Abstract
In the course of work aimed at the discovery of new pharmaceutical lead compounds from marine bacteria, a lipophilic extract of the bacterium Pseudoalteromonas rubra displayed significant cytotoxicity against SKOV-3, a human ovarian adenocarcinoma cell line. Bioassay-directed fractionation of this extract resulted in the isolation of a series of known and new prodiginine-type azafulvenes. The structure of the major metabolite was elucidated by interpretation of spectroscopic data as a 2-substituted prodigiosin, which we named 2-(p-hydroxybenzyl)prodigiosin (HBPG).
As part of a screening program aimed at the discovery of new lead compounds for the treatment of infective and neoplastic diseases, we isolated a bacterium from the surface of a nudibranch recovered by SCUBA in waters off Oahu, Hawaii, and from a sponge, Mycale armata, collected in shallow waters. Both isolates were shown to be Pseudoalteromonas rubra on the basis of sequence of the 16S rDNA ITS linker gene sequence. Extracts of this bacterium grown in marine broth consistently displayed broad spectrum biological activity against Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), methicillin-resistant Staphylococcus aureus (ATCC 43300), Candida albicans as well as the human ovarian adenocarcinoma cell line SKOV-3 (ATCC HTB-77). Therefore, this isolate was selected for scale-up and isolation of the active component. A brief optimization study of fermentation conditions indicated that agitation of the culture was detrimental and that a large surface to volume ratio was beneficial to production of the active component. The active principle was located in both the cell pellet and the medium. After extraction with a 2:3 mixture of acetone/Et2O, a combination of silica-gel flash chromatography and RP-HPLC yielded 1 in 1.3 % yield based on dry extract mass.
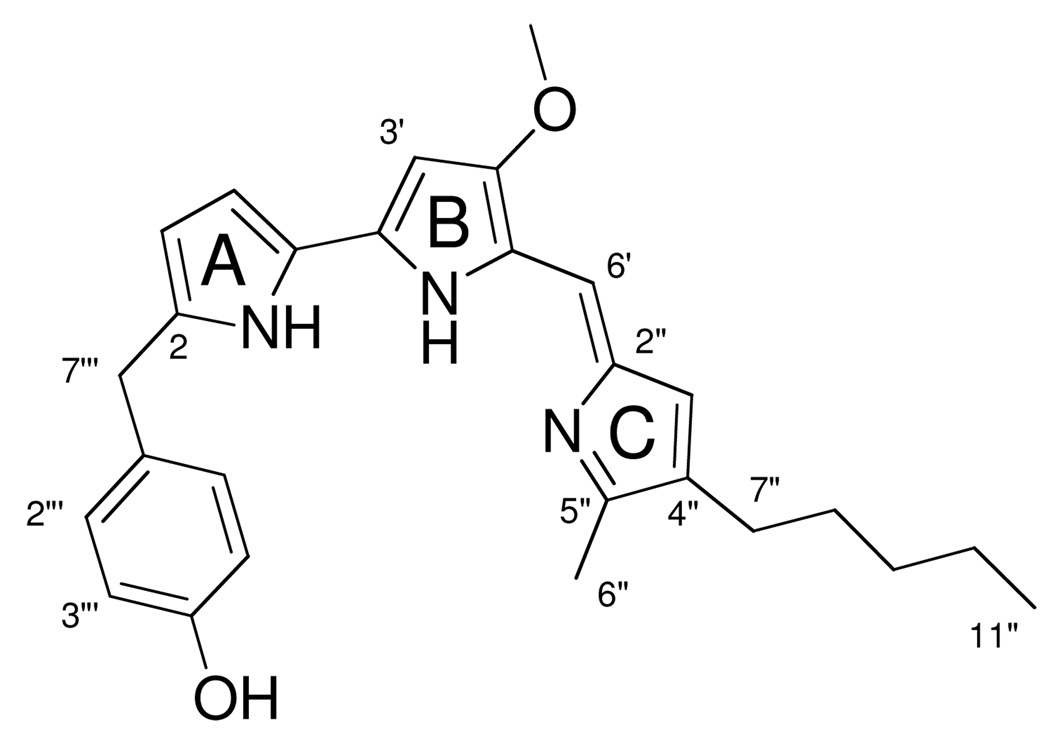
The active principle was isolated as a dark red, optically inactive solid. The molecular formula was established by high-resolution positive ion APCI-TOF MS in conjunction with 1H and 13C NMR spectroscopic data. A pseudomolecular [M+H]+ ion was observed at m/z = 430.2489, consistent with a molecular formula of C27H32N3O2 (calc. 430.2489, Δ = 0.0 ppm). Support for this molecular formula came from the analysis of 13C NMR and HSQC spectra recorded in CDCl3, which revealed the presence of 25 carbon resonances, comprising resonances for three methyl, five methylene, seven methine and ten quaternary carbon atoms. An inspection of the 1H NMR spectrum of 1 indicated the presence of a p-substituted phenyl ring. The resulting degeneracy explains the difference in carbon count between the number of the resonances in the 13C NMR spectrum and the carbon count suggested by the mass spectrum. The molecular formula requires 14 degrees of unsaturation, of which ten are accounted for by 19 resonances between 166 and 92 ppm in the 13C NMR spectrum. This suggests that the compound contains four rings.
Substructures were assembled on the basis of the interpretation of COSY spectra in conjunction with gHMBC data. In the downfield region of the 1H NMR spectrum two doublets were evident, each integrating for two protons and coupling to each other. Interpretation of gHMBC correlations from these resonances and chemical shift considerations allowed assembly of an oxygen-bearing p-substituted phenyl ring connected to a methylene group that was represented by a two-proton singlet resonating at δH 4.02.
Another easily interpretable feature of the 1H NMR spectrum was the three-proton singlet resonating at δH 3.97, which was assigned to an O-methyl group (δC 58.67) that was linked to a quaternary carbon atom resonating at δC 165.6 on the basis of gHMBC data.
Further analysis of features in the upfield portion of the spectra suggested the presence of a pentyl chain, which likely was joined to an aromatic system as indicated by the chemical shifts of the resonances for C-6″(δC 25.31) and the protons H-6″(δH 2.38). Also present was an allylic methyl group (δH 2.52; δC 12.30), which was likely attached at the 2-position of a heterocycle as suggested by the 13C NMR chemical shift. HMBC data suggested that this methyl and the pentyl chain were situated at vicinal carbon atoms.
At this point all but ten carbon atoms had been assigned whereas the three nitrogen atoms as well as three rings still remained to be accounted for. These structural elements seemed at first glance to be insufficient to explain the intense red color of 1 (λmax = 551nm, log ε= 5.0). However, both the position of the absorption maximum, its intensity and the presence of the pentyl chain as well as the methyl group at vicinal positions of a heterocyclic ring were reminiscent of prodigiosin, a member of the prodiginine class of pigments, which are characterized by a 4-methoxypyrrolyldipyrromethene. Chemical shift comparisons with literature values indeed bore this out (ΔδH ≤ 0.15 for all but H-2 (missing) and H-3; ΔδC ≤ 1.7 for all but C-2). In the parent prodigiosin H-3 appears as a broad ddd resonance at δ 6.37, whereas in 1 the equivalent resonance is a broad doublet at δ 6.02 and that for H-2 of prodigiosin is absent. This allowed the site of attachment of the p-hydroxybenzyl group to be identified unequivocally as C-2 (A-ring). The structure of 1 is therefore as shown in Figure 1.
Figure 1.
Structure of 2-(p-hydroxybenzyl)prodigiosin (HBPG).
The red-pigmented fraction also contained 4″-(n-hexyl)prodigiosin 2 and 4″-(n-heptyl)prodigiosin1 3, two prodiginines with elongated alkyl chains. Compound 2 had previously not been isolated, although its existence had been suggested on the basis of TLC and MS.2 Subsequent LC-MS/MS studies had further supported the proposed structure.3 In the present work sufficient amounts of 2 were isolated to allow full structural characterization by NMR spectroscopy.
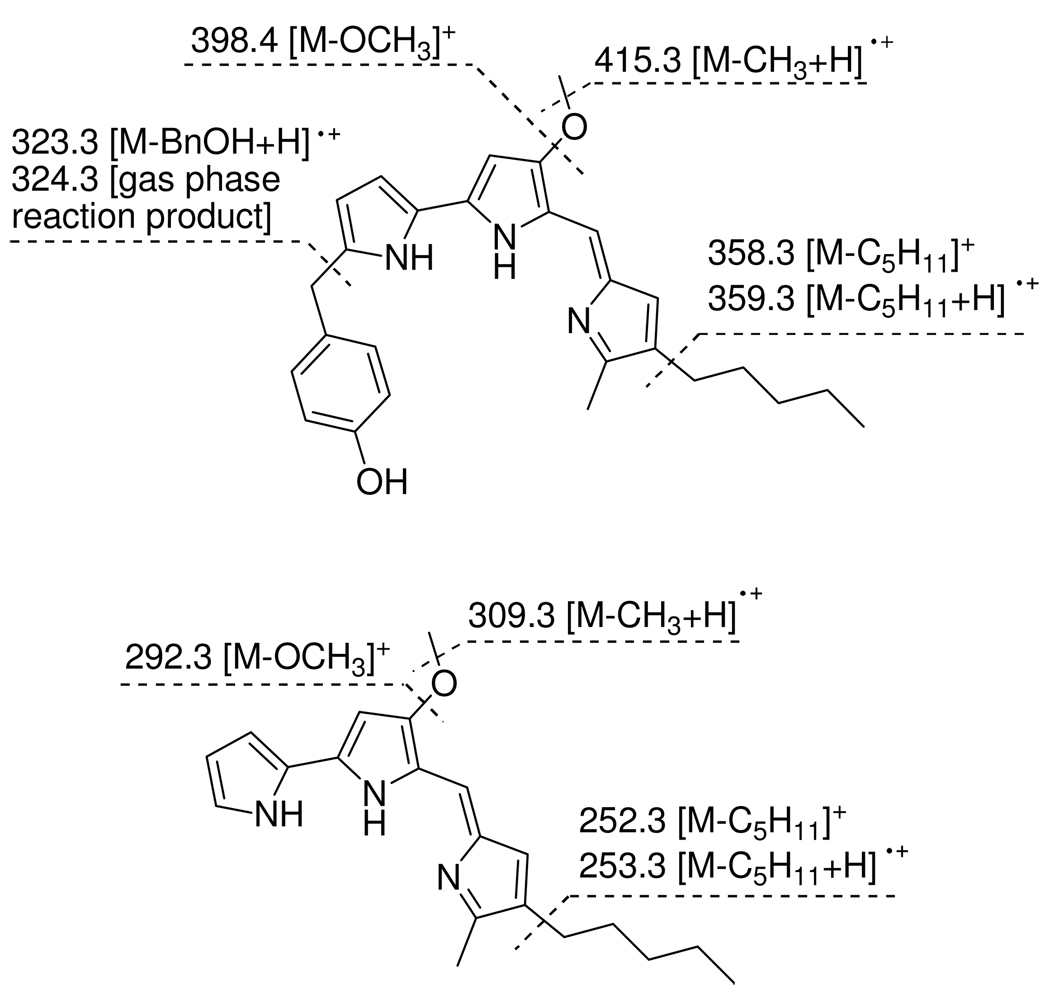
Since our LCMS data suggested the presence of additional prodiginines, we used ESI-MS/MS to examine the fragmentation patterns of 1, 2 and 3 as shown in Figure 2. The non-hydroxybenzylated series of compounds follow a distinctive fragmentation pattern characteristic for the class of prodigiosins with elongated alkyl chains, in which methyl, methoxy and alkyl loss are observed. In addition to these characteristic fragmentations compound 1 shows loss of the p-hydroxybenzyl group at C-2 by two different mechanisms, one involving a gas phase reaction leading to the major peak at m/z = 324.3, the other a collision-induced dissociation producing the minor peak at m/z = 323.3. With these data in hand an additional six prodiginines, namely cycloprodigiosin, 2-(p-hydroxybenzyl) cycloprodigiosin, 4″-(n-butyl)prodigiosin, 4″-(n-butyl)-2-(p-hydroxybenzyl)prodigiosin, 4″-(n-hexyl)-2-(p-hydroxybenzyl)prodigiosin and 4″-(n-heptyl)-2-(p-hydroxybenzyl)prodigiosin were detected by LC-APCI-HRMS and LC-ESI-MS/MS. All of these compounds display the UV/vis-maxima characteristic for the prodigiosene core. Since sufficient amounts of these minor components could not be isolated for NMR analysis MS/MS spectra were recorded to confirm their structures on the basis of the previously established fragmentation patterns (Figure 2).
Figure 2.
ESI-MS/MS fragmentation patterns of 2-(p-hydroxybenzyl)prodigiosin (HBPG) and prodigiosin (PG).
Prodiginine-type compounds display a variety of biological activities on specific cellular targets ranging from inhibition of protein phosphatase, protein kinase and topoisomerase I to DNA cleavage in the presence of copper ion as well as H+/Cl− symport.4 In addition, prodiginines induce apoptosis through caspase activation and independently of caspases through the mitochondrial apoptosis-inducing factor (AIF).5 They also display immunosuppressive effects on T-cells by blocking IL-2-dependent proliferation through down-regulation of the IL-2Rα receptor.6
Among the known natural prodiginines 1 is unusual inasmuch as it is bearing a carbon substituent at C-2 of ring A, although a few compounds with this substitution have been accessed through total synthesis.7 While the natural product nonylprodigiosin is substituted at C-2 of ring A, it is a cyclic molecule and therefore belongs to a different structural subgroup within the prodiginine class of metabolites.8 It was therefore of interest to compare the bioactivity of 1 with that of prodigiosin itself. In side-by-side comparisons 1 was indistinguishable from prodigiosin in its activity as an inhibitor of human topoisomerase I in the standard assay. Similarly, both 1 and prodigiosin displayed identical IC50 values of 1.3 µM against SKOV-3, a human adenocarcinoma cell line, in a standard cytotoxocity assay using the sulforhodamine (SRB) protocol.
Another point of interest of the structure of 1 is its place in the accepted biosynthetic scheme for the prodiginines. On the basis of feeding experiments and more recent biochemical investigations proline is known to be the precursor of ring A of prodigiosin and C-2 of ring B.9,10 There are two conceivable ways in which the p-hydroxybenzyl moiety may be introduced into the prodiginine core structures: modification of a completed prodiginine skeleton by way of a Mannich condensation once the oxidation of the pyrrolidine ring to the pyrrole has begun. Alternatively, the peptide synthetase might accept both proline and a proline bearing a p-hydroxybenzyl substituent at C-5 as a substrate. The latter could arise by condensation of tyrosine with 2-oxosuccinic acid followed by decarboxylation and reduction.
In conclusion, we have described a first member of a new class of 2-substituted prodigiosins. While this substitution pattern has interesting implications for the established biosynthetic scheme for the prodigiosins, the biological activity of the compound is indistinguishable from the parent with respect to cytotoxicity and topoisomerase I inhibition.
Expremintal Section
General experimental procedures
UV/vis spectra were recorded on a Beckman DU 7400 spectrophotometer. IR spectra were obtained on a ThermoElectron Corp. Nicolet 380 FT-IR system. NMR spectra were recorded in a Shigemi tube on a VARIAN INOVA 500 instrument equipped with a 3 mm microprobe. Positive ion HR-APCI-TOFMS data were recorded on an Agilent 6100 TOFMS instrument equipped with an Agilent 1100 chromatography module and an Agilent APCI source. MS/MS spectra were recorded on a Thermo Finnigan LCQ instrument equipped with an Agilent 1100 solvent delivery system.
Isolation and Taxonomy of Pseudoalteromonas rubra
Isolate CMMED 294 was isolated from a small piece of sponge, most likely Mycale armata, collected while snorkeling approximately one meter from the surface in Kaneohe bay off Oahu, HI. The sponge was sampled with a 10 µL bacterial inoculation loop and the recovered liquid was streaked onto a bacterial agar plate (MA2216). After incubation for 48 hours the plate was examined for growth of bacterial colonies. Colonies were picked and re-streaked onto clean MA2216 plates for isolation.
CMMED 418 - This isolate was recovered from a clear, gilled flatworm collected via SCUBA at a depth of approximately 85 feet. The animal attached to the YO257 shipwreck off Waikiki on Oahu, HI was surface sampled using a 10 µL bacterial loop. Further work-up was performed as above.
Both isolates are deposited under the accession number CMMED 294 and CMMED 418 in the culture collection of the Center for Marine Microbial Ecology and Diversity at the University of Hawaii.
Taxonomy
DNA was isolated from a confluent culture of the isolates in marine broth. An aliquot was removed, diluted 1:100 in distilled water and boiled at 98°C for 5–8 min. Amplification of the16S rDNA gene was carried out in duplicate using primers (Biosynthesis Inc.) 8f and 543r where r and f indicate reverse and forward, respectively, and numbers are based on the E. coli gene.11 The amplification was performed in 30 cycles of denaturation at 94 °C for 180 sec, annealing at 60°C for 60 sec and extension at 72 °C for 60 sec. The product was purified using the Qiagen PCR Purification Kit (Qiagen Inc.) in accordance with the manufacturer's instructions. Sequencing was performed at the Greenwood Molecular Biology Facility at the University of Hawaii. Sequences were analyzed, aligned and combined by using Sequencher version 4.8 (Gene Codes Co.). A NCBI and GenBank database search using the BLAST algorithm12 for highly similar sequences identified the culture as belonging to the Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas. Within the Pseuodalteromonas genus the culture was shown to be a 99.8% match to Pseudoalteromonas rubra ATCC 29570T (X82147). Furthermore, the culture closely resembled the previously described phenotypic characteristics of Pseudoalteromonas rubra.13
Isolation of 1
P. rubra was grown as still cultures at RT in 20 3 L Fernbach flasks containing 1 L marine broth (MA2216) resulting in a surface to volume ratio of approximately 300 cm2/L. After a 5-day growth period the red-pigmented cultures were extracted with a mixture of acetone/Et2O (2:3). The combined extract was concentrated in vacuo to yield 103 mg of dry solid. Si flash-chromatography of the extract with a stepwise gradient from EtOAc/hexanes/MeOH (100:100:1) to EtOAc/MeOH (100:1) yielded 39 mg of a red-pigmented, cytotoxic fraction. RP-C8 HPLC with a linear gradient of from 70 to 80% MeOH/H2O (w/ 0.1% TFA) gave 17.6 mg (17% dry weight) of pure prodigiosin, and compounds 1–3 as dark-red solids with masses of 1.3 mg (1.3%), 2 mg (2%), and 0.4 mg (0.4%) (all as w/w dry extract mass), respectively.
Cytotoxicity
The in vitro sulphorhodamine B (SRB) assay for cytotoxicity utilizing the SKOV-3 (ATCC HTB-77) human ovarian adenocarcinoma cell line was used for comparative analysis of the activity of prodigiosin against the new 2-(p-hydroxybenzyl)prodigiosin. Plates were inoculated at 5×105 cells per well at a volume of 200 µL and incubated overnight at 37 °C. Standard 2-fold sample dilutions were then loaded in triplicate, and incubated for an additional 48 hours at 37 °C. The cells were then fixed to the plate, dyed, and the IC50 values were determined with a Multiscan MCC/340 (Thermo Fisher Scientific Inc.) at 595 nm absorbance.
2-(p-Hydroxybenzyl)prodigiosin (1)
UV/vis (MeOH, 0.1%TFA) λmax (log ε) 551 (5.0); 522 (4.4); 398 (4.1); 379 (4.1); 304 (4.2); 283 (4.1) nm; IR (NaCl) νmax 3232, 2955, 2926, 2855, 1673, 1631, 1609, 1547, 1536, 1514, 1443, 1413, 1365, 1324, 1261, 1202, 11356, 1046, 975, 838, 787 cm−1; 1H-NMR (CDCl3, 500 MHz) δ[ppm] (carbon position, integration, multiplicity; J[Hz]) 7.20 (3″′/5″′, 2H, d; 8.1), 6.87 (6′,1H, s), 6.81 (4, 1H, brd; 3.5), 6.79 (2″′/6″′, 2H, d; 8.3), 6.63 (3″,1H, brs), 6.02 (3, 1H, brd, 3.5), 6.00 (3′, 1H, s), 4.01 (7″′, 2H, s), 3.97 (OMe, 3H, s), 2.52 (6″, 3H, brs), 2.38 (7″, 2H, t; 7.6), 1.53 (8″, 2H, m), 1.34 (10″, 2H, m), 1.31 (9″, 2H, m), 0.89 (11″, 3H, t; 7.1); 13C NMR (125 MHz, CDCl3) δ[ppm] (carbon position) 165.6 (4′), 154.4 (1″′), 147.5 (2′), 145.6 (5″), 143.1 (2), 130.5 (4″′), 130.1 (3″′/5″′), 127.9 (4″), 127.5 (3″), 124.9 (2″), 121.3 (5), 121.0 (5′), 118.5 (4), 115.5 (2″′/6″′), 114.8 (6′), 110.8 (3), 92.6 (3′), 58.7 (OCH3), 33.7 (7″′), 31.4 (9″), 29.9 (8″), 25.3 (7″), 22.5 (10″), 14.1 (11″), 12.3 (6″); HR-APCI-TOFMS [M+H]+ m/z = 430.2489 (calcd for C27H32N3O2, 430.2489); ESI MS/MS m/z = 430 ([M+H+]+ 15%), 415 (45%), 398 (4%), 358 (4%), 324 (100%), 323 (47%), 309 (3%), 266 (4%).
4″-(n-Hexyl)prodigiosin (2)
UV/vis (MeOH, 0.1%TFA) λmax (log ε) 539 (4.9), 512 (4.5), 390 (3.7); 372 (3.8); 300 (3.85) nm; IR (NaCl) νmax 3223, 2957, 2928, 2857, 1672, 1631, 1606, 1577, 1543, 1514, 1457, 1418, 1362, 1264, 1202, 1138, 1068, 1044, 994, 961, 838, 800, 743, 719 cm−1; 1H-NMR (CDCl3, 500 MHz) δ[ppm] (carbon position, integration, multiplicity; J[Hz]) 7.25 (2, 1H, s), 6.96 (6′,1H, s), 6.94 (4,1H, brs), 6.70 (3″,1H, s), 6.37 (3, 1H, ddd; 4.0/2.2/2.2), 6.10 (3′, 1H, d; 1.7), 4.01 (OMe, 3H, s), 2.41 (6″, 3H, s), 2.39 (7″, 2H, t; 7.6), 1.53 (8″, 2H, m), 1.36-1.26 (9″/10″/11″, 6H, m), 0.89 (12″, 3H, t; 6.8); 13C NMR (125 MHz, CDCl3) δ[ppm] (carbon position) 166.1 (4′), 148.5 (2′), 146.4(5″), 129.1 (4″), 128.4 (3″), 127.8 (2), 125.6 (2″), 121.9 (5), 121.1 (5′), 117.8 (4), 116.1 (6’), 111.9 (3), 93.3 (3′), 58.7 (OCH3), 31.7, 30.1, 28.9 (8″/9″/10″), 25.4 (7″), 22.6 (11″), 14.1 (12″), 12.3 (6″); HR-APCI-TOFMS [M+H]+ m/z = 338.2301 (calcd. for C21H28N3O, 338.2227, Δ 0.4 mmu); ESI-MSMS m/z = 338 ([M+H+]+, 32%), 323 (100%), 306 (10%), 252 (12%); EI-MS m/z = 337 (M+, 100%), 322 (7%), 266 (65%), 236 (3%), 168 (5%), 133 (10%).
4″-(n-Heptyl)prodigiosin (3)
UV/vis (MeOH, 0.1%TFA) λmax (log ε) 538 (5.0); 512 (4.6); 385 (3.8); 361 (3.8); 295 (4.0); 274 (3.8) nm; IR (NaCl) νmax 3223, 2955, 2927, 2856, 1676, 1631, 1605, 1578, 1546, 1513, 1453, 1414, 1360, 1263, 1201, 1137, 1068, 1043, 994, 961, 838, 801, 742, 719 cm−1; 1H-NMR (CDCl3, 500 MHz) δ[ppm] (carbon position, integration, multiplicity; J[Hz]) 7.26 (2, 1H, s), 6.97 (6′, 1H, s), 6.95 (4, 1H, brs), 6.70 (3″, 1H, s), 6.37 (3, 1H, m), 6.10 (3′, 1H, s), 4.02 (OMe, 3H, s), 2.41 (6″, 3H, s), 2.39 (7″, 2H, m), 1.53 (8″, 2H, m), 1.32-1.24 (9″/10″/11″/12″, 8H, m), 0.89 (13″, 3H, m); HR-APCI-TOFMS [M+H]+ m/z = 352.2389 (calcd. for C22H30N3O, 352.2383, Δ 0.6mmu); ESI-MSMS m/z = 352 ([M+H+]+, 35%), 337 (100%), 320 (10%), 252 (12%); EI-MS m/z = 351 (M+, 100%), 336 (7%), 266 (55%), 175 (3%), 133 (10%).
Supplementary Material
Acknowledgment
We thank Wesley Yoshida for NMR support, Charles O'Kelly for help with taxonomy and Andrea Messer for collection and isolation of the producing organism. Financial support through NOAA OHHI (NA04OAR4600206), the NSF (OCE 04-32479) and the NIEHS (ES P50 ES012740) is gratefully acknowledged.
Footnotes
Supporting Information Available. 1H and 13C NMR spectra of 1.
References and Notes
- 1.Sertan-de Guzman AA, Predicala RZ, Bernardo EB, Neilan BA, Elardo SP, Mangalindan GC, Tasdemir D, Ireland CM, Barraquio WL, Concepcion GP. FEMS Microbiol. Lett. 2007;277:188–196. doi: 10.1111/j.1574-6968.2007.00950.x. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi NM, Patell JR, Gandhi J, De Souza NJ, Kohl H. Marine Biology. 1976;34:223–227. [Google Scholar]
- 3.Kim D, Lee JS, Park YK, Kim JF, Jeong H, Oh T-K, Kim BS, Lee CH. J. Appl. Microbiol. 2007;102:937–944. doi: 10.1111/j.1365-2672.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- 4.For overviews see: Fürstner A. Angew. Chemie Int. Ed. 2003;42:582–3603. Williamson NR, Fineran PC, Leeper FJ, Salmond GPC. Nature Rev. Microbiol. 2006;4:887–899. doi: 10.1038/nrmicro1531. Pérez-Tomás R, Montaner B, Llagostera E, Soto-Cerrato V. Biochem. Pharmacol. 2003;66:1447–1452. doi: 10.1016/s0006-2952(03)00496-9.
- 5.a) Llagostera E, Soto-Cerrato V, Montaner B, Pérez-Tomás R. Ann. NY Acad. Sci. 2003;1010:178–18. doi: 10.1196/annals.1299.030. [DOI] [PubMed] [Google Scholar]; b) Soto-Cerrato V, Llagostera E, Montaner B, Scheffer GL, Pérez-Tomás R. Biochem. Pharmacol. 2004;68:1345–1352. doi: 10.1016/j.bcp.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 6.a) Mortellaro A, Songia S, Gnocchi P, Ferrari M, Fornasiero C, D'Alessio R, Isetta A, Colotta F, Golay J. J. Immunol. 1999;162:7102–7109. [PubMed] [Google Scholar]; b) Han SB, Park SH, Jeon YJ, Kim YK, Kim HM, Yang KH. J. Pharmacol. Exp. Ther. 2001;299:415–425. [PubMed] [Google Scholar]; c) D’Alessio R, Bargiotti A, Carlini O, Colotta F, Ferrari M, Gnocchi P, Isetta A, Mongelli N, Motta P, Rossi A, Rossi M, Tibolla M, Vanotti E. J. Med. Chem. 2000;43:2557–2565. doi: 10.1021/jm001003p. [DOI] [PubMed] [Google Scholar]; d) Han SB, Kim HM, Kim YH, Lee CW, Jang E-S, Son KH, Kim SU, Kim YK. Int. J. Immunopharmacol. 1998;20:1–13. doi: 10.1016/s0192-0561(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 7.Fürstner A, Grabowski J, Lehmann CW, Kataoka T, Nagai K. ChemBioChem. 2001;2:60–68. doi: 10.1002/1439-7633(20010105)2:1<60::AID-CBIC60>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 8.a) Gerber NN. Tetrahedron Lett. 1970:809–812. doi: 10.1016/s0040-4039(01)97837-2. [DOI] [PubMed] [Google Scholar]; b) Gerber NN. J. Antibiot. 1971;24:636–640. doi: 10.7164/antibiotics.24.636. [DOI] [PubMed] [Google Scholar]; c) Gerber NN. J. Heterocycl. Chem. 1973;10:925–929. [Google Scholar]
- 9.Wassermann HH, Sykes RJ, Peverada P, Shaw CK, Cushley RJ, Lipsky SR. J. Am. Chem. Soc. 1973;95:6874–6875. doi: 10.1021/ja00801a080. [DOI] [PubMed] [Google Scholar]
- 10.Thomas MG, Burkart MD, Walsh CT. Chem. Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 11.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Proc. Natl. Acad. Sci. U S A. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.a) Gauthier G, Gauthier M, Christen R. Int. J. Syst. Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]; b) Gerber NN, Gauthier MJ. Appl. Environ. Microbiol. 1979;37:1176–1. doi: 10.1128/aem.37.6.1176-1179.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.