown no evidence for substantial association between SNPs in GNRH and GNRHR [36] or LEP and LEPR [58] and alterations in pubertal timing. Variation near these genes but outside the regions typically studied in candidate gene association studies could still influence population variation in the timing of puberty, as is the case for a common variant near MC4R and obesity [74]. It is also possible that variants with low effect sizes in or near these genes could play a role in regulating pubertal timing, although such variants would be unlikely to explain much of the observed phenotypic variation and would not negate the need to look elsewhere for variants that influence the timing of puberty. More likely, though, is that unless other genes, or combinations of genes, in these pathways modulate the timing of puberty in the general population, new regulators need to be identified and studied [39,40].
The need to identify new genes and pathways is perhaps not surprising since mutations in known genes are only responsible for about 30% of the cases of HH and KS [18,49]. Indeed, although there are notable exceptions, most of the genes that have recently been identified through genome-wide association studies as being associated with various complex traits have not been prior candidate genes for the phenotypes in question.
Despite their promise, there are limitations to the approach taken in traditional association studies. Such studies have typically involved relatively small numbers of individuals and may lack the power to identify variants with small effect sizes such as those that have been identified through genome-wide association (GWA) studies [75], particularly when large sample sizes are achieved through collaboration and meta-analysis. Candidate gene association studies also examine only those regions of the genome that directly abut the gene in question, but variants associated with phenotypes may lie outside those regions [74].
Linkage analysis
One approach that does not rely upon candidate genes and that interrogates the whole genome is linkage analysis. These studies are designed to identify regions of the genome [quantitative trait loci (QTLs)] that harbor genes that modulate a specific trait. Linkage studies do not require a priori assumptions about causative genes or pathways and may, therefore, lead to discovery of novel regulatory genes. Several recent studies have used linkage to investigate the genetic basis for variation in age at menarche (AAM) in human populations [76–79], but the results have been somewhat disappointing. As can be seen from inspection of Table 1, none of the findings from the individual studies has been independently replicated, and each study describes QTLs at different genomic locations. Thus, it is possible that the results represent false positives or false negatives and replication is needed before one can conclude that linkage analyses have identified areas of the genome that modulate AAM. The lack of a statistically significant finding in a recent large linkage study [76] that involved more than 13,000 individuals and almost 5,000 sister-pairs suggests that no single QTL explains a large proportion of the variance in AAM.
Table 1. Quantitative Trait Loci (QTLs) associated with age at menarche (AAM) in linkage analyses.
Compiled data from several studies indicating significant QTLs (with LOD scores) associated with age at menarche. Those results with evidence of statistically significant linkage are shown in bold; areas with only suggestive linkage are in normal text. In Rothenbuler et al. the analyses for AAM found only nominally significant QTLs; however, when AAM was adjusted for weight standard deviation score (weight SDS) at menarche, several significant QTLs were identified. The bottom row represents data from mice showing a significant linkage with timing of vaginal opening (VO), a marker of pubertal onset in mice, on the region of mouse chromosome 6 that correlates with human chromosome 12p11–12. Table modified with permission from Kaminski BA and Palmert MR. Human Puberty: Physiology, Progression, and Genetic Regulation of Variation in Onset. In: D Pfaff, A Arnold, A Etgen, S Fahrbach, and R Rubin, editors. Hormones, Brain, and Behavior (2nd ed). Elsevier Science (USA), San Diego, CA. Copyright Elsevier 2008.
| Human Chromosome | ||||||||
|---|---|---|---|---|---|---|---|---|
| Trait (Reference) | 1 | 4 | 6 | 8 | 12 | 16 | 18 | 22 |
| AAM (Guo et al. 2006) | 22q11 (2.68) 22q13 (3.70) |
|||||||
| AAM (Long et al. 2005) | 6q25.3 (2.01) | |||||||
| AAM (Rothenbuhler et al. 2006) | ||||||||
| AAM/weight SDS (Rothenbuhler et al. 2006) | 8p12 (2.18) |
16q12 (3.12) 16q21 (3.33) 16q21 (2.46) |
||||||
| AAM (Anderson et al. 2008) | 1 (2.4), UK only | 4 (2.0), UK only | 12q (2.0) | 18 (3.2), UK only | ||||
| Age at VO (Nathan et al. 2006) | 12p11- 12 | |||||||
As a corollary to human studies, investigation using animal models may suggest new lines of investigation for human studies. As one example, a combination of linkage analysis and generation of congenic mouse strains has led to the identification and validation of a statistically significant QTL associated with timing of vaginal opening (a phenotypic marker of puberty in mice) on mouse chromosome 6 in a region that corresponds to human chromosome 12p11–12 [80]. This and other studies [40,81] may eventually provide important clues as to new candidate genes/pathways to investigate in humans.
Genome-wide association (GWA) studies
Over the past two years, great progress has been made using GWA studies to identify genes that affect susceptibility to common diseases and that modulate complex trait phenotypes, such as obesity and height [74,82–86]. GWA studies are particularly powerful because they query SNPs throughout the genome (often testing for association between a particular disease/phenotype and as many as 900,000 SNPs) and because collectively GWA studies have been performed in populations of tens or even hundreds of thousands of individuals. Therefore, these large, often highly collaborative, studies have sufficient power to detect genetic variants with relatively small effects on disease susceptibility/trait phenotype.
Results from the first GWA studies for AAM have now been published, with confirmed associations at two loci, at 6q21 (in or near the LIN28B gene) and at 9q31.2 [87–90]. These studies involved between 17,000 and 25,000 individuals all of European descent. In each case, AAM was analyzed but in one study [87] additional phenotypes (breast development in girls, voice breaking and pubic hair development in boys, and tempo of height growth in both boys and girls) were found to associate with variants in LIN28B, suggesting that control of pubertal timing in boys and girls shares some common elements. One study found that the signal at LIN28B could be split into two haplotypes, suggesting that either multiple variants may associate with AAM at this locus or that a SNP that has not yet been tested for association may represent the true association signal [88]. Effect sizes were estimated at approximately 1.2 months earlier per effect allele for LIN28B [87,90] and 9q31.2 [90]. The two loci together are estimated to explain 0.6% of the variation in AAM [89]. None of the recent GWA studies observed the previously reported association at the SPOCK locus [91], suggesting that that reported association may represent a false positive.
It is important to note that although these studies and many of the previously described studies have primarily used recalled AAM for analyses and that recalled AAM is highly correlated with actual AAM [92,93], even when recalled as many as 30 years later [93]. Categorizing AAM and/or using the extremes can both improve the accuracy of recall [93,94]. However, AAM is only one of several events that comprise puberty, and other data should also be used when available. One of the recent large GWA studies found that the same allele that was associated with earlier AAM was also associated with earlier breast development [87], supporting the known correlation between breast development and AAM [95]. Should large cohorts become available with phenotype data for other puberty-related events, they may provide additional insights into the genetics of the timing of puberty and help to determine if different genes modulate different aspects of pubertal development (i.e, adrenarche, gonadarche, menarche) [95].
Variation at LIN28B has also been associated with adult height, although the effect at this locus on AAM does not appear to be mediated in a simple manner through its effect on adult height [88]. One study found that the allele associated with earlier age at menarche in was associated with reduced adult height in the same samples [90]. More likely, this result demonstrates that the same genetic pathway can regulate both phenotypes. LIN28B has two distinct isoforms [96] and encodes a regulator of the let-7 class of microRNAs [97]. LIN28B is thought to play an important role in both cell pluripotency and cancer [97], but how this regulatory system modulates growth and the timing of puberty is unknown. The identification of this locus as a regulator of the timing of puberty will likely lead to new biology new understanding about how microRNAs regulate human developmental processes.
The biology behind the locus at 9q31.2 also remains unknown. The associated SNPs lie in an intergenic region with no obvious candidate genes nearby. The closest gene is TMEM38B, a transmembrane protein gene, which lies approximately 400kb away from the signal at 9q31.2 [90].
Because AAM has been correlated with both height [98] and body mass index (BMI) [99], and because of the LIN28B effects on both height and AAM, several of the recent GWA studies also looked for association of AAM with the known height and BMI loci. One study saw no association between height loci other than LIN28B and AAM [88], although another study found that some height loci were nominally significant for association with AAM [90]. Both studies found that many of the BMI loci were associated with AAM, with the BMI increasing allele associated with earlier AAM [88,90]. This finding suggests that increased BMI in childhood may directly decrease AAM, which is an interesting finding given the observed link in population studies between the age at puberty and BMI [99]. An important area of future research will involve investigating the extent to which genes that regulate growth, genes that regulate sex steroid production and response, and genes that regulate the central portion of the HPG axis are distinct or overlapping. As discussed above, there are already some potential hints of overlap, at LIN28b (puberty and height) and at several loci previously reported to be associated with BMI (puberty and BMI).
It is a common story in the investigation of many common diseases and complex traits that great progress has derived from the study of monogenic diseases but that less progress has derived from candidate gene approaches and from linkage analysis. In these examples, as with the timing of puberty, GWA studies have been needed to make substantial progress in identifying common genetic variants that modulate disease susceptibility/trait phenotype within the general population. It is also a common experience that the effect sizes of many of the associations identified in GWA studies are relatively small [100]. However, the small effect size does not negate the importance of the discovery. Findings from GWA studies have highlighted biological pathways involved in a variety of phenotypes, both through “rediscovery” of genes known to be important (for example, 11 of 23 associations for lipid traits are in or near key lipid metabolism proteins) and through identifying previously unsuspected pathways (for example, height and chromatin or Crohn’s disease and autophagy) [65].
Other approaches
Although the first genetic associations with the timing of puberty are now beginning to emerge, they, as discussed, represent variants with small effect sizes, consistent with findings for other complex traits such as height and obesity [75,101]. This suggests that to understand more fully the genetic variation that influences the timing of puberty, other approaches will be necessary.
Meta-analysis
The first of these will likely involve a meta-analyis of the GWA study data published by the four independent research groups. Such an analysis has the potential to involve more than 100,000 women and will, undoubtedly, identify additional loci that regulate the timing of puberty. However, it is likely that each of these loci has a smaller effect than the loci at LIN28B and at 9q32.1. Thus, even after the meta-analysis is performed, additional strategies will be needed to identify the genetic factors that explain 50–80% of the variation in pubertal timing among the general population.
Sequencing
If common variants can explain only a small portion of genetic heritability, it is possible that rare variants explain some of the variation as well. The common variant/common disease hypothesis [102,103] would suggest that common sequence variants (such as SNPs, generally present in ≥ 1% of the population), each exerting a relatively small effect on the phenotype, act in combination to influence the timing of puberty. An alternative, although not mutually exclusive, hypothesis is that larger numbers of rare variants, each present in perhaps ≤ 1 % of the population, exert modest to large individual effects that collectively explain much of the genetic variance in pubertal timing. Current GWA studies have allowed interrogation of SNPs present at ≥ 5% in the population, but newer versions of GWA studies building on data from the 1000 Genomes Project (www.1000genomes.org) will allow investigation of rarer SNPs. One approach that addresses both of these mechanisms is resequencing of candidate genes to determine if sequence variants are present at different frequencies among individuals with early or late pubertal development. We and others are actively investigating the role that sequence variation in genes responsible for HH and KS plays in modulating pubertal timing. For example, we have resequenced the exonic regions of several candidate genes, including GNRHR, GNRH1, LEP, LEPR, and FGFR1, in populations of approximately 50 individuals with late, but otherwise normal pubertal development [36,37,58]. Thus far, no variants have been identified in these genes that explain the variation in pubertal timing within the general population. However, recent studies that used resequencing to identify variants that control triglyceride levels suggest that several hundred to thousands of subjects/DNA samples may be needed to employ this strategy most effectively [104,105].
Alternative genetic mechanisms
In addition to common and rare variants, future investigation of the genetic regulation of pubertal timing should expand to include non-traditional genetic mechanisms. It is possible, for example, that rare, large copy number variants may play a role in regulation of the onset of puberty as they have in susceptibility to diseases such as schizophrenia and cancer [106]. Epigenetic mechanisms have been shown to regulate the timing of puberty and reproductive function in rodents [107], and this is another mechanism that could modulate the timing of puberty in humans and effect gene by environment interactions.
Population Variation in Pubertal Timing
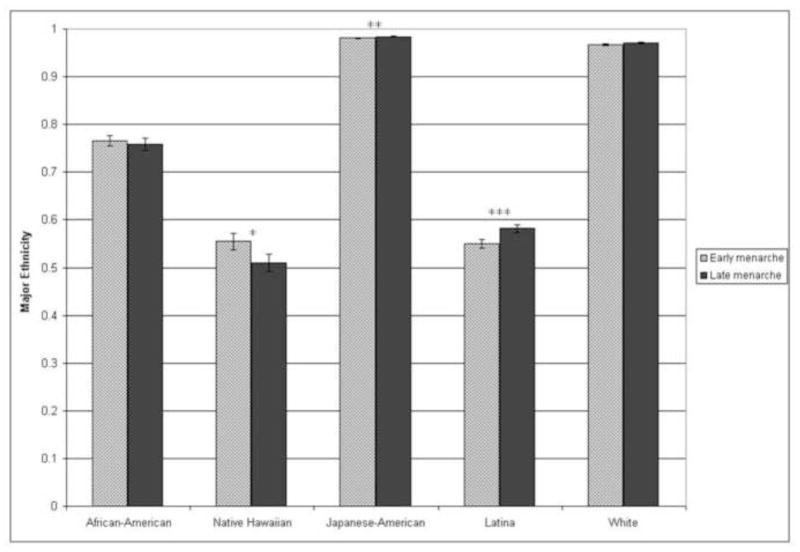
Thus far we have focused our discussion on variation in pubertal timing among individuals, but the timing of the appearance of the different secondary sexual characteristics that typify puberty varies among population groups as well [1]. For example, age at menarche is known to vary among different ethnic groups within the United States [108–114]. Given this, it seems reasonable to hypothesize that there is an association between global genetic ancestry and markers of pubertal timing. To test this hypothesis, we genotyped SNPs that show high variation in frequency in different population groups (ancestry informative markers) to estimate global genetic ancestry and used those estimates to test for an association between genetic ancestry and age at menarche among a sample of participants in the Hawai’i and Los Angeles Multiethnic Cohort. We found significant evidence of association between European ancestry and age at menarche among Latinas, with increased European ancestry and decreased Native American ancestry associated with late menarche, and suggestive evidence of association between Native Hawaiian ancestry and age at menarche in Native Hawaiians, with increased Native Hawaiian ancestry and decreased European and East Asian ancestry associated with early menarche (Figure 2). We did not see any association between estimated genetic ancestry and menarche in whites or African Americans [37]. The effect of estimated global ancestry on age at menarche is small, and it likely does not fully explain the differences observed among racial/ethnic groups. However, further study of the racial/ethnic group-specific factors that modulate the timing of puberty is another strategy that will likely inform further our understanding of the regulation of pubertal timing in the general population.
Figure 2. Genetic ancestry and age at menarche.
Mean ancestry estimates for each self-reported racial/ethnic group separated by early and late menarche. The “major ethnicity” on the y-axis represents the estimated contribution of the ancestry that has the largest contribution to a given self-reported ethnic group. For African-Americans, major ethnicity is estimated West African ancestry; for Native Hawaiians, major ethnicity is estimated Native Hawaiian ancestry; for Japanese-Americans, major ethnicity is estimated East Asian ancestry; for Latinas, major ethnicity is estimated European ancestry; for whites, major ethnicity is estimated European ancestry. ***p<0.01, **p<0.05, *p<0.1. Figure reproduced with permission from Journal of Clinical Endocrinology and Metabolism 93: 4290–4298. Copyright 2008, The Endocrine Society.
Summary
The timing of puberty varies greatly among individuals and much of this variation is due to inherited factors. However, the exact causes and mechanisms that underlie this variation remain largely unknown. There has been much progress in identifying genes underlying reproductive endocrine disorders such as KS and HH, but the genes and variants that influence the normal spectrum of pubertal timing are just beginning to emerge. It is clear that to understand the regulation of pubertal timing in the general population more fully, it will be necessary to look beyond the common variants that have been and will be identified through large-scale GWA studies. The next step will likely be new forms of GWA studies and large-scale sequencing efforts to look for rarer genetic variants than those captured by GWA arrays as well as investigation of other modes of inheritance such as copy number variants and epigenetics. It is likely that as the techniques used to investigate pubertal timing expand our understanding of the regulation of pubertal timing will expand as well.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Zofia K.Z. Gajdos, Program in Genomics and Division of Endocrinology, Children’s Hospital. Boston, Massachusetts 02115; Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115; Program in Medical and Population Genetics, Broad Institute, Cambridge, Massachusetts 02142
Katherine D. Henderson, Department of Population Sciences, Division of Cancer Etiology, City of Hope Comprehensive Cancer Center, 1500 East Duarte Road, Duarte, California 91010
Joel N. Hirschhorn, Program in Genomics and Division of Endocrinology, Children’s Hospital, Boston, Massachusetts 02115; Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115; Program in Medical and Population Genetics, Broad Institute, Cambridge, Massachusetts 02142
Mark R. Palmert, Email: mark.palmert@sickkids.ca, Division of Endocrinology, The Hospital for Sick Children, Department of Paediatrics, The University of Toronto, 555 University Avenue, Toronto, Ontario, M5G 1X8, Canada, Phone: 416-813-6217, Fax: 416-813-6304
References
- 1.Parent AS, Teilmann G, Juul A, Skakkebaek NE, Toppari J, Bourguignon JP. The Timing of Normal Puberty and the Age Limits of Sexual Precocity: Variations around the World, Secular Trends, and Changes after Migration. Endocr Rev. 2003;24:668–693. doi: 10.1210/er.2002-0019. [DOI] [PubMed] [Google Scholar]
- 2.Gajdos ZK, Hirschhorn JN, Palmert MR. What controls the timing of puberty? An update on progress from genetic investigation. Curr Opin Endocrinol Diabetes Obes. 2009;16:16–24. doi: 10.1097/MED.0b013e328320253c. [DOI] [PubMed] [Google Scholar]
- 3.Hodges CA, Palmert MR. Genetic Regulation of the Variation in Pubertal Timing. In: Pescovitz OH, Walvoord EC, editors. When Puberty is Precocious: Scientific and Clinical Aspects. Humana Press, Inc; 2007. pp. 83–102. [Google Scholar]
- 4.Palmert MR, Hirschhorn JN. Genetic approaches to stature, pubertal timing, and other complex traits. Mol Genet Metab. 2003;80:1–10. doi: 10.1016/s1096-7192(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 5.Plant TM, Barker-Gibb ML. Neurobiological mechanisms of puberty in higher primates. Hum Reprod Update. 2004;10:67–77. doi: 10.1093/humupd/dmh001. [DOI] [PubMed] [Google Scholar]
- 6.Towne B, Czerwinski SA, Demerath EW, Blangero J, Roche AF, Siervogel RM. Heritability of age at menarche in girls from the Fels Longitudinal Study. Am J Phys Anthropol. 2005;128:210–219. doi: 10.1002/ajpa.20106. [DOI] [PubMed] [Google Scholar]
- 7.Fischbein S. Onset of puberty in MX and DZ twins. Acta Genet Med Gemellol (Roma) 1977;26:151–158. doi: 10.1017/s0001566000009946. [DOI] [PubMed] [Google Scholar]
- 8.Sklad M. The rate of growth and maturing of twins. Acta Genet Med Gemellol (Roma) 1977;26:221–237. doi: 10.1017/s0001566000009703. [DOI] [PubMed] [Google Scholar]
- 9.Kaprio J, Rimpela A, Winter T, Viken RJ, Rimpela M, Rose RJ. Common genetic influences on BMI and age at menarche. Hum Biol. 1995;67:739–753. [PubMed] [Google Scholar]
- 10.van den Berg SM, Boomsma DI. The Familial Clustering of Age at Menarche in Extended Twin Families. Behav Genet. 2007 doi: 10.1007/s10519-007-9161-4. [DOI] [PubMed] [Google Scholar]
- 11.Parent AS, Rasier G, Gerard A, Heger S, Roth C, Mastronardi C, Jung H, Ojeda SR, Bourguignon JP. Early onset of puberty: tracking genetic and environmental factors. Horm Res. 2005;64 (Suppl 2):41–47. doi: 10.1159/000087753. [DOI] [PubMed] [Google Scholar]
- 12.Demerath EW, Towne B, Chumlea WC, Sun SS, Czerwinski SA, Remsberg KE, Siervogel RM. Recent decline in age at menarche: the Fels Longitudinal Study. Am J Hum Biol. 2004;16:453–457. doi: 10.1002/ajhb.20039. [DOI] [PubMed] [Google Scholar]
- 13.Euling SY, Herman-Giddens ME, Lee PA, Selevan SG, Juul A, Sorensen TI, Dunkel L, Himes JH, Teilmann G, Swan SH. Examination of US puberty-timing data from 1940 to 1994 for secular trends: panel findings. Pediatrics. 2008;121 (Suppl 3):S172–191. doi: 10.1542/peds.2007-1813D. [DOI] [PubMed] [Google Scholar]
- 14.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 15.Palmert MR, Boepple PA. Variation in the timing of puberty: clinical spectrum and genetic investigation. J Clin Endocrinol Metab. 2001;86:2364–2368. doi: 10.1210/jcem.86.6.7603. [DOI] [PubMed] [Google Scholar]
- 16.Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5:569–576. doi: 10.1038/nrendo.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhangoo A, Jacobson-Dickman E. The genetics of idiopathic hypogonadotropic hypogonadism:unraveling the biology of human sexual development. Pediatr Endocrinol Rev. 2009;6:395–404. [PubMed] [Google Scholar]
- 18.Herbison AE. Genetics of puberty. Horm Res. 2007;68 (Suppl 5):75–79. doi: 10.1159/000110583. [DOI] [PubMed] [Google Scholar]
- 19.Kalantaridou SN, Chrousos GP. Clinical review 148: Monogenic disorders of puberty. J Clin Endocrinol Metab. 2002;87:2481–2494. doi: 10.1210/jcem.87.6.8668. [DOI] [PubMed] [Google Scholar]
- 20.Silveira LF, MacColl GS, Bouloux PM. Hypogonadotropic hypogonadism. Semin Reprod Med. 2002;20:327–338. doi: 10.1055/s-2002-36707. [DOI] [PubMed] [Google Scholar]
- 21.Achermann JC, Ozisik G, Meeks JJ, Jameson JL. Genetic causes of human reproductive disease. J Clin Endocrinol Metab. 2002;87:2447–2454. doi: 10.1210/jcem.87.6.8622. [DOI] [PubMed] [Google Scholar]
- 22.de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 23.De Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, Thresher RR, Malinge I, Lomet D, Carlton MB, et al. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc Natl Acad Sci U S A. 2005;102:1761–1766. doi: 10.1073/pnas.0409330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 26.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Increased hypothalamic GPR54 signaling: a potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci U S A. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedecarrats GY, Linher KD, Janovick JA, Beranova M, Kada F, Seminara SB, Michael Conn P, Kaiser UB. Four naturally occurring mutations in the human GnRH receptor affect ligand binding and receptor function. Mol Cell Endocrinol. 2003;205:51–64. doi: 10.1016/s0303-7207(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 28.Bedecarrats GY, Linher KD, Kaiser UB. Two common naturally occurring mutations in the human gonadotropin-releasing hormone (GnRH) receptor have differential effects on gonadotropin gene expression and on GnRH-mediated signal transduction. J Clin Endocrinol Metab. 2003;88:834–843. doi: 10.1210/jc.2002-020806. [DOI] [PubMed] [Google Scholar]
- 29.Wolczynski S, Laudanski P, Jarzabek K, Mittre H, Lagarde JP, Kottler ML. A case of complete hypogonadotropic hypogonadism with a mutation in the gonadotropin-releasing hormone receptor gene. Fertil Steril. 2003;79:442–444. doi: 10.1016/s0015-0282(02)04667-8. [DOI] [PubMed] [Google Scholar]
- 30.Karges B, Karges W, Mine M, Ludwig L, Kuhne R, Milgrom E, de Roux N. Mutation Ala(171)Thr stabilizes the gonadotropin-releasing hormone receptor in its inactive conformation, causing familial hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2003;88:1873–1879. doi: 10.1210/jc.2002-020005. [DOI] [PubMed] [Google Scholar]
- 31.Bhagavath B, Ozata M, Ozdemir IC, Bolu E, Bick DP, Sherins RJ, Layman LC. The prevalence of gonadotropin-releasing hormone receptor mutations in a large cohort of patients with hypogonadotropic hypogonadism. Fertil Steril. 2005;84:951–957. doi: 10.1016/j.fertnstert.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 32.Beranova M, Oliveira LM, Bedecarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, et al. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2001;86:1580–1588. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 33.Chan YM, de Guillebon A, Lang-Muritano M, Plummer L, Cerrato F, Tsiaras S, Gaspert A, Lavoie HB, Wu CH, Crowley WF, Jr, et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2009;106:11703–11708. doi: 10.1073/pnas.0903449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouligand J, Ghervan C, Tello JA, Brailly-Tabard S, Salenave S, Chanson P, Lombes M, Millar RP, Guiochon-Mantel A, Young J. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med. 2009;360:2742–2748. doi: 10.1056/NEJMoa0900136. [DOI] [PubMed] [Google Scholar]
- 35.Lin L, Conway GS, Hill NR, Dattani MT, Hindmarsh PC, Achermann JC. A homozygous R262Q mutation in the gonadotropin-releasing hormone receptor presenting as constitutional delay of growth and puberty with subsequent borderline oligospermia. J Clin Endocrinol Metab. 2006;91:5117–5121. doi: 10.1210/jc.2006-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedlmeyer IL, Pearce CL, Trueman JA, Butler JL, Bersaglieri T, Read AP, Clayton PE, Kolonel LN, Henderson BE, Hirschhorn JN, et al. Determination of Sequence Variation and Haplotype Structure for the Gonadotropin-Releasing Hormone (GnRH) and GnRH Receptor Genes: Investigation of Role in Pubertal Timing. J Clin Endocrinol Metab. 2005;90:1091–1099. doi: 10.1210/jc.2004-0649. [DOI] [PubMed] [Google Scholar]
- 37.Gajdos ZKZ, Butler JL, Henderson KD, He C, Supelak PJ, Egyud M, Price A, Reich D, Clayton PE, Le Marchand L, et al. Association studies of common variants in ten hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab. 2008:jc.2008–0981. doi: 10.1210/jc.2008-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seminara SB. Converging at puberty’s hub. Endocrinology. 2007;148:5145–5146. doi: 10.1210/en.2007-0953. [DOI] [PubMed] [Google Scholar]
- 40.Roth CL, Mastronardi C, Lomniczi A, Wright H, Cabrera R, Mungenast AE, Heger S, Jung H, Dubay C, Ojeda SR. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology. 2007;148:5147–5161. doi: 10.1210/en.2007-0634. [DOI] [PubMed] [Google Scholar]
- 41.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, et al. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal pathfinding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 43.Hardelin JP, Levilliers J, Blanchard S, Carel JC, Leutenegger M, Pinard-Bertelletto JP, Bouloux P, Petit C. Heterogeneity in the mutations responsible for X chromosome-linked Kallmann syndrome. Hum Mol Genet. 1993;2:373–377. doi: 10.1093/hmg/2.4.373. [DOI] [PubMed] [Google Scholar]
- 44.Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 45.Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 46.Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000;14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- 48.Miura K, Acierno JS, Jr, Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH) J Hum Genet. 2004;49:265–268. doi: 10.1007/s10038-004-0137-4. [DOI] [PubMed] [Google Scholar]
- 49.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitteloud N, Acierno JS, Jr, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, Plummer L, Jacobson-Dickman EE, Mellon PL, Zhou QY, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, et al. Mutations in Prokineticin 2 (PROK2) and PROK2 Receptor (PROKR2) in Human Gonadotrophin-Releasing Hormone Deficiency: Molecular Genetics and Clinical Spectrum. J Clin Endocrinol Metab. 2008 doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jongmans MC, van Ravenswaaij-Arts CM, Pitteloud N, Ogata T, Sato N, Claahsen-van der Grinten HL, van der Donk K, Seminara S, Bergman JE, Brunner HG, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome--the clinical overlap with CHARGE syndrome. Clin Genet. 2009;75:65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitteloud N, Acierno JS, Jr, Meysing AU, Dwyer AA, Hayes FJ, Crowley WF., Jr Reversible kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 57.Raivio T, Falardeau J, Dwyer A, Quinton R, Hayes FJ, Hughes VA, Cole LW, Pearce SH, Lee H, Boepple P, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357:863–873. doi: 10.1056/NEJMoa066494. [DOI] [PubMed] [Google Scholar]
- 58.Banerjee I, Trueman JA, Hall CM, Price DA, Patel L, Whatmore AJ, Hirschhorn JN, Read AP, Palmert MR, Clayton PE. Phenotypic variation in constitutional delay of growth and puberty: relationship to specific leptin and leptin receptor gene polymorphisms. Eur J Endocrinol. 2006;155:121–126. doi: 10.1530/eje.1.02184. [DOI] [PubMed] [Google Scholar]
- 59.Jackson RS, Creemers JW, Ohagi S, Raffin-Sanson ML, Sanders L, Montague CT, Hutton JC, O’Rahilly S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 60.Ligon AH, Moore SD, Parisi MA, Mealiffe ME, Harris DJ, Ferguson HL, Quade BJ, Morton CC. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–348. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menten B, Buysse K, Zahir F, Hellemans J, Hamilton SJ, Costa T, Fagerstrom C, Anadiotis G, Kingsbury D, McGillivray BC, et al. Osteopoikilosis, short stature and mental retardation as key features of a new microdeletion syndrome on 12q14. J Med Genet. 2007;44:264–268. doi: 10.1136/jmg.2006.047860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Florez JC, Jablonski KA, Kahn SE, Franks PW, Dabelea D, Hamman RF, Knowler WC, Nathan DM, Altshuler D. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes. 2007;56:531–536. doi: 10.2337/db06-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 65.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 66.Reindollar RH, McDonough PG. Delayed sexual development: common causes and basic clinical approach. Pediatr Ann. 1981;10:30–39. [PubMed] [Google Scholar]
- 67.Wehkalampi K, Silventoinen K, Kaprio J, Dick DM, Rose RJ, Pulkkinen L, Dunkel L. Genetic and environmental influences on pubertal timing assessed by height growth. Am J Hum Biol. 2008;20:417–423. doi: 10.1002/ajhb.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wehkalampi K, Widen E, Laine T, Palotie A, Dunkel L. Patterns of inheritance of constitutional delay of growth and puberty in families of adolescent girls and boys referred to specialist pediatric care. J Clin Endocrinol Metab. 2008;93:723–728. doi: 10.1210/jc.2007-1786. [DOI] [PubMed] [Google Scholar]
- 69.Toublanc JE, Roger M, Chaussain JL. Etiologies of late puberty. Horm Res. 1991;36:136–140. doi: 10.1159/000182147. [DOI] [PubMed] [Google Scholar]
- 70.Sperlich M, Butenandt O, Schwarz HP. Final height and predicted height in boys with untreated constitutional growth delay. Eur J Pediatr. 1995;154:627–632. doi: 10.1007/BF02079065. [DOI] [PubMed] [Google Scholar]
- 71.Sedlmeyer IL, Hirschhorn JN, Palmert MR. Pedigree analysis of constitutional delay of growth and maturation: determination of familial aggregation and inheritance patterns. J Clin Endocrinol Metab. 2002;87:5581–5586. doi: 10.1210/jc.2002-020862. [DOI] [PubMed] [Google Scholar]
- 72.Sedlmeyer IL, Palmert MR. Delayed puberty: analysis of a large case series from an academic center. J Clin Endocrinol Metab. 2002;87:1613–1620. doi: 10.1210/jcem.87.4.8395. [DOI] [PubMed] [Google Scholar]
- 73.de Vries L, Kauschansky A, Shohat M, Phillip M. Familial central precocious puberty suggests autosomal dominant inheritance. J Clin Endocrinol Metab. 2004;89:1794–1800. doi: 10.1210/jc.2003-030361. [DOI] [PubMed] [Google Scholar]
- 74.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 76.Anderson CA, Zhu G, Falchi M, van den Berg SM, Treloar SA, Spector TD, Martin NG, Boomsma DI, Visscher PM, Montgomery GW. A genome-wide linkage scan for age at menarche in three populations of European descent. J Clin Endocrinol Metab. 2008;93:3965–3970. doi: 10.1210/jc.2007-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Long JR, Xu H, Zhao LJ, Liu PY, Shen H, Liu YJ, Xiong DH, Xiao P, Liu YZ, Dvornyk V, et al. The oestrogen receptor alpha gene is linked and/or associated with age of menarche in different ethnic groups. J Med Genet. 2005;42:796–800. doi: 10.1136/jmg.2004.028381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo Y, Shen H, Xiao P, Xiong DH, Yang TL, Guo YF, Long JR, Recker RR, Deng HW. Genomewide linkage scan for quantitative trait loci underlying variation in age at menarche. J Clin Endocrinol Metab. 2006;91:1009–1014. doi: 10.1210/jc.2005-2179. [DOI] [PubMed] [Google Scholar]
- 79.Rothenbuhler A, Fradin D, Heath S, Lefevre H, Bouvattier C, Lathrop M, Bougneres P. Weight-adjusted genome scan analysis for mapping quantitative trait Loci for menarchal age. J Clin Endocrinol Metab. 2006;91:3534–3537. doi: 10.1210/jc.2006-0150. [DOI] [PubMed] [Google Scholar]
- 80.Nathan BM, Hodges CA, Supelak PJ, Burrage LC, Nadeau JH, Palmert MR. A Quantitative Trait Locus on Chromosome 6 Regulates the Onset of Puberty in Mice. Endocrinology. 2006 doi: 10.1210/en.2006-0745. [DOI] [PubMed] [Google Scholar]
- 81.Ojeda SR, Lomniczi A, Mastronardi C, Heger S, Roth C, Parent AS, Matagne V, Mungenast AE. Minireview: the neuroendocrine regulation of puberty: is the time ripe for a systems biology approach? Endocrinology. 2006;147:1166–1174. doi: 10.1210/en.2005-1136. [DOI] [PubMed] [Google Scholar]
- 82.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 83.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson N, Lettre G, Usala G, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007 doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009 doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009 doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 89.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009 doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009 doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu YZ, Guo YF, Wang L, Tan LJ, Liu XG, Pei YF, Yan H, Xiong DH, Deng FY, Yu N, et al. Genome-wide association analyses identify SPOCK as a key novel gene underlying age at menarche. PLoS Genet. 2009;5:e1000420. doi: 10.1371/journal.pgen.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koprowski C, Coates RJ, Bernstein L. Ability of young women to recall past body size and age at menarche. Obes Res. 2001;9:478–485. doi: 10.1038/oby.2001.62. [DOI] [PubMed] [Google Scholar]
- 93.Must A, Phillips SM, Naumova EN, Blum M, Harris S, Dawson-Hughes B, Rand WM. Recall of early menstrual history and menarcheal body size: after 30 years, how well do women remember? Am J Epidemiol. 2002;155:672–679. doi: 10.1093/aje/155.7.672. [DOI] [PubMed] [Google Scholar]
- 94.Cooper R, Blell M, Hardy R, Black S, Pollard TM, Wadsworth ME, Pearce MS, Kuh D. Validity of age at menarche self-reported in adulthood. J Epidemiol Community Health. 2006;60:993–997. doi: 10.1136/jech.2005.043182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van den Berg SM, Setiawan A, Bartels M, Polderman TJ, van der Vaart AW, Boomsma DI. Individual differences in puberty onset in girls: Bayesian estimation of heritabilities and genetic correlations. Behav Genet. 2006;36:261–270. doi: 10.1007/s10519-005-9022-y. [DOI] [PubMed] [Google Scholar]
- 96.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 97.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Onland-Moret NC, Peeters PH, van Gils CH, Clavel-Chapelon F, Key T, Tjonneland A, Trichopoulou A, Kaaks R, Manjer J, Panico S, et al. Age at menarche in relation to adult height: the EPIC study. Am J Epidemiol. 2005;162:623–632. doi: 10.1093/aje/kwi260. [DOI] [PubMed] [Google Scholar]
- 99.Kaplowitz PB. Link between body fat and the timing of puberty. Pediatrics. 2008;121 (Suppl 3):S208–217. doi: 10.1542/peds.2007-1813F. [DOI] [PubMed] [Google Scholar]
- 100.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 101.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 103.Hirschhorn JN. Genetic approaches to studying common diseases and complex traits. Pediatr Res. 2005;57:74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- 104.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Topol EJ, Frazer KA. The resequencing imperative. Nat Genet. 2007;39:439–440. doi: 10.1038/ng0407-439. [DOI] [PubMed] [Google Scholar]
- 106.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–350. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 107.Cameron NM, Shahrokh D, Del Corpo A, Dhir SK, Szyf M, Champagne FA, Meaney MJ. Epigenetic programming of phenotypic variations in reproductive strategies in the rat through maternal care. J Neuroendocrinol. 2008;20:795–801. doi: 10.1111/j.1365-2826.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- 108.Anderson SE, Dallal GE, Must A. Relative weight and race influence average age at menarche: results from two nationally representative surveys of US girls studied 25 years apart. Pediatrics. 2003;111:844–850. doi: 10.1542/peds.111.4.844. [DOI] [PubMed] [Google Scholar]
- 109.Wu T, Mendola P, Buck GM. Ethnic differences in the presence of secondary sex characteristics and menarche among US girls: the Third National Health and Nutrition Examination Survey, 1988–1994. Pediatrics. 2002;110:752–757. doi: 10.1542/peds.110.4.752. [DOI] [PubMed] [Google Scholar]
- 110.Biro FM, Huang B, Crawford PB, Lucky AW, Striegel-Moore R, Barton BA, Daniels S. Pubertal correlates in black and white girls. J Pediatr. 2006;148:234–240. doi: 10.1016/j.jpeds.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 111.Chumlea WC, Schubert CM, Roche AF, Kulin HE, Lee PA, Himes JH, Sun SS. Age at menarche and racial comparisons in US girls. Pediatrics. 2003;111:110–113. doi: 10.1542/peds.111.1.110. [DOI] [PubMed] [Google Scholar]
- 112.Herman-Giddens ME, Kaplowitz PB, Wasserman R. Navigating the recent articles on girls’ puberty in Pediatrics: what do we know and where do we go from here? Pediatrics. 2004;113:911–917. doi: 10.1542/peds.113.4.911. [DOI] [PubMed] [Google Scholar]
- 113.Herman-Giddens ME, Slora EJ, Wasserman RC, Bourdony CJ, Bhapkar MV, Koch GG, Hasemeier CM. Secondary sexual characteristics and menses in young girls seen in office practice: a study from the Pediatric Research in Office Settings network. Pediatrics. 1997;99:505–512. doi: 10.1542/peds.99.4.505. [DOI] [PubMed] [Google Scholar]
- 114.Sun SS, Schubert CM, Chumlea WC, Roche AF, Kulin HE, Lee PA, Himes JH, Ryan AS. National estimates of the timing of sexual maturation and racial differences among US children. Pediatrics. 2002;110:911–919. doi: 10.1542/peds.110.5.911. [DOI] [PubMed] [Google Scholar]