Abstract
Purpose of Review
Families with multiple individuals affected with chronic lymphocytic leukemia (CLL) and other related B-cell tumors have been described in the literature and strong familial aggregation has been seen in population studies. However, predisposing germ line mutations have not been identified. We will discuss the spectrum of conditions associated with CLL in families and the advances in identifying the underlying susceptibility genes.
Recent Findings
Familial CLL does not appear to differ substantially from sporadic CLL in terms of prognostic markers and clinical outcome, although it may be associated with more indolent disease. The precursor condition, monoclonal B-cell lymphocytosis (MBL) also aggregates in CLL families. Linkage studies have been conducted in high-risk CLL families to screen the whole genome for susceptibility loci but no gene mutations have yet been identified by this method. Association studies of candidate genes have implicated several genes as being important in CLL but more studies are needed. Results from whole genome association studies are promising.
Summary
The ability to conduct large scale genomic studies in unrelated CLL cases and in high risk CLL families will play an important role in detecting susceptibility genes for CLL over the next few years and thereby help to delineate etiologic pathways.
Keywords: chronic lymphocytic leukemia, monoclonal b-cell lymphocytosis, familial, germ line genes
Introduction
Chronic lymphocytic leukemia (CLL) is a malignancy characterized by the accumulation of small, mature-appearing lymphocytes in the bone marrow, blood, and lymphoid tissues. CLL accounts for 34% of adult leukemias in the United States.1 The latest WHO classification scheme considers CLL as a mature B-cell neoplasm and does not distinguish it from small lymphocytic lymphoma (SLL).2 Data from the United States Surveillance, Epidemiology, and End Results (SEER) Registry estimate the U.S. incidence in the period 2002–2006 to be 4.1 per 100,000 with a median age at diagnosis of 72 years.3 Incidence rates in men are nearly twice as high as in women. Although advanced age, white ancestry, and family history of hematologic malignancies are risk factors, the etiology of CLL is unknown.4 This report will review what is known about the familial aggregation of CLL and other lymphoid malignancies, the association of CLL with a precursor condition, monoclonal B-cell lymphocytosis (MBL), and the evidence for specific genes associated with CLL.
Familial vs. Sporadic CLL
Since CLL is uncommon, a CLL patient with at least one affected relative is considered “familial”. In population-based samples, approximately 5% of patients with CLL reported a family history of leukemia.5 Studies involving CLL cases from multiplex families have generally shown familial CLL to have an earlier age at diagnosis compared to sporadic CLL.6 However, larger population-based studies have not found differences in age at diagnosis between sporadic and familial cases.7 Some studies have found a higher proportion of female cases among familial compared to sporadic CLL.8,9 The molecular features of CLL have been compared in familial and sporadic patients. One study reported that shorter telomeres or CD38 positivity was similar in familial and sporadic cases.10 The frequency of IgVH mutated CLL was found to be higher in a series of 327 familial CLL cases compared to 724 sporadic cases although the IgVH gene usage patterns were similar.9 Ng et al found that 12/14 familial cases studied by FISH had a chromosome 13q deletion,11 contrasting with a rate of 50–60% in a large case series.12 A recent study found that B-lymphocyte stimulator levels (BlyS, (also known as B-cell activating factor, BAFF) were higher in familial CLL cases than in sporadic cases or controls.13 Another small study found familial cases to have higher BAFF levels than sporadic cases but the combined group of CLL patients had significantly lower values than controls.14 Overall, published data do not suggest large differences between familial and sporadic CLL patients. Most importantly, in one large study, familial patients did not have an adverse prognosis compared to sporadic patients.8
High Risk Families
We have conducted clinical and genetic studies in a series of CLL pedigrees collected at the National Cancer Institute.6 These efforts have been expanded in a multi-center effort, the Genetic Epidemiology of CLL Consortium (GEC). Figures 1 and 2 show examples of some of the more striking pedigrees in our study in terms of aggregation of lymphoproliferative (LP) tumors. These pedigrees show CLL and other LP conditions (including the precursor condition, MBL) segregating in relatives. Figure 2 shows a family segregating CLL and Waldenström’s macroglobulinemia (WM). There is no consistent pattern of illness that can be explained by a simple mode of genetic transmission in high risk families but it is likely that there are shared genes explaining the variant B-cell tumors in these families. Some investigators have noted that there is anticipation in age of onset in multigenerational pedigrees, where younger generations have an earlier age of onset than older generations.15 However, it is hard to eliminate bias based on the criteria for ascertaining families in these studies.
Figure 1. Pedigree with multiple cases of CLL as well as the precursor, MBL.
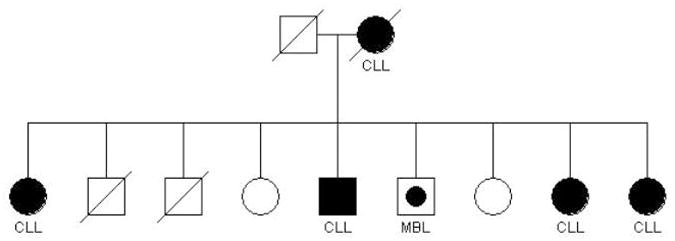
Figure 2. Pedigree with both CLL and WM cases, as well as the precursor MGUS.
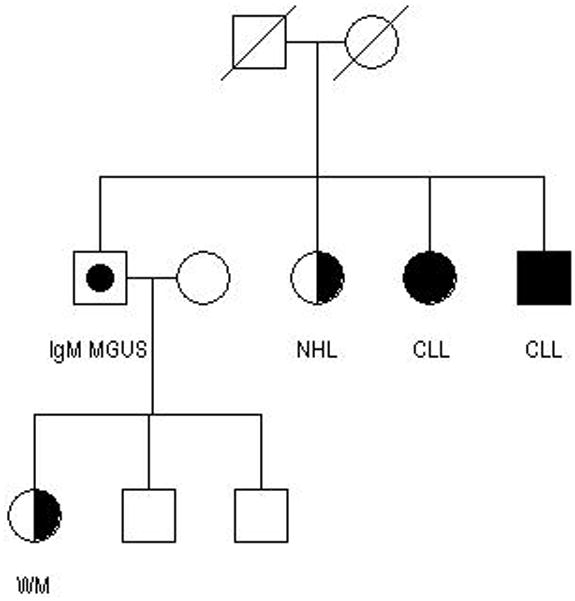
Reproduced with permission Lynn R. Goldin, Ola Landgren, Gerald E. Marti, Neil E. Caporaso, Familial aspects of chronic lymphocytic leukemia, monoclonal b-cell lymphocytosis, and related lymphomas, European Journal of Clinical and Medical Oncology, 2009
Population and Registry Studies
Early population-based case-control studies of CLL reported family history of lymphoproliferative (LP) tumors to be a significant risk factor.16 More recently, a family history analysis of case-control samples pooled from the InterLymph consortium examined family history of LP by histological subtype of lymphoma in the cases.17 Because of the large number of studies that were pooled, they were able to consider CLL/SLL cases separately and found that family history of “any LP” or family history of “leukemia only” were both significant predictors of risk for CLL/SLL.
We have published a series of studies based on linked registry data from Sweden and Denmark to quantify familial aggregation of CLL and other lymphomas.7,18–23 These studies were conducted by linking population-based registries that contain parent-offspring links to the cancer registries. These are the largest studies to date that are population-based, the goal being to quantify risks to relatives of LP tumors in a comprehensive manner. Our analyses were based on cancer outcomes in relatives of LP patients compared to that in relatives of matched population controls. We applied a survival analysis method that accounted for correlations among family members.24 In our most recent study of CLL using Swedish registry data, we evaluated outcomes in 26,947 first-degree relatives of 9,717 CLL patients (diagnosed 1958–2004) compared with 107,223 first-degree relatives of 38,159 matched controls.7 The results for CLL and other LPDs are shown in Table 1. Compared to relatives of controls, relatives of CLL patients had an increased risk for CLL (RR=8.5, 6.1–11.7) and other non-Hodgkin lymphomas (RR=1.9, 1.5–2.3). We found first-degree relatives of LPL/WM patients to have a significantly higher risk of developing LPL/WM, NHL, CLL, and MGUS, respectively.22 In another study, we reported that relatives of patients with MGUS had an increased risk of MGUS, MM, LPL/WM, and CLL.25 Table 2 shows relative risks for a more detailed panel of lymphoma subtypes among relatives of CLL patients. We found a striking excess of indolent B-cell NHL, specifically lymphoplasmacytic lymphoma (LPL)/WM and hairy cell leukemia (HCL). No excesses of aggressive B-cell or T-cell lymphomas were found. There was no statistical excess of Hodgkin lymphoma (HL), multiple myeloma (MM), or the precursor condition, monoclonal gammopathy of undetermined significance (MGUS), among CLL relatives. In summary, our studies have shown that some indolent subtypes (CLL, LPL/WM, HCL) aggregate together and may share a component of common etiology.
Table 1.
Relative risks of LP tumors in first-degree relatives of LP probands compared to first-degree relatives of controls**
Table 2.
Relative risks of LP tumor in first-degree relatives of CLL cases compared to first-degree relatives of controls. Obtained from Haematologica Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin’s lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009;94:647–653 (ref 7)
| Outcome | RR (95% CI) |
|---|---|
| B-cell NHL | 1.8 (1.3–2.5) |
| Indolent B-cell NHL | 2.2 (1.5–3.2) |
| Folicular lymphoma | 1.6 (0.87–2.8) |
| Mantle cell lymphoma | 1.1 (0.24–5.5) |
| Hairy cell leukemia | 3.3 (1.0–10.9) |
| LPL/WM | 4.0 (2.0–8.2) |
| Aggressive B-cell NHL | 1.0 (0.41–2.5) |
| T-cell NHL | 1.3 (0.36–4.9) |
One can argue that the strong familial aggregation of CLL and related lymphomas is a result of relatives sharing environmental risk factors. Epidemiological studies have not consistently identified environmental risk factors associated with CLL.4 CLL does show substantial geographic variation worldwide with a 40-fold difference in rates (the highest among Caucasians in North America and Europe and very low in Asians.)26 The rates of CLL in the United States have been relatively stable over time.3 Studies of Asian populations in the U.S. have reported rates similar to their counterparts in Asia.27 Thus, risk of CLL does not seem to increase in individuals who migrate to the United States from low risk countries. This supports a stronger role for genetic factors in etiology.
Monoclonal B-cell Lymphocytosis (MBL) and CLL
MBL is an asymptomatic hematologic condition characterized by small B-cell clones with a surface phenotype similar to that of CLL.28,29 These clones are detectable at low cell numbers in otherwise healthy individuals using sensitive 6 or 8 color flow cytometry analysis. The condition was recognized when this technology revealed clonal B-cell expansions in adults in a series of environmental health studies28 and in studies of asymptomatic members of CLL kindreds.30,31 We and others have reported MBL in 13–18% of first degree relatives of CLL patients in high risk families.30,31 This can be compared to 3–5% in the general population using comparable laboratory detection methods and suggests that MBL is a marker of inherited predisposition to CLL.32–34 Several studies have addressed factors that influence the prognosis of MBL. 35–37 These studies distinguish MBL diagnosed in the clinic after referral for lymphocytosis (“clinical” MBL) and MBL detected in asymptomatic individuals from investigational screening studies. Individuals with clinical MBL progress to need CLL-specific treatment at a rate of about 1.1% per year. 36 However, individuals with “low count” MBL found through screening studies (i.e. MBL detected in an individual with a normal B-absolute lymphocyte count) have not been observed to progress over the course of several years suggesting that progression is associated with higher B-cell counts.
An important question related to the recognition of MBL is whether all CLL cases are preceded by MBL, or if CLL cases commonly develop de novo. To address this question, we conducted a prospective cohort study based on 77,469 healthy adults who were enrolled in the nationwide, population-based Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial.38 We identified 45 subjects with peripheral whole-blood collection and a diagnosis of CLL up to 6.4 years after blood collection draws. Using six-color flow cytometry and immunoglobulin heavy-chain gene rearrangement (IGHV) by an RT-PCR assay, we found evidence of prediagnostic monoclonality among B-cells (by either method) in 44/45 patients (up to 6.4 years before the initial diagnosis. The presence of IGHV genes was determined in 35 of 45 prediagnosticclones and 27/35 were mutated. This study suggests that virtually all cases of CLL (both with mutated and unmutated IGHV genes) are preceded by MBL.
In another recent study (unpublished data), we conducted flow cytometry on 430 relatives from 132 CLL families from the Genetic Epidemiology of CLL Consortium. The overall rate of MBL was substantially higher among first degree relatives compared to the general population and also increased with age. If MBL is an early step in the process of development of CLL, then germ line genes are likely to be acting early in leukemogenesis with more oncogenic events required before CLL develops.
The recent adjustment of the CLL diagnosis criteria39 will focus more attention on MBL. Nieto et al 40 have shown that with sensitive detection, the population prevalence of MBL is higher than previously thought. Matos et al.41 reported that 4% of first-degree relatives of sporadic CLL cases had MBL, with prevalence increasing with age. Other recent studies have attempted to describe the immunoglobulin repertoire32 and the molecular characteristics42 of low count MBL.
Genetic Susceptibility of CLL
Linkage studies of CLL localize genes using the co-inheritance of genetic markers and disease (i.e., CLL) in families, and are based on the idea that the occurrence of multiple individuals with CLL from the same family is a result of shared “genetic” exposure. These studies are powered to identify rare genetic variants with large CLL risks. To date, linkage studies of CLL have largely been unsuccessful in indentifying genetic determinants.43 Reasons for this include the limited power to detect linkage due to small number of pedigrees or small pedigree size (the majority of the families in the previous studies had only two affected CLL individuals genotyped due to the late age of onset). With the emerging evidence that MBL is a precursor lesion for CLL and occurs more often in CLL families, more informative pedigrees may be obtained by including MBL in the analyses.
Genetic association studies are epidemiological studies that are powerful for detecting common genetic variants of modest CLL risk. Over 50 genetic association studies of CLL have been conducted (reviewed in 44,45) and most of these studies focused on a set of genes that have biological plausibility in CLL etiology. Several promising genes have been recently identified including CD38 46 in which the expression of CD38 is a well-established prognostic marker for CLL. Other genes include CCNH, APF1, IL16, and CASP8.47 These novel findings need validation in independent studies. In contrast to candidate-gene studies, two genome-wide association studies (GWAS) of CLL have been conducted.48,49 GWAS are agnostic, in that one looks for association across the entire genome without any biological assumptions. Di Bernardo et al.48 identified 7 genetic variants located on chromosomes 2q13 (rs17483466), 2q37.1 (rs13397985), 6p25.3 (rs872071, rs9378805), 11q24.1 (rs735665), 15q23 (rs7176508), and 19q13.32 (rs11083846) in a sample of 517 CLL cases and 1,438 controls. They replicated the findings through two large internal validation samples. The second GWAS 49 included 148 CLL and 592 controls. No findings met genome-wide significance; however, 3 of the 7 variants (rs735665, rs13397985, and rs872071) identified by Di Bernardo et al., showed evidence of increased risk in this second study. Finally, in our own genetic association study of 400 CLL case and 300 controls using GEC resources,50 we evaluated these 7 variants and were able to replicate all but rs11083846 on chromosome 19q13.32. A few additional loci have been identified in a follow-up study by the Houlston group.51 The next step is combine data from all available CLL GWAS, thereby offering greater power in the identification of novel susceptibility genes for CLL.
There are other possible germ line changes that could lead to carcinogenesis and CLL. For example, differences in gene regulation have been studied in relation to familial CLL. Calin et al.52 have described germ line mutations in a microRNA gene (miR-16-1) located in the commonly deleted region on 13q14 in 2/75 CLL patients tested, and one of these patients had a family history of CLL. Raval et al.53 have recently described a germ line change near the DAPK1 gene on chromosome 9 that was associated with increased methylation of the gene and decreased allelic expression in CLL cases from one family. Other rare variants (possibly copy number changes in genes) could play a role in disease susceptibility. If several rare variants account for familial CLL, then these will be hard to detect by linkage or association studies. Large scale gene sequencing may be able to identify these rare changes and then one can test for associations of rare variants with CLL at the population level.
Clinical Applications
As described earlier, first-degree relatives of CLL patients have an 8.5- fold relative risk for developing CLL and are also at increased risk for developing other indolent forms of NHL compared to relatives of matched controls.7 However, because the baseline risk of these conditions in the population is low, the absolute risk to a relative of a CLL patient developing either CLL or a related malignancy is still very low and early detection of CLL is not likely to affect outcome since patients are generally not treated until symptoms become evident.39 Relatives of CLL cases from high risk families are at increased risk for developing MBL but studies have shown that these are mostly “low count” MBL and are at low risk for progression. Currently, there is some controversy over whether or not matched relatives with MBL should be disqualified as hematopoetic stem cell transplant donors.54,55 Future studies are needed to clarify these issues.
Conclusions
There is significant familial aggregation of CLL, with evidence pointing to a greater importance of inheritance rather than shared environments. There are likely to be specific genes associated with CLL but also genes common to CLL and other LP tumors. However, the failure to identify any specific mutation with a large effect suggests that common genetic variants with smaller effects or multiple rare genes with large effects account for the familial risk. These genes may be harder to identify but the advances in large scale genomics methods that can be applied to population samples or high risk families offer promise that specific genes causing susceptibility to CLL and other LP will be identified in the near future. As our knowledge of germ line changes that lead to susceptibility for CLL expands, we will obtain a better understanding of the etiologic pathways relevant to both familial and sporadic CLL. This will lead to better treatment approaches to this still incurable tumor.
Acknowledgments
This material is based upon work supported by the Intramural Program of the National Cancer Institute, National Institutes of Health, Bethesda, Maryland and by NIH grant CA118444.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe ES, Harris NL, Stein H, Isaacson PG. Classification of lymphoid neoplasms: the microscope as a tool for disease discovery. Blood. 2008;112:4384–4399. doi: 10.1182/blood-2008-07-077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horner MJ, Ries LAG, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2006. National Cancer Institute; Bethesda, MD: 2009. [Google Scholar]
- 4.Linet MS, Schubauer-Berigan MK, Weisenburger DD, et al. Chronic lymphocytic leukaemia: an overview of aetiology in light of recent developments in classification and pathogenesis. Br J Haematol. 2007;139:672–686. doi: 10.1111/j.1365-2141.2007.06847.x. [DOI] [PubMed] [Google Scholar]
- 5.Houlston RS, Sellick G, Yuille M, Matutes E, Catovsky D. Causation of chronic lymphocytic leukemia--insights from familial disease. Leuk Res. 2003;27:871–876. doi: 10.1016/s0145-2126(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 6.Ishibe N, Sgambati MT, Fontaine L, et al. Clinical characteristics of familial B-CLL in the National Cancer Institute Familial Registry. Leuk Lymphoma. 2001;42:99–108. doi: 10.3109/10428190109097681. [DOI] [PubMed] [Google Scholar]
- 7*.Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin’s lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009;94:647–653. doi: 10.3324/haematol.2008.003632. This study used large population-based registries in Sweden to compare 26,947 relatives of 9,717 CLL patients compared to relatives of matched controls. They demonstrated significant familial aggregation of CLL and related conditions, especially other indolent lymphomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mauro FR, Giammartini E, Gentile M, et al. Clinical features and outcome of familial chronic lymphocytic leukemia. Haematologica. 2006;91:1117–1120. [PubMed] [Google Scholar]
- 9*.Crowther-Swanepoel D, Wild R, Sellick G, et al. Insight into the pathogenesis of chronic lymphocytic leukemia (CLL) through analysis of IgVH gene usage and mutation status in familial CLL. Blood. 2008;111:5691–5693. doi: 10.1182/blood-2008-03-142349. This study found no difference in repertoire and frequency of IgVH usage between 327 familal CLL patients and 724 sporadic CLL patients. Familial CLL patients were more likely to be IgVH mutated. [DOI] [PubMed] [Google Scholar]
- 10.Ishibe N, Prieto D, Hosack DA, et al. Telomere length and heavy-chain mutation status in familial chronic lymphocytic leukemia. Leuk Res. 2002;26:791–794. doi: 10.1016/s0145-2126(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 11.Ng D, Toure O, Wei MH, et al. Identification of a novel chromosome region, 13q21.33–q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood. 2007;109:916–925. doi: 10.1182/blood-2006-03-011825. [DOI] [PubMed] [Google Scholar]
- 12.Winkler D, Dohner H, Stilgenbauer S. Genetics, gene expression, and targeted therapies in chronic lymphocytic leukemia. Curr Drug Targets. 2006;7:1313–1327. doi: 10.2174/138945006778559184. [DOI] [PubMed] [Google Scholar]
- 13.Novak AJ, Grote DM, Ziesmer SC, et al. Elevated serum B-lymphocyte stimulator levels in patients with familial lymphoproliferative disorders. J Clin Oncol. 2006;24:983–987. doi: 10.1200/JCO.2005.02.7938. [DOI] [PubMed] [Google Scholar]
- 14.Molica S, Digiesi G, Mauro F, et al. Increased serum BAFF (B-cell activating factor of the TNF family) level is a peculiar feature associated with familial chronic lymphocytic leukemia. Leuk Res. 2009;33:162–165. doi: 10.1016/j.leukres.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Goldin LR, Sgambati M, Marti GE, Fontaine L, Ishibe N, Caporaso N. Anticipation in familial chronic lymphocytic leukemia. Am J Hum Genet. 1999;65:265–269. doi: 10.1086/302458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sgambati M, Linet MS, Devesa SS. Chronic lymphocytic leukemia epidemiological, familial, and genetic aspects. In: Cheson BD, editor. Chronic lymphoid leukemias Basic and clinical oncology. 2. Vol. 26. New York: Marcel Dekker; 2001. pp. 33–62. [Google Scholar]
- 17.Wang SS, Slager SL, Brennan P, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;109:3479–3488. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009;146:91–94. doi: 10.1111/j.1365-2141.2009.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldin LR, Landgren O, McMaster ML, et al. Familial aggregation and heterogeneity of non-Hodgkin lymphoma in population-based samples. Cancer Epidemiol Biomarkers Prev. 2005;14:2402–2406. doi: 10.1158/1055-9965.EPI-05-0346. [DOI] [PubMed] [Google Scholar]
- 20.Goldin LR, Pfeiffer RM, Gridley G, et al. Familial aggregation of Hodgkin lymphoma and related tumors. Cancer. 2004;100:1902–1908. doi: 10.1002/cncr.20189. [DOI] [PubMed] [Google Scholar]
- 21.Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–1854. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- 22*.Kristinsson SY, Bjorkholm M, Goldin LR, McMaster ML, Turesson I, Landgren O. Risk of lymphoproliferative disorders among first-degree relatives of lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia patients: a population-based study in Sweden. Blood. 2008;112:3052–3056. doi: 10.1182/blood-2008-06-162768. The authors used linked registries in Sweden along with outpatient records on individuals with MGUS to evaluate the risk of cancer and MGUS in more than 6000 relatives of 2144 LPL/WM patients compared to more than 24,000 relatives of controls. The study demonstrated strong familial aggregation of WM/LPL and also co-aggregation with other NHL, CLL and MGUS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landgren O, Linet MS, McMaster ML, Gridley G, Hemminki K, Goldin LR. Familial characteristics of autoimmune and hematologic disorders in 8,406 multiple myeloma patients: a population-based case-control study. Int J Cancer. 2006;118:3095–3098. doi: 10.1002/ijc.21745. [DOI] [PubMed] [Google Scholar]
- 24.Pfeiffer RM, Goldin LR, Chatterjee N, et al. Methods for testing familial aggregation of diseases in population-based samples: application to Hodgkin lymphoma in Swedish registry data. Ann Hum Genet. 2004;68:498–508. doi: 10.1046/j.1529-8817.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- 25*.Landgren O, Kristinsson SY, Goldin LR, et al. Risk of plasma cell and lymphoproliferative disorders among 14621 first-degree relatives of 4458 patients with monoclonal gammopathy of undetermined significance in Sweden. Blood. 2009;114:791–795. doi: 10.1182/blood-2008-12-191676. The authors used linked registries in Sweden along with outpatient records on individuals with MGUS to evaluate the risk of MGUS and other hematologic tumors in a large number of relatives of patients with MGUS compared to relatives of controls. The study demonstrated strong familial aggregation of MGUS, MM, LPL/WM and CLL among relatives of MGUS patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linet MS, Devesa SS, Morgan GJ. The Leukemias. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. 3. New York: Oxford University Press; 2006. pp. 841–871. [Google Scholar]
- 27.Pang JWY, Cook LS, Schwartz SM, Weiss NS. Incidence of leukemia in Asian migrants to the United States and their descendents. Cancer Causes and Control. 2002;13:791–795. doi: 10.1023/a:1020608328969. [DOI] [PubMed] [Google Scholar]
- 28.Marti G, Abbasi F, Raveche E, et al. Overview of monoclonal B-cell lymphocytosis. Br J Haematol. 2007;139:701–708. doi: 10.1111/j.1365-2141.2007.06865.x. [DOI] [PubMed] [Google Scholar]
- 29.Marti GE, Rawstron AC, Ghia P, et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005;130:325–332. doi: 10.1111/j.1365-2141.2005.05550.x. [DOI] [PubMed] [Google Scholar]
- 30.Marti GE, Carter P, Abbasi F, et al. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2003;52:1–12. doi: 10.1002/cyto.b.10013. [DOI] [PubMed] [Google Scholar]
- 31.Rawstron AC, Yuille MR, Fuller J, et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002;100:2289–2290. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 32**.Dagklis A, Fazi C, Sala C, et al. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)-like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood. 2009;114:26–32. doi: 10.1182/blood-2008-09-176933. This is a study of MBL in 1725 healthy adults from a genetically isolated region of Northern Italy. The authors found an overall 7.4% prevalence of MBL and have introduced the term, “low-count MBL” to refer to the MBL in the presence of total lymphocyte and B-cell counts in the normal range. CLL like MBL cases clustered in families, consistent with the strong familial risk seen in CLL. Importantly, the IGVH repertoire in this low-count MBL was different from what is typically seen in CLL and very few cases showed stereotyped HCDR3, indicating that these clones do not reflect a pre-leukemic state. [DOI] [PubMed] [Google Scholar]
- 33.Ghia P, Prato G, Scielzo C, et al. Monoclonal CD5+ and CD5- B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004;103:2337–2342. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 34.Rawstron AC, Green MJ, Kuzmicki A, et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002;100:635–639. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 35.Fung SS, Hillier KL, Leger CS, et al. Clinical progression and outcome of patients with monoclonal B-cell lymphocytosis. Leuk Lymphoma. 2007;48:1087–1091. doi: 10.1080/10428190701321277. [DOI] [PubMed] [Google Scholar]
- 36**.Rawstron AC, Bennett FL, O’Connor SJ, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–583. doi: 10.1056/NEJMoa075290. This study found MBL in 5.1% of subjects with normal blood count and 13.9% of subjects with lymphocytosis, respectively. In a 6.7 year median follow-up, CLL requiring treatment occurred at a rate of 1.1% per year in those with lymphocytosis. The total lymphocyte count at diagnosis was the most significant risk factor for progression. [DOI] [PubMed] [Google Scholar]
- 37*.Shanafelt TD, Kay NE, Rabe KG, et al. Brief Report: Natural History of Individuals With Clinically Recognized Monoclonal B-Cell Lymphocytosis Compared With Patients With Rai 0 Chronic Lymphocytic Leukemia. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.21.2704. This study demonstrates that B-cell count is the best predictor for progression of MBL to CLL requiring treatment although the rate of progression at five years is low. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Landgren O, Albitar M, Ma W, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–667. doi: 10.1056/NEJMoa0806122. In a large cohort study, the investigators obtained stored blood samples from 45 CLL patients up to 6.4 years before they were diagnosed with CLL. Using six-color flow cytomety and PCR, evidence of monoclonal B-cells was found in 44/45 individuals before they were diagnosed with CLL. This suggests that nearly all CLL is preceded by an MBL state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. This is an updated review of recommendations for management of CLL patients. Based on the indolent nature of early stage CLL, this paper proposes a change in the blood count requirement for diagnosing CLL to 5000 B cells per μl where the 1996 guidelines set the count to 5000 lymphocytes per μl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieto WG, Almeida J, Romero A, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009;114:33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 41.Matos DM, Ismael SJ, Scrideli CA, de Oliveira FM, Rego EM, Falcao RP. Monoclonal B-cell lymphocytosis in first-degree relatives of patients with sporadic (non-familial) chronic lymphocytic leukaemia. Br J Haematol. 2009;147:339–346. doi: 10.1111/j.1365-2141.2009.07861.x. [DOI] [PubMed] [Google Scholar]
- 42*.Lanasa MC, Allgood SD, Volkheimer AD, et al. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia. 2009;24:133–140. doi: 10.1038/leu.2009.192. The authors examined several MBL cases with low B-cell counts from CLL families. Four of six cases were oligoclonal and the clones tended to be IgVH mutated, and often showed loss of 13q14. These findings imply that MBL at low counts is indolent and during CLL leukemogenesis, a single dominant clone arises from several existing clones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellick GS, Goldin LR, Wild RW, et al. A high-density SNP genome-wide linkage search of 206 families identifies susceptibility loci for chronic lymphocytic leukemia. Blood. 2007;110:3326–3333. doi: 10.1182/blood-2007-05-091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slager SL, Kay NE, Fredericksen ZS, et al. Susceptibility genes and B-chronic lymphocytic leukaemia. Br J Haematol. 2007;139:762–771. doi: 10.1111/j.1365-2141.2007.06872.x. [DOI] [PubMed] [Google Scholar]
- 45.Zintzaras E, Kitsios GD. Synopsis and synthesis of candidate-gene association studies in chronic lymphocytic leukemia: the CUMAGAS-CLL information system. Am J Epidemiol. 2009;170:671–678. doi: 10.1093/aje/kwp201. [DOI] [PubMed] [Google Scholar]
- 46.Jamroziak K, Szemraj Z, Grzybowska-Izydorczyk O, et al. CD38 gene polymorphisms contribute to genetic susceptibility to B-cell chronic lymphocytic leukemia: evidence from two case-control studies in Polish Caucasians. Cancer Epidemiol Biomarkers Prev. 2009;18:945–953. doi: 10.1158/1055-9965.EPI-08-0683. [DOI] [PubMed] [Google Scholar]
- 47.Enjuanes A, Benavente Y, Bosch F, et al. Genetic variants in apoptosis and immunoregulation-related genes are associated with risk of chronic lymphocytic leukemia. Cancer Res. 2008;68:10178–10186. doi: 10.1158/0008-5472.CAN-08-2221. [DOI] [PubMed] [Google Scholar]
- 48**.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. This is the first whole genome association study conducted in CLL. The authors included an initial sample and two replication samples for a total of 1529 CLL patients and 3115 controls. Six genes were associated with CLL, with SNPs in the IRF4 gene showing the strongest association. [DOI] [PubMed] [Google Scholar]
- 49.Skibola CF, Bracci PM, Halperin E, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slager SL, Goldin LR, Strom SS, et al. Genetic susceptibility variants for chronic lymphoctyic leukemia. Cancer Epidemiol Biomarkers Prev. doi: 10.1158/1055-9965.EPI-09-1217. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 53.Raval A, Tanner SM, Byrd JC, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Giudice I, Mauro FR, De Propris MS, et al. Identification of monoclonal B-cell lymphocytosis among sibling transplant donors for chronic lymphocytic leukemia patients. Blood. 2009;114:2848–2849. doi: 10.1182/blood-2009-06-228395. [DOI] [PubMed] [Google Scholar]
- 55.Hardy NM, Grady C, Pentz R, et al. Bioethical considerations of monoclonal B-cell lymphocytosis: donor transfer after haematopoietic stem cell transplantation. Br J Haematol. 2007;139:824–831. doi: 10.1111/j.1365-2141.2007.06862.x. [DOI] [PubMed] [Google Scholar]


