Abstract
The Mediator complex interacts extensively with the RNA polymerase II enzyme and regulates its ability to express protein-coding genes. The mechanisms by which Mediator regulates gene expression remain poorly understood, in part because Mediator structure and even its composition can change, depending upon the promoter context. Combined with the sheer size of the human Mediator complex (26 subunits, 1.2 MDa), this structural adaptability bestows seemingly unlimited regulatory potential within the complex. Recent efforts to understand Mediator structure and function have identified expanded roles that include control of both pre- and post-initiation events; it is also evident that Mediator performs both general and gene-specific roles to regulate gene expression.
Introduction
Mediator was first discovered in yeast by the Young and Kornberg labs [1,2]; isolation of a human Mediator complex occurred a few years later [3]. Since that time, biochemical, genetic, and biophysical studies have revealed some similarities between yeast and human Mediator, yet a number of clear structural and functional differences have also been identified. We are still in the early stages of understanding how human Mediator functions to regulate both general and gene-specific events in transcription. One reason for this is that Mediator is a 1.2 MDa, 26-subunit complex, which precludes recombinant expression of the entire assembly. Instead, Mediator must be purified directly from human cells; however this presents a challenge because Mediator is a low-abundance complex that exists in multiple, biochemically-distinct forms. Another reason Mediator remains poorly-understood is that the function of its subunits cannot be inferred with bioinformatics: Mediator sequences contain almost no predicted functional motifs. Furthermore, among the components of the general transcription machinery (e.g. TFIID and RNA polymerase II; pol II), Mediator sequences have evolved most rapidly relative to their yeast counterparts [4]. Bioinformatics studies have, however, revealed an unusually high percentage of intrinsically disordered regions within Mediator subunits [5], and these domains likely contribute to Mediator structural plasticity and its vast potential for protein–protein interactions.
Mediator is apparently specific to eukaryotic organisms, as evidence for Mediator or a Mediator-like activity in microbes is lacking. Similarly, most of the so-called general transcription factors (GTFs)—including TFIIE, TFIIH, and TFIID—are not present in microbial genomes: like Mediator, these factors emerged in eukaryotic lineages. (Archaea possess orthologs of TFIIB and TBP; domains within TFIIF are homologous to bacterial sigma factors.) The apparent co-evolution of Mediator with most GTFs suggests these complexes might coordinately function to regulate expression of protein-coding genes. This idea is strongly supported by biochemical data that reveal a functional cooperativity between human Mediator and most GTFs, including TFIIB, TFIIE, TFIID, TFIIH, and pol II itself. This functional cooperativity will be described in this review, along with other recent discoveries that have further established Mediator’s role as a primary regulator of pol II-dependent transcription.
Mediator controls assembly and activity of the transcription machinery
Human Mediator was first isolated as an activity that restored activator-dependent transcription in cell-free, in vitro assays [3,6–9]. As a result, Mediator was considered a co-activator of transcription. A co-activator function for Mediator is also implied because Mediator is a general target of DNA-binding transcription factors (i.e. activators) that play a role in recruiting Mediator to appropriate regulatory sites throughout the genome. Although Mediator clearly plays essential roles in activator-dependent transcription, numerous studies have also pointed to activator-independent functions. For instance, human Mediator can stimulate even basal (activator-independent) transcription [10,11], suggesting that Mediator might control fundamental events in transcription initiation. Furthermore, it is evident that Mediator significantly enhances pol II recruitment and stabilizes transcription complexes at the promoter [12,13]. Studies in yeast have revealed that Mediator is as important as pol II itself for expression of protein-coding genes [14], and Mediator subunits localize to promoters on a genome-wide scale [15,16]. Indeed, it is evident from the available literature that Mediator should in fact be considered a general transcription factor.
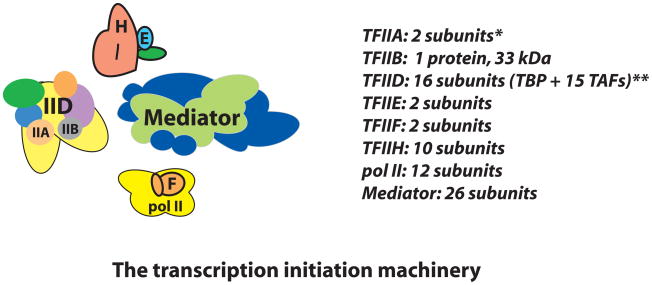
The transcription initiation machinery (Box 1) is an elaborate assembly of proteins and protein complexes that is over 3.5 MDa in size [17]. Collectively, this assembly has been called the Pre-Initiation Complex (PIC); however, recent revelations have prompted a re-evaluation of this terminology. Many metazoan genes appear to be regulated at stages after pol II recruitment, in which limited initiation may or may not have occurred. Therefore the PIC terminology can be slightly misleading and instead the more general term Pre-Elongation Complex (PEC) is proposed (Box 1). In short, the term PEC can be considered equivalent to PIC except that it also accounts for promoter-bound, stalled pol II complexes that have not initiated productive elongation.
Box 1. The Pre-Elongation Complex (PEC).
Human gene expression can be regulated in many different ways (e.g. via changes in nuclear architecture, alterations in chromatin state, factor occupancy and activity, etc.) and at multiple times during the activation process (e.g. prior to pol II recruitment, after pol II recruitment, during pol II elongation, etc.). In human cells, a significant fraction of genes appear to be regulated at a “post-recruitment” stage of transcription [68,69]. That is, at a stage in which pol II has been recruited to the promoter but has not initiated productive elongation [70]. These inactive, promoter-bound polymerases might exist in a number of distinct functional states [71]. Some pol II complexes might adopt a “closed” state in which promoter melting and initiation has not occurred, despite complete assembly of the transcription machinery (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Mediator, and pol II). Such a functional state can be accurately described as a Pre-initiation Complex [17]. However, a distinct population of pol II complexes might have engaged the promoter (via promoter melting and open complex formation) and initiated transcription (perhaps through 20–50 bp). Pol II complexes that have initiated transcription but have not transitioned to a productively-elongating state are thought to be “stalled” because they have the capacity—given the proper biochemical signals—to transition off the promoter and generate a full-length transcript. In light of the relatively recent discovery that pol II stalling is widespread in metazoans [72,73], the term Pre-Elongation Complex (PEC) is proposed as a more broad descriptor of the factors involved in transcription initiation. These factors include TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Mediator, and pol II. Implicit in the PEC terminology is that pol II has not initiated productive elongation and might exist in a variety of functional states (e.g. a “closed” complex or a stalled pol II complex). Note that at this stage, however, the exact composition of PECs containing stalled pol II complexes has not been thoroughly examined.
Box 1, Figure I.
The transcription initiation machinery. Each PEC component is shown at a scale that approximates the relative sizes of each factor. *TFIIA can be proteolytically processed into 3 subunits; **the subunit composition of TFIID is variable.
Data from several laboratories suggested Mediator might play a role in post-recruitment activation of pol II. Using immobilized template assays responsive to the hepatocyte nuclear factor 4 (HNF4) transcription factor, the Roeder lab observed that HNF4 (which interacts directly with Mediator) was not required for pol II recruitment, but rather correlated with transcription activation [18]. These results suggested that HNF4—potentially via its interactions with Mediator—might promote post-recruitment activation of transcription. While studying Ets-like gene 1 (ELK1)-dependent activation of early growth response 1 (Egr1), the Berk lab observed that Mediator recruitment resulted in a 3-fold increase in pol II occupancy, but a 13-fold increase in transcription. This finding provided clear evidence that Mediator performs additional roles in transcription activation beyond just pol II recruitment [19].
So how might Mediator trigger post-recruitment activation of pol II? Although it is not known precisely how Mediator works within the PEC, the large size and shape of Mediator provides an extensive surface area that facilitates multiple protein–protein interactions. Accordingly, human Mediator has been shown to physically and/or functionally interact with most PEC components, including pol II [20–22], TFIIB [13], TFIID [23], TFIIE [13], and TFIIH [24]. Furthermore, as Mediator serves as a central scaffold about which the PEC assembles (Box 1), structural shifts within Mediator might work to control PEC structure and function. In support of this, a recent study in our laboratory revealed that specific structural shifts within Mediator—triggered by the p53 activation domain—were critical for activation of promoter-bound, stalled pol II enzymes [24]. This p53-Mediator structural shift not only correlated with pol II activation, it also affected TFIIH-dependent pol II CTD phosphorylation within the PEC. Thus, p53-directed structural shifts appear to coordinate the activity of Mediator, pol II, and TFIIH at the promoter. Although such structure-function links have only been shown with p53, similar conformational shifts are observed in other activator-bound Mediator structures (Figure 1), suggesting a general means by which activators might indirectly regulate pol II activity. Additional support for a Mediator structural shift controlling pol II promoter escape stems from follow-up studies of ELK1-dependent Egr1 activation [25]. In its unphosphorylated form, ELK1 can interact with Mediator to facilitate pol II recruitment to Egr1. However, transcription activation does not occur until ELK1 becomes phosphorylated, and this phosphorylation event alters and enhances ELK1–Mediator interactions. As postulated by Berk and co-workers, this modified activator–Mediator interface might be coupled to structural shifts within Mediator that trigger post-recruitment pol II activation [25].
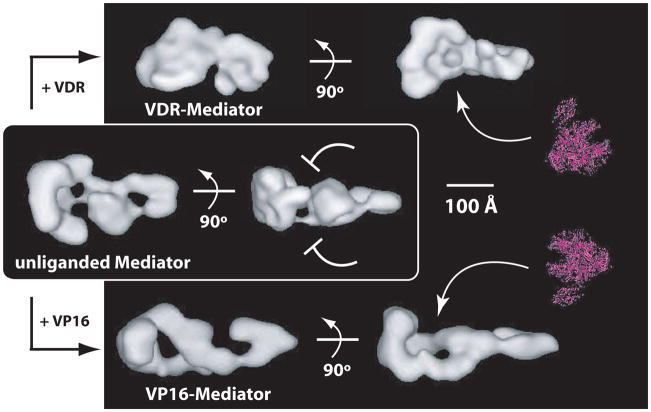
Figure 1.
A universal mechanism for activated transcription? Shown are Mediator structures without an activator bound (unliganded Mediator), or bound to the activation domain of the vitamin D receptor (VDR–Mediator) or VP16 (VP16–Mediator). The pol II enzyme is shown to scale in red. Note that activator binding causes major structural re-organization within Mediator and that VDR induces a distinct structural state relative to VP16. Despite the structural differences between VDR-Mediator and VP16-Mediator, a prominent, shared feature is a large “pocket” domain (arrows) that is of sufficient size and shape to accommodate the pol II enzyme. In fact, pol II binds at this site within yeast and human Mediator [24,27]. In contrast to the activator-bound structures, the unliganded Mediator structure lacks this pocket domain. Interestingly, the VDR–Mediator and VP16–Mediator structures can each strongly activate transcription [31,79], whereas the unliganded Mediator structure does not [24], indicating that activator-dependent formation of the pol II “pocket” domain within Mediator might represent a common means by which activators activate transcription. It is currently not known why the pocket domain forms in different orientations within Mediator (e.g. pol II binds from a different face with VDR–Mediator compared with VP16–Mediator). One simple explanation is that it could enable divergent transcription (sense and anti-sense) from gene promoters [68,69].
Biochemical studies with human Mediator link its occupancy at the promoter with PEC assembly [12]. Although structural information for the human PEC is far from complete, it is likely that Mediator represents a central scaffold around which the PEC assembles [26]. Much of the existing structural data for the Mediator–pol II assembly has been generated with yeast Mediator and yeast pol II. These studies indicate that pol II—and likely the rest of the PEC—assembles around the head module of Mediator [27,28]. The extensive interactions observed in the yeast Mediator–pol II assembly, coupled with well-established interactions between human Mediator and the pol II CTD [20], provide a clear means by which Mediator recruits and stabilizes pol II within the PEC. Paradoxically, Mediator can also block pol II incorporation into the PEC. Mediator interaction with the cyclin-dependent kinase 8 (CDK8) subcomplex (Figure 2) triggers a structural shift within Mediator that renders it incapable of simultaneously interacting with pol II [29]. Thus, Mediator can control even the composition of the PEC: Mediator can promote pol II incorporation to activate transcription or, when bound to the CDK8 submodule, Mediator can block pol II assembly and repress transcription.
Figure 2.
Structural differences between Mediator and CDK8-Mediator. Shown are different views of the Mediator (blue) or CDK8-Mediator structures (white), and also an overlay of the structures. Each structure is bound to the activation domain of VP16. The 600 kDa CDK8 submodule comprises the “foot” domain within CDK8-Mediator [29]. Upon binding to Mediator, the CDK8 subcomplex also induces a structural shift within the head/body region (arrows). Within Mediator itself, this region corresponds to the pol II binding site (i.e. the “pocket” domain that interfaces extensively with the pol II enzyme); however, upon binding the CDK8 submodule, this site becomes occluded and pol II does not bind [29].
The CDK8 submodule: negative and positive roles in transcription
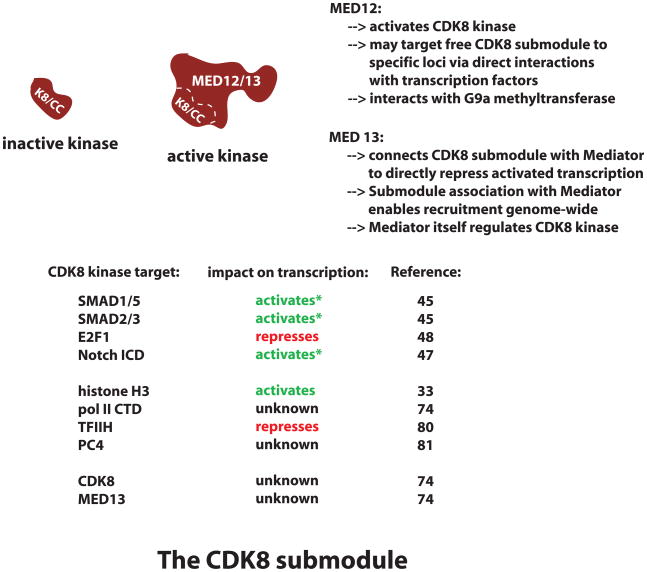
One of the most misunderstood aspects of human Mediator function relates to the CDK8 submodule, a 600 kDa assembly that can reversibly associate with Mediator and contains the proteins CDK8, cyclin C, MED12, and MED13. One reason for confusion is that early preparations of human Mediator (when Mediator was provisionally called TRAP, ARC, or DRIP) actually contained a mixture of Mediator and CDK8-Mediator [3,6–8,30]. Due to the presence of Mediator, these preparations exhibited co-activator function in reconstituted transcription assays. Only upon biochemical separation of CDK8-Mediator and Mediator itself was it established that Mediator was responsible for activator-dependent transcription, whereas CDK8-Mediator was unable to activate transcription [31]. Subsequent studies indicated the CDK8 submodule represses activated transcription by binding to Mediator and inhibiting its ability to recruit and activate pol II [29]. The Boyer lab identified another means by which the CDK8 submodule can repress transcription: through interactions with the G9a methyltransferase, which catalyzes histone H3K9 methylation, a repressive chromatin mark. G9a-dependent repression is particularly sensitive to the CDK8 submodule component MED12, which controls the recruitment of G9a to a subset of silenced neuronal genes in HeLa cells [32]. Notably, MED12 knockdown does not appear to impact global levels of H3K9 methylation, suggesting a gene-selective role for MED12 in these cells.
The results linking MED12 to the G9a methyltransferase provided a key link between CDK8-Mediator and histone modifications; another link was revealed upon isolation and biochemical characterization of a distinct CDK8-Mediator complex that contained the TRRAP and GCN5L polypeptides [33]. This “T/G-Mediator” complex directly modified chromatin templates, yet its chromatin-modifying activity does not simply result from the GCN5L acetyltransferase. Rather, CDK8 itself possesses intrinsic chromatin-modifying activity as a histone H3S10 kinase. In fact, CDK8 and GCN5L function synergistically within T/G-Mediator to phosphorylate and acetylate histone H3 (H3S10P/K14Ac) in vitro and in cells; CDK8 knockdown resulted in global reduction in H3 S10P/K14Ac levels, suggesting T/G-Mediator is a widespread regulator of this H3 mark [33]. Coupled H3 S10/K14 phosphorylation and acetylation correlates with activation of at least a subset of genes [34–37]. Thus, these data identified a biochemical role for CDK8 in the activation of transcription: as an H3S10 kinase, CDK8 likely helps to establish a chromatin environment favorable for transcription. Potentially, the chromatin-modifying activity of T/G-Mediator might function together with chromatin remodelers, such as SWI/SNF, to help remodel the promoter prior to transcription initiation. Given the existence of multiple H3S10 kinases in mammalian cells [38–43], it is currently unclear how many genes might rely upon CDK8-dependent H3S10 phosphorylation for their expression.
CDK8 has also been shown to function as a positive regulator at specific p53-regulated genes such as p21 and HDM2 [44]. Upon p53 activation by UVC, Mediator subunits occupy, but do not activate, the p21 promoter. By contrast, stimulation of p53 with the drug nutlin 3 strongly induces p21 expression. Mediator occupancy at p21 remains constant under these different conditions (UVC or nutlin 3), yet a significant increase in CDK8 submodule occupancy is observed only after nutlin 3 treatment [44]. Interestingly, changes in CDK8 occupancy at p21 (UVC vs. nutlin 3 treatment) also correlate with changes in occupancy of other PEC factors. These results suggest the CDK8 submodule might promote PEC assembly under some conditions, perhaps through an indirect mechanism involving its kinase activity.
Additional evidence that CDK8 can serve as a co-activator derives from studies that focused on SMAD-dependent transcriptional activation in response to bone morphogenetic protein (SMAD1/5) or transforming growth factor beta (SMAD2/3) signaling. CDK8-dependent phosphorylation of the linker region within SMAD1/5 or SMAD2/3 complexes appears to activate these transcription factor assemblies [45]. However, CDK8-dependent phosphorylation also targets SMAD complexes for proteasomal degradation, thereby also restraining transcriptional activation [45,46]. A similar mechanism appears to operate with transcriptional activation by the Notch intracellular domain (ICD) enhancer complex. CDK8 phosphorylation of the Notch ICD activates this assembly while concomitantly targeting it for degradation [47]. CDK8 also phosphorylates the E2F1 transcription factor; unlike the SMADs or the Notch enhancer complex, however, CDK8-dependent modification is linked to repression of E2F1 activity [48]. As E2F1, in turn, represses β-catenin activity, this observation helped to identify CDK8 as a key colorectal cancer oncoprotein that regulates the β-catenin pathway [48,49]. Importantly, oncogenesis is linked specifically to CDK8 kinase activity, as kinase-dead CDK8 mutants cannot transform cells or activate β-catenin target genes. These examples (SMAD1/5, SMAD2/3, Notch ICD, E2F1) reveal that the kinase activity of CDK8 is capable of positive or negative regulation of transcription factor activity (Box 2). The number of functionally diverse targets for CDK8 suggests that regulation of its kinase activity must be strictly enforced. Recent insights regarding CDK8 kinase regulation are described in Box 2.
Finally, a role for the CDK8 submodule in regulating transcription elongation was recently reported [50]. Interestingly, this novel regulatory function for CDK8 requires positive transcription elongation factor b (P-TEFb). The CDK8 submodule appears to coordinate P-TEFb loading and activity at a number of serum-response genes, including FOS, EGR1, EGR2, and EGR3. At these genes, CDK8 helps control pol II CTD phosphorylation and elongation, most likely by regulating P-TEFb kinase activity. Significantly, these results provide the first clear evidence that CDK8 can impact pol II CTD phosphorylation (albeit perhaps indirectly, through P-TEFb) in human cells. Indeed, cellular knockdown of CDK8 yielded no detectable change in global pol II CTD phosphorylation levels (S5P or S2P), suggesting that the CDK8-dependent effect on pol II CTD phosphorylation is gene-specific [50]. Precisely how CDK8 affects P-TEFb-directed pol II CTD phosphorylation at serum response genes remains unknown; regulation might occur via CDK8-dependent phosphorylation of P-TEFb or simply by CDK8 subcomplex–P-TEFb association. CDK8 and P-TEFb might also cooperatively regulate SMAD1/5 or SMAD 2/3 activity and stability, as both CDK8 and P-TEFb can modify the same regulatory sites within SMAD transcription factor complexes [45].
Given the multiple and functionally diverse roles for the CDK8 submodule in gene regulation, a model consistent with the existing data involves the CDK8 submodule serving as a checkpoint for transcription (Figure 3). A general feature of this model is that CDK8-Mediator provides a platform from which a variety of functionally divergent outcomes can be initiated. The reversible association of the CDK8 submodule with Mediator provides a facile means to regulate pol II initiation events, perhaps even resulting in long-term silencing of transcription. The ability of CDK8 to regulate transcription factor stability also implies a checkpoint role in that it would limit the duration that an activator remains stably bound at the promoter. These two distinct regulatory mechanisms result in similar functional outcomes in that each serves to check and restrain activated transcription. In each circumstance, re-activation of transcription might involve i) dissociation of the CDK8 submodule from Mediator, enabling subsequent recruitment of another pol II enzyme, or ii) subsequent binding of a newly-translated transcription factor to the promoter.
Figure 3.
A framework for understanding how Mediator might function to regulate transcription. A) CDK8-Mediator provides a platform for multiple, functionally divergent events. Upon binding the TRRAP and GCN5L polypeptides, this assembly (provisionally called T/G-Mediator) can modify chromatin templates [33], perhaps to facilitate recruitment of additional activities such as chromatin remodeling complexes or STAGA [52], that help establish an environment favorable for transcription and PEC assembly. Note that CDK8 might also phosphorylate transcription factors (green) at this stage, which might coordinately activate the transcription factor and target it for subsequent degradation [45,47]. B) At some point prior to full PEC assembly, the CDK8 submodule (red) and perhaps other bound factors (e.g. TRRAP/GCN5L) must dissociate. Mediator can then serve as the central scaffold about which the PEC assembles. C) Further remodeling of the promoter region is required, together with assembly of the PEC (Mediator, pol II, TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH). The precise mechanisms by which this occurs are not known. At this stage, pol II can exist in a number of possible functional states, including a stalled state in which pol II has engaged the promoter but has not initiated elongation [70]. D) With the appropriate signal (e.g. Mediator structural shift, post-translational modifications, etc.), pol II clears the promoter and initiates productive elongation. The remaining scaffold assembly can then recruit additional pol II complexes and re-initiate transcription. Alternatively, the CDK8 submodule could bind at this stage to block re-initiation [29]. At a later time, CDK8 might dissociate from Mediator to again allow re-activation of transcription (red arrow). At a subset of genes, the CDK8 submodule might also positively impact pol II elongation events via functional interactions with elongation factors such as P-TEFb [50]. E) If CDK8-Mediator occupancy is sustained (i.e. dissociation of the CDK8 submodule no longer occurs), activated transcription can be shut down. At this stage, dis-assembly of other PEC factors (e.g. TFIID, TFIIH) might occur and/or transcription factors might dissociate from the promoter and be degraded. The chromatin architecture at the promoter might also be altered, returning the locus to a more basal state. F) Long-term repression might be further enforced via CDK8-Mediator interactions with chromatin remodeling/modifying factors such as the G9a methyltransferase [32]. Note that many regulatory factors are not included in this scheme for clarity and also to better highlight Mediator-specific regulatory functions; this schematic also does not formally propose that each regulatory event shown occurs at every protein-coding gene.
Mediator: a global regulator of transcription with gene-selective functions
Among the diverse functions thus far described for the human Mediator complex, several likely operate at most, if not all, protein-coding genes. These general regulatory roles probably include i) pol II recruitment and activation, ii) coordination of PEC assembly, iii) control of TFIIH-dependent pol II CTD phosphorylation within the PEC, and iv) sustained or transient repression of transcription initiation via Mediator–CDK8 submodule interactions. Recent studies have also identified physical or functional interactions between human Mediator, STAGA (a histone acetyltransferase complex), or the co-activator p300 [51,52]. Because both p300 and STAGA help regulate a large percentage of genes, their interactions with Mediator also likely reflect widespread roles in gene regulation.
Recent work has defined a growing number of gene-selective regulatory roles for Mediator [53,54], with many involving the Mediator subunit MED1. MED1 represents the largest subunit within Mediator (220 kDa) and MED1 is a general target for nuclear receptors. Several labs have shown that MED1 can reversibly associate with the human Mediator complex [22,55]. Notably, Fondell and co-workers observed that MED1 can be phosphorylated by extracellular signal-regulated kinase (ERK), which stabilizes MED1 within the Mediator complex; Mediator complexes containing phosphorylated MED1 show enhanced ability to activate transcription at thyroid hormone receptor (TR)-regulated promoters [56,57]. The Roeder lab identified a physical and functional cooperativity between MED1 and the PGC-1α co-activator that was critical for TRα/RXRα-dependent uncoupling protein 1 (UCP1) expression [58]. MED1 also appears to function together with the CCAR1 co-activator to express a number of estrogen receptor (ER) and glucocorticoid receptor (GR) target genes [59]. In addition, MED1 plays a key role in directing expression of the TR-regulated CRABP1 gene and the AR-regulated PSA gene. Interestingly, in both of these cases, MED1 was not only required for gene expression but was also important for formation of a chromatin loop between the enhancer and the proximal promoter [60,61]. Later, a similar link between MED1-dependent gene expression and chromatin loop formation was made during analysis of PDK4 (a PPARδ target gene) activation in HEK293 cells [62]. The results at the nuclear receptor-regulated PSA, CRABP1, and PDK4 genes imply a novel role for Mediator as a chromatin architectural factor within the nucleus. Further studies, however, will be required to confirm this hypothesis.
Combined with the other gene-selective roles described in the previous sections (e.g. MED12–G9a regulation of a subset of neuronal genes; CDK8-dependent co-activation of serum response genes) it is becoming clear that despite being generally required for expression of protein-coding genes, Mediator activity is controlled in different ways by different co-regulatory factors at different genes.
Concluding remarks
Although far from comprehensive, a number of areas for future studies are outlined below. i) As promoter-proximal pol II stalling appears to be a common intermediate in transcription activation, it will be important to define the composition of human PECs that contain stalled polymerases. Although Mediator almost certainly remains promoter-associated in this context (Mediator has already been linked to activation of stalled pol II within the PEC), it is not known whether other GTFs (e.g. TFIIB) similarly occupy PECs with stalled pol II. ii) How exactly does Mediator control pol II activity at the promoter? Activator-induced structural shifts within Mediator clearly play a role, but how do these structural shifts translate into pol II activation? What Mediator subunits are specifically involved in pol II interactions? iii) How might Mediator functionally interact with other factors within the PEC? Mediator controls the kinase activity of TFIIH; however does it also control TFIIH helicase function? And what about the other GTFs? As a central scaffold around which the rest of the transcription initiation machinery assembles, it is likely that Mediator coordinates multiple activities within the PEC. iv) Different activators can induce distinct structural shifts within Mediator upon binding the complex (e.g. Figure 1). Do these structural shifts unlock potential gene-specific functions? v) The CDK8 submodule was recently implicated in controlling elongation at serum response genes by influencing the activity of P-TEFb [50]. Does Mediator and/or the CDK8 submodule help regulate other processes in transcription elongation? vi) What factors control CDK8 submodule–Mediator association at the promoter? How dynamic is this association at actively-transcribing genes? As CDK8 submodule → Mediator interactions prevent transcription initiation (whereas dissociation of the CDK8 submodule from Mediator enables productive Mediator–pol II interactions), it is important to identify how their association is regulated in human cells. Past work indicates that poly (ADP-ribose) polymerase 1 (PARP1) can release the CDK8 submodule from Mediator, but this appears to affect only a subset of genes [63]. vii) Does Mediator help control formation of gene loops in human cells? Recent data point to a possible role for the Mediator subunit MED1 in this process. However, the genes in which loop formation was evident had a general MED1 requirement for Mediator recruitment. Thus, the apparent MED1 dependence may simply reflect reduced Mediator occupancy at these genes. viii) Does CDK8 phosphorylate a wide variety of human transcription factors? If so, what are the functional consequences of these modifications? ix) Mediator contains 26 subunits and none of these possess any predicted enzymatic function, based upon their sequences. Does Mediator harbor any enzymatic activities? x) Ablation of the Med17 subunit is known to make yeast Mediator structurally unstable, and conditional MED17 knockouts are unable to express protein-coding genes on a genome-wide scale [64]. However, a handful of genes are expressed under these conditions; their activation is not Mediator-independent, but instead required only a subset of Mediator proteins that evidently formed a stable sub-assembly [65,66]. Recent studies suggest that in differentiated cells, human Mediator adopts a far simpler state (perhaps 6–8 subunits), whereas within cancer cells or stem cells, Mediator appears to operate as a 26-subunit assembly [67]. How might the structure and function of Mediator be altered in distinct cell types?
Box 2. How is CDK8 kinase activity regulated?
Given its oncogenic properties and its potential widespread role as a controller of transcription factor activity and/or stability, it is important to understand how CDK8 kinase activity might be regulated in human cells. Although the mechanisms that regulate the CDK8 kinase remain poorly-understood, a few regulatory factors have been identified. One major regulator of CDK8 kinase activity is the MED12 protein; in fact, MED12 appears to be required for CDK8 kinase activity [74]. This key biochemical role for MED12 has several mechanistic implications. For one, it likely ensures strict control over CDK8 kinase targets. MED12 dimerizes with another 250 kDa protein—MED13—which, together with CDK8–cyclin C, comprises the 600 kDa CDK8 submodule. Although relatively little is known about the MED12 and MED13 proteins, it is evident these large polypeptides enable CDK8 submodule binding to specific nuclear targets. For instance, MED12 is known to interact with multiple different transcription factors [75–78], and MED13 is responsible for targeting the CDK8 submodule to Mediator itself [29]. Thus, a MED12 requirement for CDK8 kinase activity ensures CDK8 becomes active only within the context of the CDK8 submodule or CDK8-Mediator. This likely prevents the CDK8–cyclin C dimer from promiscuous, unregulated substrate modification in cells. Another regulator of the CDK8 kinase is Mediator itself: CDK8 cannot phosphorylate chromatin templates unless it is Mediator-associated, whereas free histone octamers can be modified by either CDK8-Mediator or the free CDK8 submodule [74]. Interestingly, CDK8 phosphorylates itself, and this or other post-translational modifications might also control CDK8 function. The varied and apparently widespread roles for the CDK8 kinase suggest many other mechanisms exist to control its activity and substrate specificity.
Box 2, Figure I.
MED12 and MED13 subunits
Box 2, Table I.
The CDK8 submodule
| CDK8 kinase target |
Impact on transcription |
Refs |
|---|---|---|
| SMAD1/5 | activatesa | 45 |
| SMAD2/3 | activatesa | 45 |
| E2F1 | represses | 48 |
| Notch ICD | activatesa | 47 |
| Histone H3 | activates | 33 |
| pol II CTD | unknown | 74 |
| TFIIH | represses | 80 |
| PC4 | unknown | 81 |
| CDK8 | unknown | 74 |
| MED13 | uknown | 74 |
CDK8-dependent phosphorylation correlates with both activation and degradation of these factors.
Acknowledgments
I thank Dr. Matthew Knuesel for assistance with Figure 2. The Taatjes lab is grateful for support from the NIH, NCI, the American Cancer Society, the American Heart Association, and the Ellison Medical Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thompson CM, et al. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y, et al. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 3.Fondell JD, et al. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 5.Toth-Petroczy A, et al. Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer TG, et al. Mammalian Srb/Mediator complex is targeted by adenovirus E1a protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 7.Naar AM, et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 9.Ryu S, et al. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 10.Mittler G, et al. Novel critical role of a human mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baek HJ, et al. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantin GT, et al. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc Natl Acad Sci U S A. 2003;100:12003–12008. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek HJ, et al. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 14.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 15.Andrau J, et al. Genome-wide location of the coactivator Mediator: binding without activation and transient cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Zhu X, et al. Genome-wide occupancy profile of Mediator and the srb8-11 submodule reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik S, et al. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Naar AM, et al. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes & Development. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Malik S, et al. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson KM, et al. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes & Development. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer KD, et al. p53 activates transcription by directing structural shifts within Mediator. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1816. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balamotis MA, et al. Complexity in transcription control at the activation domain-Mediator interface. Sci Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chadick JZ, Asturias FJ. Structure of eukaryotic Mediator complexes. TIBS. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Davis JA, et al. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 28.Takagi Y, et al. Head module control of Mediator interactions. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Knuesel MT, et al. The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes & Development. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun X, et al. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 31.Taatjes DJ, et al. Structure, Function, and Activator-Induced Conformations of the CRSP Coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 32.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer KD, et al. Cooperative activity of CDK8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 35.Clayton AL, et al. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre B, et al. Phosphorylation of histone H3 is functionally linked to retinoic acid receptor beta promoter activation. EMBO Rep. 2002;3:335–340. doi: 10.1093/embo-reports/kvf066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson S, et al. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell. 2001;8:1231–1241. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- 38.Hirota T, et al. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 39.Sassone-Corsi P, et al. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto Y, et al. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 41.Zippo A, et al. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 42.Soloaga A, et al. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anest V, et al. A nucleosomal function for IκB kinase-α in NFκB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 44.Donner AJ, et al. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-b pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao S, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fryer CJ, et al. Mastermind recruits CycC:Cdk8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Morris EJ, et al. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donner AJ, et al. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black JC, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, et al. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W, et al. Med14 and Med1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20:560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 54.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 55.Taatjes DJ, Tjian R. Structure and Function of CRSP/Med2: a promoter-selective transcriptional co-activator complex. Mol Cell. 2004;14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Belakavadi M, et al. MED1 phosphorylation promotes its association with Mediator: implications for nuclear receptor signaling. Mol Cell Biol. 2008;28:3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandey PK, et al. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol Cell Biol. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen W, et al. Dynamic interactions and cooperative functions of PGC-1α and MED1 in TRα-mediated activation of the brown fat-specific UCP-1 gene. Mol Cell. 2009;35:755–768. doi: 10.1016/j.molcel.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JH, et al. CCAR1, a key regulator of Mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q, et al. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 61.Park SW, et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Degenhardt T, et al. Population-level transcription cycles derive from stochastic timing of single-cell transcription. Cell. 2009;138:489–501. doi: 10.1016/j.cell.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 63.Pavri R, et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 64.Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci U S A. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- 66.McNeil JB, et al. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deato MDE, et al. MyoD targets TAF3/TRF3 to activate Myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 72.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knuesel MT, et al. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of Mediator. Mol Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S, et al. Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 76.Tutter AV, et al. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284:3709–3718. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- 77.Zhou R, et al. SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Research. 2002;30:3245–3452. doi: 10.1093/nar/gkf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gwack Y, et al. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi’s sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol Cell Biol. 2003;23:2055–2067. doi: 10.1128/MCB.23.6.2055-2067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taatjes DJ, et al. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat Struct Mol Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 80.Akoulitchev S, et al. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 81.Gu W, et al. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]