Abstract
Voltage-gated sodium (Na) channels contribute to the regulation of cellular excitability due to their role in the generation and propagation of action potentials. They are composed of a pore-forming α subunit and are modulated by at least two of four distinct β subunits (β1–4). Recent studies have implicated a role for the intracellular domain of β subunits in modulating Na channel gating and trafficking. In β3, the intracellular domain contains a serine residue at position 161 that is replaced by an alanine in β1. In this study, we have probed the functional importance of β3S161 for modulating Na channel gating. Wild-type β3 and point mutations β3S161A or β3S161E were individually co-expressed in HEK 293 cells stably expressing human Nav1.2. WTβ3 expression increased Na current density, shifted steady-state inactivation in a depolarized direction, and accelerated the kinetics of recovery from inactivation of the Na current. Analogous effects were observed with β3S161E co-expression. In contrast, β3S161A abolished the shifts in steady-state inactivation and recovery from inactivation of the Na current, but did increase Na current density. Immunocytochemistry and Western blot experiments demonstrate membrane expression of WTβ3, β3S161E, and β3S161A, suggesting that the differences in Na channel gating were not due to disruptions in β subunit trafficking. These studies suggest that modification of β3S161 may be important in modulating Na-channel gating.
Keywords: Sodium channels, Electrophysiology, Auxiliary β subunits, Site directed mutagenesis, Immunohistochemistry
Introduction
Voltage-gated sodium (Na) channels are responsible for the generation and conduction of action potentials and, thus, are important regulators of cellular excitability [39]. They consist of a pore-forming α subunit of approximately 220–260 kDa of which ten distinct isoforms have been cloned (Nav1.1–Nav1.9, Nax), with varying tissue distribution [1]. While the α subunit is sufficient to induce Na currents in heterologous expression systems, in vivo, it is associated with at least two of four different auxiliary β subunit isoforms, β1–β4 [11, 12, 25, 40]. β subunits are approximately 30–40 kDa in size and are comprised of a large immunoglobin-like extracellular domain at the N-terminus, a single transmembrane alpha helix and a short intracellular segment at the C-terminus [10].
β1 and β3 are structurally similar with 57% sequence identity. They interact with the α subunit in a noncovalent manner and modulate activation, inactivation, and recovery from inactivation gating parameters of Na channels [20, 38]. Structure/function studies have identified the P loop of domain IV of the Na channel α subunit as a likely interaction point for the extracellular domain of β1, accounting for the fine tuning of Na channel activation and inactivation gating [28]. Disruptions in this interaction, as a result of mutations in either the α subunit or the β subunits, are known to result in changes in cellular excitability and the development of cardiac arrhythmias or epileptic seizures [18, 36, 37]. For example, mutations in the extracellular region of β1 have been linked to GEFS+, an inherited form of epilepsy [23] while mutations in the extracellular domain of β3 and β4 have both been linked to cardiac arrhythmias, idiopathic ventricular fibrillation, and Long-QT syndrome, respectively [24, 33].
More recently, an important modulatory interaction between the intracellular domain of the β subunit and the Na channel has been suggested. Yeast-two hybrid experiments have demonstrated an interaction between the C terminus of β1 and β3 and the C terminus of the Nav1.1 α subunit isoform [31]. Disruption of this association, as a result of a mutation in the cytoplasmic domain of Nav1.1, leads to generalized epilepsy [31]. The intracellular segments of the β1 and β3 subunits also have proven important for trafficking of both α and β subunits to the membrane. Complete deletion of the intracellular domain of β3 leads to the retention of the β subunit within the endoplasmic reticulum (ER) [38]. Physical interaction between β3 and the Nav1.8 α subunit has been shown to mask an ER retention signal on the α subunit, leading to an increase in Na channel density [41]. Additionally, a sequence conserved in the intracellular domain of both β1 and β3, YLAI, is a site for ankyrin G recruitment [19, 22]. This sequence fits the consensus for an internalization site potentially recognized by clathrin-coated pit adaptors [25]. Phosphorylation of the tyrosine residue increases Na channel cell surface density by preventing ankyrin G recruitment and subsequent channel internalization [19, 21].
The intracellular domain of β3 has a serine residue at position 161 that is surrounded by a series of basic residues, suggestive of a potential site for phosphorylation. In contrast, β1 has a non-phosphorylatable alanine at this position. In the present study, we have investigated the role of this residue in modulating the gating of Nav1.2, a neuronal Na channel isoform, by generating a phosphorylation mimic (S161E) and a non-phosphorylatable residue (S161A). Our findings suggest that β3S161 is important for the modulation of Nav1.2 inactivation-gating parameters and recovery from inactivation. Our studies suggest that modulation of Na channel gating by β3 may be unique in comparison to β1 due to the functional importance of the S161 residue.
Materials and methods
Cell culture
Human embryonic kidney (HEK) cells stably expressing human Nav1.2 were a kind gift from H.A. Hartmann (University of Maryland) and were grown in Dulbecco's Modified Eagle Medium/F12 medium (Invitrogen, Corp, CA, USA) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 μg/ml), and G418 (500 μg/ml; Sigma, MO, USA). Cells were grown in a humidified atmosphere with 5% CO2 at 37°C.
Transient transfection
Rat β3 was engineered into the vector pEGFP N1 (Clontech) as previously described [38]. The mutations β3S161A and β3S161E were constructed using the QuikChange XL site-directed mutagenesis kit (Stragene), and cells were transiently transfected (10 μg DNA) using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. HEK293 cells were incubated in the transfection mixture for 4–6 h after which the medium was changed to DMEM/F12 containing 20% FBS for a further 15 h. Cells were returned to medium containing 10% FBS for an additional 24 h prior to electrophysiology and immunocytochemistry.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 10 min, washed with PBS, permeabilized for 60 min in PBS blocking solution (PBS-B; 5% fish skin gelatin, 5% serum of the secondary antibody host animal, 0.25% Triton X-100, and 0.65% w/v bovine serum albumin (BSA)), and incubated in PBS-B with primary antibody overnight at 4°C. Cells were then washed with PBS, incubated with PBS-B for 60 min, and incubated in secondary antibody in PBS-B for 45 min. Cells were then washed with PBS, treated with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) nuclei stain for 5 min, and washed for a final time with PBS. They were then viewed on a Zeiss LSM 510 confocal microscope using a 40×1.3 NA oil immersion objective. The primary antibody used was mouse anti-Nav1.2 (NeuroMab, K69/3). The secondary antibody used was goat anti-mouse Alexa 594 (Invitrogen).
Membrane isolation and western blotting
WTβ3, β3S161A, and β3S161E transfected and nontransfected cells, all stably expressing Nav1.2, were harvested and prepared for membrane biotinylation using the EZ-link NHS-SS-biotin cell surface isolation kit according to the manufacturer's instructions (Pierce) using an EDTA-free protease inhibitor cocktail (Roche Applied Sciences). After biotinylation, the surface proteins were selectively precipitated by incubation with avidin beads. SDS-PAGE 4–20% Tris–HCl readymade gels (Biorad) were loaded with 20 μl of sample per well and run at a constant current of 20 mA for ~1.5 h at room temperature. Proteins were transferred to a PVDF membrane (Biorad) at a constant current of 350 mA for 2 h at 4°C. Nonspecific binding was blocked with 5% nonfat dry milk in PBS-Tween 20 overnight at 4°C. Standard western blotting conditions were used to probe for target proteins using the following antibodies and concentrations. An affinity-purified polyclonal rabbit antiserum was raised against the peptide sequence SENKENSVVPVEE, corresponding to residues 178–191 of rat β3[25] (BioGenes GmbH, Berlin), and was used at 1:1,000 for western blotting. Rabbit anti-Pan Nav (Alomone) antibody was used at 1:200 to detect Nav1.2 in HEK 293 cells. Mouse anti-human transferrin receptor (Invitrogen) was used at a concentration of 1:500 as a loading control. Rabbit anti-phosphoserine (Abcam) was used at 1:500. Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse (Sigma) (1:2,000 for pan Nav and 1:5,000 for all other antibodies) were used for visualization. Antibody binding was detected with the ECL western blotting detection system (Pierce) and exposed using Kodak Biomax MS film (Kodak). Exposure was varied to avoid overdevelopment. In some experiments, blots were stripped using Re-Blot Plus solution (Millipore) for 15 min and then blocked in either 5% nonfat dry milk in PBS-Tween 20 for β3 antibody or 5% BSA/0.015% gelatin in PBS-Tween for phosphoserine antibody overnight at 4°C.
Electrophysiology studies
Transfected cells were identified using a fluorescent microscope (Olympus XI70). Na currents were recorded using the whole-cell configuration of the patch clamp recording technique with an Axopatch 200 amplifier (Molecular Devices). All voltage protocols were applied using pCLAMP 9 software (Molecular Devices) and a Digidata 1322A (Molecular Devices). Currents were amplified, low pass filtered (2 kHz), and sampled at 33 kHz. Borosilicate glass pipettes were pulled using a Brown-Flaming puller (model P97, Sutter Instruments) and heat polished to produce electrode resistances of 1.5–2.0 MΩ when filled with the following electrode solution (in mM): CsCl 130, MgCl2 1, MgATP 5, BAPTA 10, HEPES 5 (pH adjusted to 7.2 with CsOH). Cells were plated on glass coverslips and superfused with a solution containing (in mM): NaCl 130, KCl 4, CaCl2 1, MgCl2 5, HEPES 5, and glucose 5 (pH adjusted to 7.4 with NaOH). All Na channel current experiments were performed at room temperature (20–22°C). After establishing whole-cell patch, a minimum series resistance compensation of 75% was applied, and cells were held at –100 mV for 2–3 min to account for any equilibrium-gating shifts. Capacitive and leak currents were corrected for using standard P/4 protocols except during steady-state inactivation and use-dependent block protocols.
Conductance as a function of voltage was derived from the current–voltage relationship using the equation:
where V is the test potential and ENa is the reversal potential. The voltage dependence of activation and steady state inactivation at 1,000 ms, and 10-s prepulse data were fit by a single Boltzmann equation:
where y is the normalized conductance (g/gmax) or the normalized current for activation and inactivation, respectively, V1/2 is voltage of half-maximal activation or inactivation, and k is the slope factor. Time constants for recovery from inactivation were obtained using a triple exponential function:
where A1, A2, and A3 are the coefficients for the phases of exponential growth, t is time (ms), and τ1, τ2, τ3 are the time constants. The percentage of the current represented by the fast time constant was calculated from the equation:
Time constants for decay of the macroscopic current were obtained using a single-exponential function:
where A1 is the coefficient for the exponential, t is time (ms), and τ1 is the time constant.
Data analysis
Electrophysiology data analysis was performed using Clampfit software (v9, Molecular Devices) and Origin (v6, OriginLab Corp). Statistical analyses were performed using the standard one-way ANOVA followed by Tukey's or Dunn's post hoc test (SigmaStat, SPSS Inc.). Averaged data are presented as means ± standard error of the mean (SEM). Statistical significance was set at P<0.05.
Results
β-subunit co-expression increases current density without modulating activation parameters
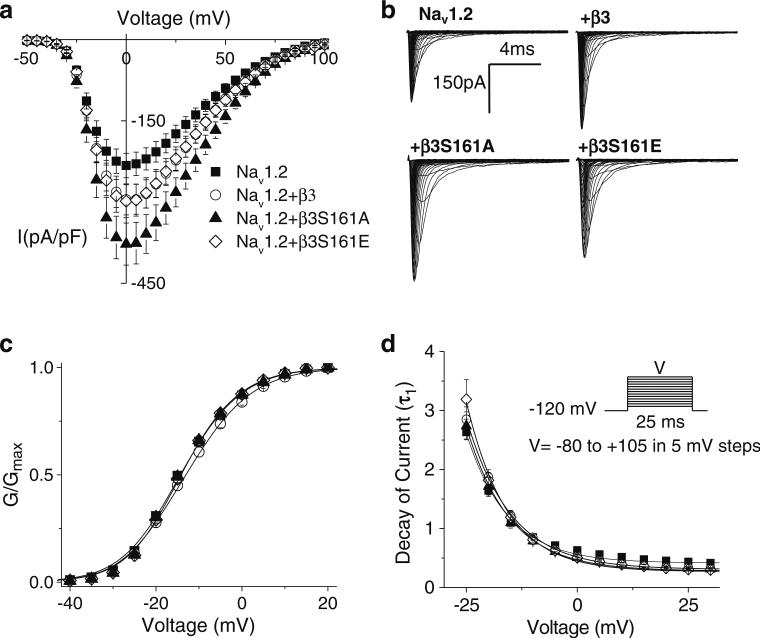
To determine the effects of WTβ3, β3S161E, and β3S161A on current density, kinetics of activation, and decay of the macroscopic current, Na currents were elicited by applying a 25-ms voltage step ranging from –80 to +105 mV in 5-mV steps from a holding potential of –120 mV (Fig. 1). Both WTβ3 and β3S161E co-expression increased the current density compared to nontransfected cells (Fig. 1a). Mean current density values were Nav1.2=–235.3±22.5 pA/pF, (n=48); β3=–306.1±29.3 pA/pF (n=57), and β3S161E=–303.4±30.4 pA/pF (n=39). Co-expression of β3S161A resulted in a significant increase in current density to –389.9±34.5 pA/pF (n=51; P<0.05 vs Nav1.2). Representative families of currents are shown in Fig. 1b. The increases in current density were not accompanied by shifts in channel conductance (V1/2) or slopes of activation (k) (Fig. 1c and Table 1). Time constants for the decay of the macroscopic currents also were not different among any of the subunits (Fig. 1d).
Fig. 1.
β3 co-expression increases current density without modulation of activation parameters. a Current density, assessed as the peak of the current voltage relationship for each cell divided by cell capacitance, was amplified by β-subunit co-expression. Currents were elicited by depolarizing steps of 25 ms from a holding potential of –120 mV to test pulses in the range of –80 to +105 mV in 5-mV increments. Co-expression of β3(β3–306.1±29.3 pA/pF, n=57) or β3S161E (–303.4±30.4 pA/pF, n=39) increased the median current density as compared to non-transfected cells (Nav1.2=–235.3±22.5 pA/pF, n=48). Co-expression of β3S161A resulted in a significantly greater median current density than non-transfected cells (–389.9±34.5 pA/pF, n=44, Dunn's test p<0.05). b Representative traces of whole-cell Na currents evoked in nontransfected HEK293 cells stably expressing Nav1.2 and cells co-transfected with WTβ3, β3S161A, and β3S161E. c Conductance plot shows no effects on activation gating parameters with any of the β-subunit constructs. Smooth lines correspond to the least squares fit when average conductance data were fit with a single Boltzmann equation. d Decay of macroscopic currents was fit to double exponential function and was not significantly different across the three expression systems. Data shown are means ± SEM
Table 1.
Steady-state activation parameters
| V1/2(mV) | k(mV) | Parameter (n) | |
|---|---|---|---|
| Nav1.2 | –14.0±0.6 | –6.3±0.2 | 49 |
| β3 | –12.9±0.6 | –6.2±0.1 | 53 |
| β3S161A | –13.7±0.6 | –5.8±0.2 | 53 |
| β3S161E | –13.7±0.6 | –6.0±0.2 | 39 |
β3S161A mutant abolishes the depolarizing shift in steady-state inactivation parameters induced by WTβ3
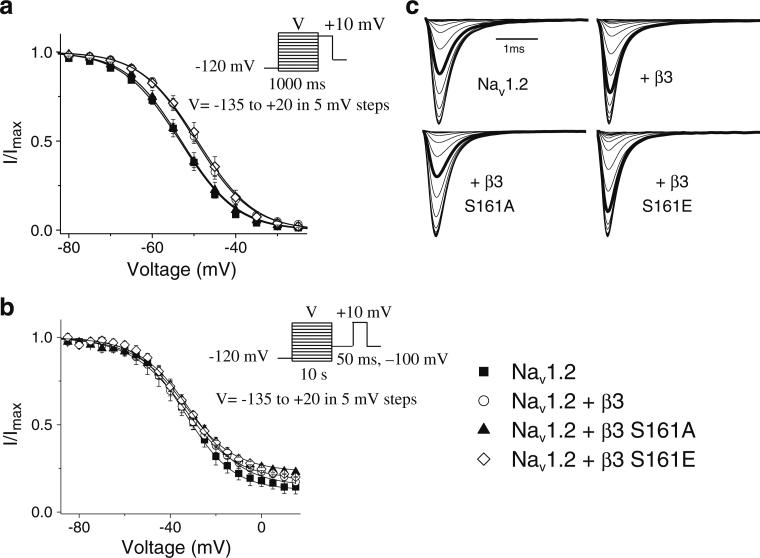
β subunits are known to modulate the fraction of channels that are available to open by influencing the voltage dependence of inactivation. We determined the effects of WTβ3, β3S161A, and β3S161E on the kinetics of inactivation using two different depolarizing pre-pulse durations, 1,000 ms and 10 s, to assess changes in intermediate and slow inactivation parameters respectively (Fig. 2). Cells were initially held at –120 mV then subjected to a pulse at voltages ranging from –135 to +20 mV. This was followed by a test pulse of +10 mV in order to establish the extent of channel inactivation. To evaluate slow inactivation, fast-inactivated channels were recovered using a 50-ms pulse at –100 mV prior to the test pulse.
Fig. 2.
Steady-state inactivation. Steady-state inactivation parameters were determined from a holding potential of –120 mV using conditioning pulses of either 1,000 ms (a) or 10 s (b) at voltages ranging from –135 to +20 mV. A test pulse of +10 mV was then used to assess channel availability. To evaluate slow inactivation, fast-inactivated channels were recovered using a 50-ms pulse at –100 mV prior to the test pulse. Smooth lines correspond to the average of least squares fits when data were fitted with a single Boltzmann equation. c Representative steady state inactivation traces for 1,000 ms pre-pulse. The bold trace representing steady state inactivation at a voltage step of –50 mV is shown for comparison purposes
Inactivation kinetics using a 1,000-ms pre-pulse was best fit with a single Boltzmann function (Table 2). Under these conditions, both WTβ3 and β3S161E significantly (P< 0.05) shifted the V1/2 in the depolarized direction from Nav1.2 alone (Table 2). This shift was abolished with co-expression of β3S161A and was no longer different from nontransfected cells expressing Nav1.2 alone. Slope factors (k) were not affected. The shifts in inactivation observed with WTβ3 and β3S161E compared with β3S161A and nontransfected cells were abolished when using a longer 10-s pre-pulse, suggesting that β subunits do not modulate slow inactivated Na channels (Table 2).
Table 2.
Steady-state inactivation parameters
| 1,000ms pre-pulse |
10s pre-pulse |
|||||
|---|---|---|---|---|---|---|
| V1/2(mV) | k(mV) | n | V1/2(mV) | k(mV) | n | |
| Nav1.2 | –53.4±0.7 | 6.2±0.1 | 20 | –33.2±0.8 | 10.2±0.4 | 6 |
| WTβ3 | –49.5±1.0* | 6.2±0.2 | 9 | –34.3±2.2 | 11.2±0.9 | 6 |
| β3S161A | –53.0±1.1 | 6.3±0.5 | 6 | –34.0±1.0 | 11.5±1.4 | 5 |
| β3S161E | –49.2±1.3* | 6.2±0.1 | 7 | –31.1±1.2 | 11.4±0.8 | 4 |
p<0.05 vs Nav1.2, Tukey Test
WTβ3 and β3S161E accelerate the rate of recovery from inactivation in comparison to β3S161A
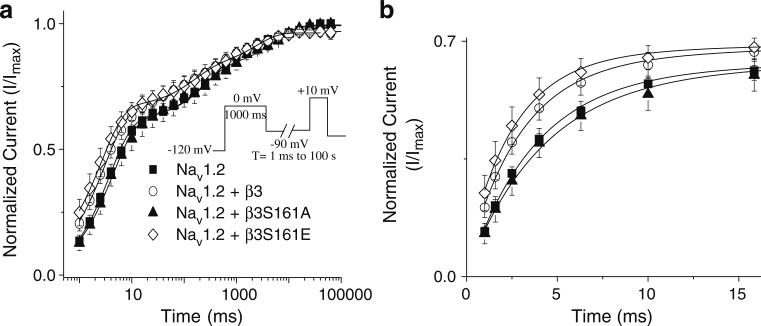
β subunits have been shown to modulate kinetics of recovery from inactivation [38]. To evaluate the effects of WTβ3 and the importance of β3S161 on recovery from inactivation kinetics, cells were held at –120 mV for 1,000 ms and depolarized to 0 mV for 1,000 ms to inactivate the Na channels. Na channels were subsequently recovered at –90 mV for a variable length of time (1 ms to 100 s) and subjected to a test pulse of +10 mV to determine the extent of recovery (Fig. 3). Data were normalized to the peak current amplitude recorded at 100 s and best fit using a triple exponential function (Table 3). Co-expression of WTβ3 or β3S161E significantly decreased (P<0.05) the fast time constant (τ1), accelerating recovery kinetics compared with β3S161A. In Fig. 3b, recovery from inactivation is shown on an expanded time scale to further illustrate the faster recovery from inactivation of WTβ3 and β3S161E compared with β3S161A and Nav1.2.
Fig. 3.
Recovery from inactivation at –90 mV. Recovery from inactivation was assessed at –90 mV using a two-pulse protocol. A pre-pulse from –120 to 0 mV was applied for 1,000 ms. Cells were then held at –90 mV for variable lengths of time (1 ms–100 s) to allow for channels to recover (a). WTβ3 and β3S161E both showed significantly faster rates of recovery of the first phase than did β3S161A. In b, an expanded time course further demonstrates the faster recovery trend for cells co-expressing WTβ3 and β3S161E when compared with Nav1.2 alone and β3S161A. Data points represent means ± SEM
Table 3.
Recovery from inactivation at –90 mV
| τ1(ms) | τ2(ms) | τ3(s) | %A1 | (n) | |
|---|---|---|---|---|---|
| Nav1.2 | 3.7±0.4 | 314.6±75.8 | 7.4±1.8 | 62.6±2.4 | 5 |
| WTβ3 | 2.7±0.5* | 166.1±55.0 | 5.5±1.8 | 59.7±4.7 | 6 |
| β3S161A | 4.3±0.6 | 233.0±38.4 | 6.2±1.2 | 60.9±5.0 | 5 |
| β3S161E | 2.6±0.4* | 276.2±97.5 | 7.9±4.0 | 65.0±3.6 | 8 |
p<0.05 vs β3S161A; Tukey Test
β3S161E and WTβ3 attenuate frequency-dependent inhibition of Nav1.2
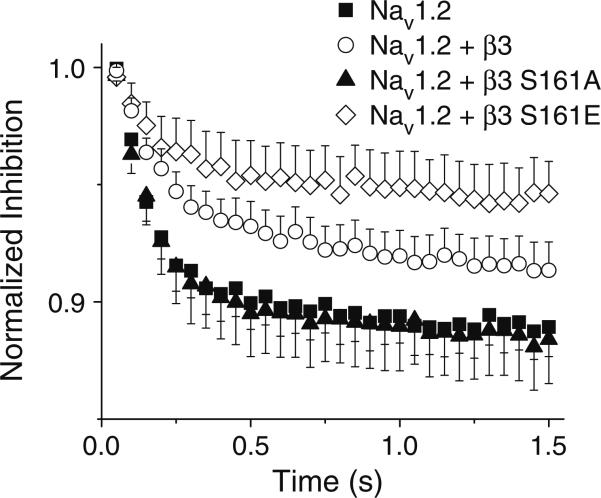
In view of the fact that WTβ3 and β3S161E delayed the entry of Nav1.2 into the inactivated state and also enhanced their recovery from inactivation, it is likely that during high-frequency stimulation channel accumulation in the inactivated state would also be reduced. To determine the effects of WTβ3 and the role of S161 in accumulating inactivated channels, a series of depolarizing pulses from a holding potential of –120 mV to +10 mV at a frequency of 20 Hz were applied (Fig. 4). In cells expressing Nav1.2 alone, stimulation at 20 Hz reduced the current amplitude by 11.1±1.2%, and this effect was not changed by co-expression of β3S161A (10.9±1.8%). In contrast, both WTβ3 and β3S161E attenuated this frequency-dependant inhibition by 8.7±1.2% and 5.4±1.4%, respectively. The differences, however, were only significant at P=0.09 and were indicative of an overall trend of impaired inactivation by WTβ3 and βS161E co-expression.
Fig. 4.
WTβ3 and β3S161E attenuates frequency dependent inhibition compared to β3S161A and Nav1.2 alone. Frequency-dependent inhibition was determined using a depolarizing pulse to +10 mV from a holding potential of –120 mV for 12 ms at a frequency of 20 Hz. Peak current was normalized to the first pulse in each experiment. Data represent mean ± SEM
Trafficking of β3S161A and β3S161E is not different from WTβ3
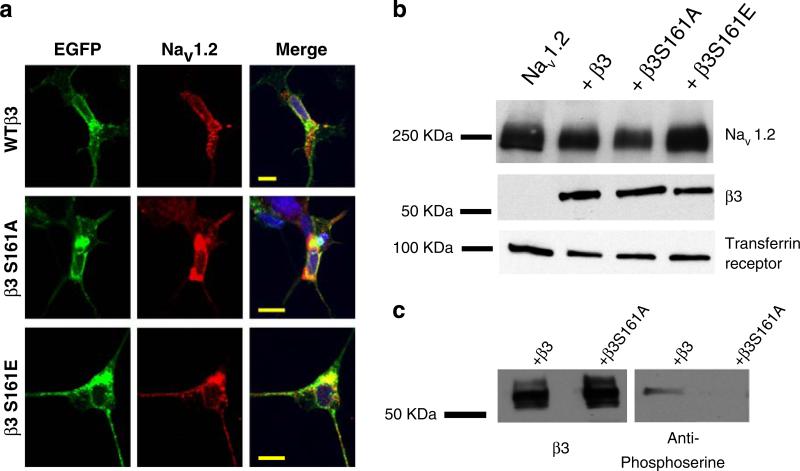
To determine if the lack of any effect of β3S161A, compared with β3 and β3S161E, were due to a complete absence of β3S161A from within the plasma membrane, immunocytochemistry and Western blot experiments were performed. Since all of the constructs were tagged with a short C-terminal EGFP, we were able to follow the cellular localization of the β subunits. WTβ3, β3S161A, and β3S161E (green) were individually colocalized with Nav1.2 (red) (Fig. 5a). Although some intracellular staining was observed, as a result of normal trafficking through the secretory pathway, overall membrane expression of β3S161A was not different from that observed for WTβ3 or β3S161E.
Fig. 5.
Immunocytochemistry and Western blot analysis. HEK293 cells stably expressing Nav1.2 were transfected with GFP-tagged WTβ3, β3S161A, or β3S161E. Staining of Nav1.2 (red) was co-localized with WTβ3, β3S161A, and β3S161E (green) within the plasma membrane (a). Blue staining represents DAPI staining of nuclei. Scale bar represents 10 μm. Western blot analysis (b and c) of membrane proteins isolated through biotinylation and neutravidin purification. WTβ3, β3S161A, and β3S161E were all detected in the plasma membrane of HEK293 cells. Membrane expression of Nav1.2 was also detected in the membrane for all experimental conditions. In c, blots of WTβ3 and β3S161A were initially probed for β3, stripped, and then reprobed for phosphoserine. Probing with phosphoserine revealed a band for WTβ3, but not β3S161A. Identical results were observed when first probing with anti-phosphoserine, stripping, and reprobing with anti-β3
Western blot analysis using an anti-phosphoserine antibody
Western blot analysis confirmed robust expression of β3S161A as well as WTβ3 and β3S161E in the membrane (Fig. 5b). Membrane expression levels of Nav1.2 were similar for all experimental conditions. The predicted molecular weight for Nav1.2 was 220–260 kDa; EGFP-labeled WTβ3 and mutants, 55–70 kDa; and the standard, transferrin receptor, 100 kDa. In another experiment, WTβ3 and β3S161A were first probed with anti-β3 antibody and then stripped and re-probed with anti-phosphoserine antibody (Fig. 5c). Membrane expression of both the WTβ3 and the β3S161A subunits were detected by the β3 antibody (Fig. 5c). In contrast, when stripped and re-probed with anti-phosphoserine antibody, only the WTβ3 subunit signal was apparent, with no detectable signal with the β3S161A subunit. The same effect was observed when first probing with the phosphoserine antibody, stripping, and re-probing with the β3 antibody. These findings suggest that membrane expressed WTβ3 is phosphorylated, while the β3S161A mutant is not phosphorylated.
Scansite 2.0 analysis of S161 as a phosphorylation site
Using the Scansite 2.0 algorithm (www.scansite.mit.edu), the probability for phosphorylation of the β3S161 residue was calculated. Scansite ranks the likelihood that a specific residue is phosphorylated against the probability of other iterations of the residue in the molecule. Lower scores correspond to a greater probability of phosphorylation of the relevant residue as has been previously shown for known phosphorylation sites within Na channels [2]. For example, S687 of the Nav1.2, a confirmed protein kinase A phosphorylation site [29], was calculated to have a score of 2.92%. In comparison, the score of β3S161 phosphorylation by protein kinase A (1.15%) was lower than that of S687, suggesting that β3S161 is more likely than Nav1.2 S687 to be phosphorylated. The positioning and surrounding residues of β3S161 make this residue favorable for phosphorylation that could provide an additional method of regulation for β3.
Secondary structure prediction for the β3 intracellular domain
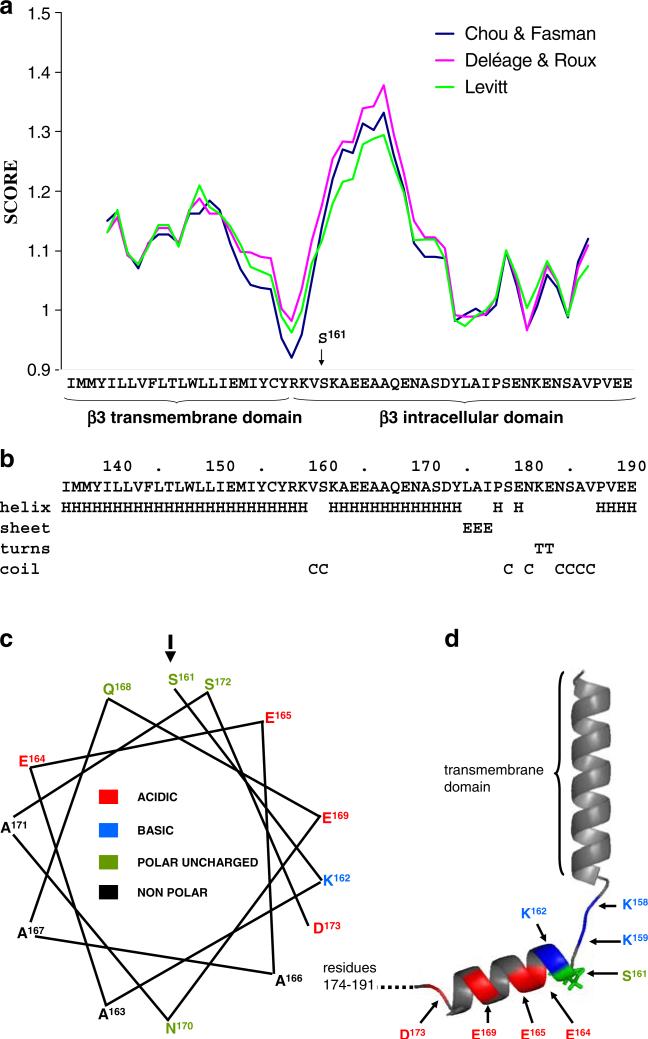
We used four separate well-established secondary structure prediction algorithms to examine the structural context within which the S161 residue is likely to lie. These programs were the methods of Garnier [7], Deléage and Roux [6], Chou and Fasman [4], and Levitt [17]. We note that all four programs predict similar secondary structure for the intracellular domain of β3 (Fig. 6a, b). The transmembrane domain alpha-helix extends between residues 136 and 157 [25] with residues R158 and K159 representing the stop transfer sequence [16]. There is a significant reduction in alpha helical potential for these amino acids immediately abutting the transmembrane domain, but this is followed by a second, strong alpha helical potential from approximately S161 to Y174 (Fig. 6). Interestingly, this putative alpha helix is amphipathic. One face is predominantly hydrophobic; the other face is charged and dominated by a stripe of acidic residues, with S161 lying at the start of the polar face (Fig. 6c and d). This suggests that S161 occupies a strategic location within the intracellular domain sequence where it could influence secondary structure elements within the intracellular domain. Furthermore, this striking structural feature has been conserved during vertebrate evolution suggesting functional importance [3]. The remaining C-terminal region of the β3 intracellular domain mostly has a low alpha helical potential (Fig. 6a and b), possibly due to the presence of prolines within the sequence [25].
Fig. 6.
Secondary structure prediction of β3 transmembrane and intracellular domains. a The propensity of the putative transmembrane and intracellular domains of human β3 (residues I136 to E191 of the mature protein) to fold into an α-helix were predicted using a window size of nine residues and the algorithms of Deléage and Roux [6], Chou and Fasman [4], and Levitt [17]. b Secondary structure prediction using the method of Garnier et al. [7]. Residues K163 to D174 are strongly predicted to fold into an α-helix comprising hydrophobic residues with changed residues at every third or fourth position. c Helical wheel projection of the putative intracellular domain helix (amino acids 161–173) showing its amphipathic nature and the position of the potentially phosphorylated S161. d Structural model of the transmembrane domain and the intracellular domain of the β3 emphasizing short disordered sequence separating helices. The model was built using pyMol
Discussion
Auxiliary β subunits modulate not only the electrophysiological gating parameters of voltage gated sodium channels [9, 13–15], but also membrane expression levels [21]. Structure/function studies have implicated the extracellular domain of β1 subunits in mediating the changes in channel gating through an interaction with the P loop of the Na channel [28]. More recently, studies have suggested an important interaction between the intracellular domain of β subunits and the Na channel α subunit that affects both channel gating and trafficking [22, 38]. In this study, we propose that phosphorylation of S161, located within the intracellular region of β3, is required for the β subunit to shift the voltage dependence of channel inactivation gating and to accelerate recovery from inactivation.
β3 co-expression increases current density
When compared with Nav1.2 alone, a trend of increased Na channel current density was observed with co-expression of WTβ3 or β3S161E with Nav1.2. A greater increase in Na channel current density occurred with co-expression of the non-phosphorylatable residue β3S161A. These data suggest that β3S161 may not play a major role in the β subunit associated trafficking of the α subunit to the membrane. The increases in current density could arise from either increases in single channel conductance or open channel probability or increases in the membrane expression of the channel. Since β subunits do not affect single-channel parameters [27], i.e., they do not increase single-channel conductance or open channel probability, a possible explanation for the observed increase in whole-cell currents is an increase in the membrane expression of Nav1.2. In agreement, β subunits have been reported to increase Na channel expression in membrane [22]. The precise mechanisms for the increase are not known, but it may involve a direct interaction between the α and β subunits and/or combination with cell-adhesion molecules and cytoskeletal components, including contactin and ankyrin [5, 21]. Evidence supports the interaction of β subunits and Na channel α subunits in the ER, enabling trafficking of both subunits to the membrane together [42]. A recent study with Nav1.8 revealed a mechanism in which β3 increases membrane expression of the α subunit. Mutational analysis suggests that β3 interacts with Nav1.8 in such a way that the RRR ER retention signal of the α subunit is physically covered, allowing the α and β subunits to be inserted into the membrane in tandem [41]. A decrease in the surface expression of the Na channel Nav1.1 has been implicated in generalized epilepsy with febrile seizures plus (GEFS+). This decrease in channel surface expression can be rescued by co-expression of any of the four β subunits, providing support for a role of β subunits in controlling membrane expression levels [30]. Regulation of ion channel expression levels by auxiliary subunits has been proposed for the calcium channel isoform Cav2.1 [34]. Modulation of the auxiliary β2a encouraged Cav2.1 channel interaction with cellular trafficking proteins, facilitating insertion in the plasma membrane.
Modulation of inactivation gating parameters
A major finding of our studies is the importance of S161 in modulating channel availability, shifting the voltage at which the Na channels inactivate in a depolarized direction and accelerating the rates of recovery from inactivation. Since the shifts in inactivation gating and recovery kinetics were abolished by co-expression of β3S161A, these findings suggest that post-translational modification of β3S161 could allow for an additional mechanism by which sodium channel gating can be modulated. The depolarized shifts in sodium channel inactivation and faster recovery rates observed with WTβ3 and β3S161E would allow more Na channels to be available at a given voltage, reducing the threshold for action potential firing and potentially, increasing neuronal excitability. It is tempting to speculate that dephosphorylation of β3 would be a means to then reduce neuronal excitability.
It is not surprising that inactivation parameters were not changed when using a long 10-s inactivating pre-pulse since these channels were likely to have entered the slow inactivation state, a state that involves movement of the S6 transmembrane segments within the channel pore and not the III to IV linker [32]. Since a shift in the steady-state inactivation curve was observed only when using a short 1,000-ms inactivating pre-pulse, and not when using a longer 10-s pre-pulse, we propose that β3 interacts with the regions of the Na channel involved in the modulation of fast inactivation. The III to IV linker of the pore forming α subunit is known to physically block the pore allowing for fast inactivation of the Na channel [8]. The C-terminus of the Na channel interacts with the III to IV linker and also with the C-terminus of the β1 and β3 subunits providing a potential mechanistic link between β subunits and their modulation of inactivation gating [26, 31].
β3 subunit modulation is likely regulated by a phosphorylation site in its intracellular domain
The similarity of the modulatory properties of WTβ3 and of our phosphomimic mutant β3S161E suggests that β3 may be in a phosphorylated state in the membrane. In support of this, western blot analysis of membrane proteins using an anti-phosphoserine antibody detected an appropriate sized band in cells expressing WTβ3. The intracellular domain of β3 contains two serine residues; however, the basic residues surrounding S161 make that residue a favored site for protein kinase A. The Scansite 2.0 program analysis predicts that S161 is more likely to be phosphorylated by PKA than 98.85% of the other serine/threonine residues in the subunit. This percentage was favorably comparable to the propensity for phosphorylation of the confirmed phosphorylation site Y181 in β1 [19].
Currently, there is nothing known about the secondary structure of the intracellular domains of any Nav β subunits. But we note the potential for a well-conserved amphipathic alpha helix in the membrane-proximal region of the β3 intracellular domain directly downstream from S161. Interestingly, short polar amino acids such as serine are known to stabilize alpha helices when located at the helix N terminus, and indeed, sequences of the form SXXE are particularly favorable [35]. An amphipathic helix would make a good candidate for a specific binding site, and it is tempting to propose that phosphorylation of the S161 residue modulates the interaction of β3 with the Na channel C-terminus, perhaps by altering the stability of this amphipathic helix or by changing the charge distribution around it. It will now be important to examine the secondary structure of the intracellular domain in this light.
In summary, our results show that S161 in the β3 subunit is important for modulating channel inactivation gating parameters and rates of recovery from inactivation. Since in β1 this residue is replaced by a non-phosphorylatable alanine, S161 may allow for specific modulation of β3 versus β1, particularly in cells that are known to express both β subunits. Since β subunits fine-tune the electrophysiological gating properties of Na channels, phosphorylation of β3 may be yet another mechanism for precision control of cellular excitability.
Acknowledgments
This work was supported in part by Award No. 08-1 from the Commonwealth of Virginia's Alzheimer's and Related Diseases Research Award Fund, administered by the Virginia Center on Aging, Virginia Commonwealth University (M.K.P. & E.C.M.), University of Virginia Center for Undergraduate Excellence Harrison Research Grant (E.C.M), National Institutes of Health grant NIH RO1 088554 (B.E.I.), GM31184 (J.J.S.), the American Heart Science Development Grant (B.E.I.), and a CASE studentship from the Biotechnology and Biological Sciences Research Council, U.K. (F.S.C.). We thank Dr. Luca Pellegrini for helpful discussions on the structure of β3.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Ellen C. Merrick, Department of Anesthesiology, University of Virginia Health System, Box 800710, Charlottesville, VA 22908-0710, USA
Christopher L. Kalmar, Department of Anesthesiology, University of Virginia Health System, Box 800710, Charlottesville, VA 22908-0710, USA
Sandy L. Snyder, Department of Anesthesiology, University of Virginia Health System, Box 800710, Charlottesville, VA 22908-0710, USA
Fiona S. Cusdin, Department of Biochemistry, University of Cambridge, Cambridge, England CB2 1QW, UK
Ester J. Yu, Department of Biochemistry, University of Cambridge, Cambridge, England CB2 1QW, UK
Julianne J. Sando, Department of Anesthesiology, University of Virginia Health System, Box 800710, Charlottesville, VA 22908-0710, USA
Brant E. Isakson, Department of Molecular and Biological Physics and Robert M. Berne Cardiovascular Research Center, University of Virginia Health System, Charlottesville, VA 22908, USA
Antony P. Jackson, Department of Biochemistry, University of Cambridge, Cambridge, England CB2 1QW, UK
Manoj K. Patel, Department of Anesthesiology, University of Virginia Health System, Box 800710, Charlottesville, VA 22908-0710, USA
References
- 1.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Yu FH, Sharp EM, Beacham D, Scheuer T, Catterall WA. Functional properties and differential neuromodulation of Na(v)1.6 channels. Mol Cell Neurosci. 2008;38:607–615. doi: 10.1016/j.mcn.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra SS, Watanabe H, Zhong TP, Roden DM. Molecular cloning and analysis of zebrafish voltage-gated sodium channel beta subunit genes: implications for the evolution of electrical signaling in vertebrates. BMC Evol Biol. 2007;7:113. doi: 10.1186/1471-2148-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Cusdin FS, Clare JJ, Jackson AP. Trafficking and cellular distribution of voltage-gated sodium channels. Traffic. 2008;9:17–26. doi: 10.1111/j.1600-0854.2007.00673.x. [DOI] [PubMed] [Google Scholar]
- 6.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 7.Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 8.Goldin AL. Mechanisms of sodium channel inactivation. Curr Opin Neurobiol. 2003;13:284–290. doi: 10.1016/s0959-4388(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 9.Grieco TM, Malhotra JD, Chen C, Isom LL, Raman IM. Open-channel block by the cytoplasmic tail of sodium channel beta4 as a mechanism for resurgent sodium current. Neuron. 2005;45:233–244. doi: 10.1016/j.neuron.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- 11.Isom LL, De Jongh KS, Patton DE, et al. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 12.Isom LL, Ragsdale DS, De Jongh KS, et al. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 13.Isom LL, Scheuer T, Brownstein AB, Ragsdale DS, Murphy BJ, Catterall WA. Functional co-expression of the beta 1 and type IIA alpha subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- 14.Johnson D, Bennett ES. Isoform-specific effects of the beta2 subunit on voltage-gated sodium channel gating. J Biol Chem. 2006;281:25875–25881. doi: 10.1074/jbc.M605060200. [DOI] [PubMed] [Google Scholar]
- 15.Ko SH, Lenkowski PW, Lee HC, Mounsey JP, Patel MK. Modulation of Na(v)1.5 by beta1– and beta3-subunit co-expression in mammalian cells. Pflugers Arch. 2005;449:403–412. doi: 10.1007/s00424-004-1348-4. [DOI] [PubMed] [Google Scholar]
- 16.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. Systematic analysis of stop-transfer sequence for microsomal membrane. J Biol Chem. 1991;266:9251–9255. [PubMed] [Google Scholar]
- 17.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 18.Makita N, Shirai N, Wang DW, et al. Cardiac Na(+) channel dysfunction in Brugada syndrome is aggravated by beta(1)-subunit. Circulation. 2000;101:54–60. doi: 10.1161/01.cir.101.1.54. [DOI] [PubMed] [Google Scholar]
- 19.Malhotra JD, Koopmann MC, Kazen-Gillespie KA, Fettman N, Hortsch M, Isom LL. Structural requirements for interaction of sodium channel beta 1 subunits with ankyrin. J Biol Chem. 2002;277:26681–26688. doi: 10.1074/jbc.M202354200. [DOI] [PubMed] [Google Scholar]
- 20.McCormick KA, Isom LL, Ragsdale D, Smith D, Scheuer T, Catterall WA. Molecular determinants of Na+ channel function in the extracellular domain of the beta1 subunit. J Biol Chem. 1998;273:3954–3962. doi: 10.1074/jbc.273.7.3954. [DOI] [PubMed] [Google Scholar]
- 21.McEwen DP, Meadows LS, Chen C, Thyagarajan V, Isom LL. Sodium channel beta1 subunit-mediated modulation of Nav1.2 currents and cell surface density is dependent on interactions with contactin and ankyrin. J Biol Chem. 2004;279:16044–16049. doi: 10.1074/jbc.M400856200. [DOI] [PubMed] [Google Scholar]
- 22.Meadows L, Malhotra JD, Stetzer A, Isom LL, Ragsdale DS. The intracellular segment of the sodium channel beta 1 subunit is required for its efficient association with the channel alpha subunit. J Neurochem. 2001;76:1871–1878. doi: 10.1046/j.1471-4159.2001.00192.x. [DOI] [PubMed] [Google Scholar]
- 23.Meadows LS, Malhotra J, Loukas A, et al. Functional and biochemical analysis of a sodium channel beta1 subunit mutation responsible for generalized epilepsy with febrile seizures plus type 1. J Neurosci. 2002;22:10699–10709. doi: 10.1523/JNEUROSCI.22-24-10699.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medeiros-Domingo A, Kaku T, Tester DJ, et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan K, Stevens EB, Shah B, et al. beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci USA. 2004;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motoike HK, Liu H, Glaaser IW, Yang AS, Tateyama M, Kass RS. The Na+ channel inactivation gate is a molecular complex: a novel role of the COOH-terminal domain. J Gen Physiol. 2004;123:155–165. doi: 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuss HB, Chiamvimonvat N, Perez-Garcia MT, Tomaselli GF, Marban E. Functional association of the beta 1 subunit with human cardiac (hH1) and rat skeletal muscle (mu 1) sodium channel alpha subunits expressed in Xenopus oocytes. J Gen Physiol. 1995;106:1171–1191. doi: 10.1085/jgp.106.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Y, Rogers JC, Chen SF, McCormick KA, Scheuer T, Catterall WA. Functional roles of the extracellular segments of the sodium channel alpha subunit in voltage-dependent gating and modulation by beta1 subunits. J Biol Chem. 1999;274:32647–32654. doi: 10.1074/jbc.274.46.32647. [DOI] [PubMed] [Google Scholar]
- 29.Rossie S, Catterall WA. Phosphorylation of the alpha subunit of rat brain sodium channels by cAMP-dependent protein kinase at a new site containing Ser686 and Ser687. J Biol Chem. 1989;264:14220–14224. [PubMed] [Google Scholar]
- 30.Rusconi R, Scalmani P, Cassulini RR, et al. Modulatory proteins can rescue a trafficking defective epileptogenic Nav1.1 Na+ channel mutant. J Neurosci. 2007;27:11037–11046. doi: 10.1523/JNEUROSCI.3515-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spampanato J, Kearney JA, de Haan G, et al. A novel epilepsy mutation in the sodium channel SCN1A identifies a cytoplasmic domain for beta subunit interaction. J Neurosci. 2004;24:10022–10034. doi: 10.1523/JNEUROSCI.2034-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulbricht W. Sodium channel inactivation: molecular determinants and modulation. Physiol Rev. 2005;85:1271–1301. doi: 10.1152/physrev.00024.2004. [DOI] [PubMed] [Google Scholar]
- 33.Valdivia C, Mereudos-Domingo A, Algiers TJ, Ackerman MJ, Makielski JC. Identification and Characterization of a Novel Susceptibility Gene, SCN3B, for Idiopathic Venricular Fibrillation. Circulation. 2008;118:S526. [Google Scholar]
- 34.Viard P, Butcher AJ, Halet G, et al. PI3K promotes voltage-dependent calcium channel trafficking to the plasma membrane. Nat Neurosci. 2004;7:939–946. doi: 10.1038/nn1300. [DOI] [PubMed] [Google Scholar]
- 35.Vijayakumar M, Qian H, Zhou HX. Hydrogen bonds between short polar side chains and peptide backbone: prevalence in proteins and effects on helix-forming propensities. Proteins. 1999;34:497–507. [PubMed] [Google Scholar]
- 36.Wallace RH, Wang DW, Singh R, et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 37.Xu R, Thomas EA, Gazina EV, et al. Generalized epilepsy with febrile seizures plus-associated sodium channel beta1 subunit mutations severely reduce beta subunit-mediated modulation of sodium channel function. Neuroscience. 2007;148:164–174. doi: 10.1016/j.neuroscience.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 38.Yu EJ, Ko SH, Lenkowski PW, Pance A, Patel MK, Jackson AP. Distinct domains of the sodium channel beta3-subunit modulate channel-gating kinetics and subcellular location. Biochem J. 2005;392:519–526. doi: 10.1042/BJ20050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2007;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu FH, Westenbroek RE, Silos-Santiago I, et al. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang ZN, Li Q, Liu C, Wang HB, Wang Q, Bao L. The voltage-gated Na+ channel Nav1.8 contains an ER-retention/ retrieval signal antagonized by the beta3 subunit. J Cell Sci. 2008;121:3243–3252. doi: 10.1242/jcs.026856. [DOI] [PubMed] [Google Scholar]
- 42.Zimmer T, Biskup C, Bollensdorff C, Benndorf K. The beta1 subunit but not the beta2 subunit colocalizes with the human heart Na+ channel (hH1) already within the endoplasmic reticulum. J Membr Biol. 2002;186:13–21. doi: 10.1007/s00232-001-0131-0. [DOI] [PubMed] [Google Scholar]