Abstract
Biosynthesis of the highly toxic and carcinogenic aflatoxins in select Aspergillus species from the common intermediate O-methylsterigmatocystin (OMST) has been postulated to require only the cytochrome P450 monooxygenase, OrdA (AflQ). We now provide evidence that the aryl alcohol dehydrogenase NorA (AflE) encoded by the aflatoxin biosynthetic gene cluster in A. flavus affects the accumulation of aflatoxins in the final steps of aflatoxin biosynthesis. Mutants with inactive norA produced reduced quantities of aflatoxin B1 (AFB1), but elevated quantities of a new metabolite, deoxyAFB1. To explain this result, we suggest that, in the absence of NorA, the AFB1 reduction product, aflatoxicol, is produced and is readily dehydrated to deoxyAFB1 in the acidic medium, enabling us to observe this otherwise minor toxin produced in wild-type A. flavus.
Keywords: Aspergillus flavus, aflatoxin biosynthesis, gene disruption, mass spectrometry, aryl alcohol dehydrogenase, aflatoxicol
Introduction
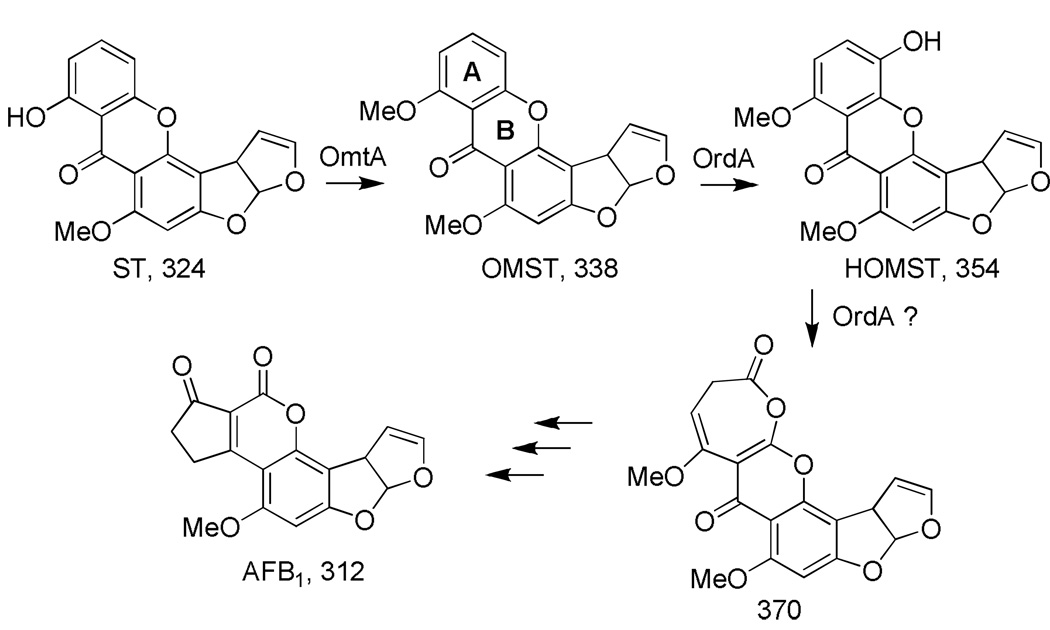
Many species of Aspergillus produce the xanthone metabolite sterigmatocystin (ST) (Scheme 1), but only a few are capable of converting ST to the far more toxic and carcinogenic aflatoxins (AFs: AFB1, AFB2, AFG1, AFG2) (Frisvad, et al., 2007). Because Aspergillus species are common agricultural contaminants and because ingestion of aflatoxins can lead to hepatocellular carcinoma, a better understanding of the final steps of aflatoxin biosynthesis is needed. For AFB1 biosynthesis, ST must first be methylated by an O-methyltransferase (OmtA) unique to AF biosynthesis (Bhatnagar, et al., 1987). The resulting methylated intermediate, O-methyl-ST (OMST) (Yu, et al., 1998) is then oxidized by the cytochrome P450 monooxygenase, OrdA (AflQ). Since AFB1 was produced when either OMST or its presumptive initial oxidation product, 11-hydroxy-OMST (HOMST), was fed to yeast cells expressing the A. parasiticus cytochrome P450 monooxygenase OrdA (Prieto, et al., 1996, Udwary, et al., 2002), it was proposed that OrdA is the only enzyme required for the conversion of OMST to AFB1. To be consistent with the yeast-feeding experiment, OrdA must also introduce an oxygen atom into HOMST (Scheme 1). The subsequent conversion steps require hydration, ring-opening, cyclization, decarboxylation, and demethylation to produce AFB1.
Scheme 1.
Known or previously postulated intermediates in the conversion of ST to AFB1. Molecular weights are shown below each compound.
The oxidative ring cleavage and rearrangement necessary for the formation of the coumarin ring system in AFB1 must be consistent with the following observations: a) NADPH is utilized in the conversion (Singh & Hsieh, 1976); b) an “NIH hydride shift” occurs so that the C-11 hydrogen is retained (Simpson, et al., 1983); c) an oxygen atom and carbon-11 in the A-ring of OMST are lost as carbon dioxide (Chatterjee & Townsend, 1994); and d) an oxygen atom incorporated into the B-ring (Scheme 1) is retained (Watanabe & Townsend, 1996).
The role of the putative aryl alcohol dehydrogenase NorA (AflE) in AF biosynthesis has not been definitively ascribed, although it was originally thought to function in the reduction of norsolorinic acid to averantin (hence the name “Nor”) (Cary, et al., 1996, Yu, et al., 2004). NorA shares more than 60% amino acid identity with NorB (AflF), an aryl alcohol dehydrogenase shown to be involved in formation of AFG1 (Ehrlich, et al., 2008). Genes encoding both enzymes are part of the AF biosynthesis gene cluster. The ST gene cluster of A. nidulans possesses only one of these genes, stcV (Brown, et al., 1996). Based on BLAST searches of genome sequence databases, genes encoding aryl alcohol dehydrogenases are common in many filamentous fungi and yeast. In the A. flavus AF gene cluster, the promoter and translation start codon of norB are missing due to a large DNA deletion in this region (Ehrlich, et al., 2004). Therefore the role of norA is most easily examined in A. flavus. We now provide evidence that A. flavus, lacking a functional copy of norA, accumulates a new metabolite, deoxyAFB1, a shunt metabolite that most likely is formed by dehydration of aflatoxicol (AFOH) in the acidic culture medium.
Materials and methods
Preparation of norA mutants in A. flavus AF13
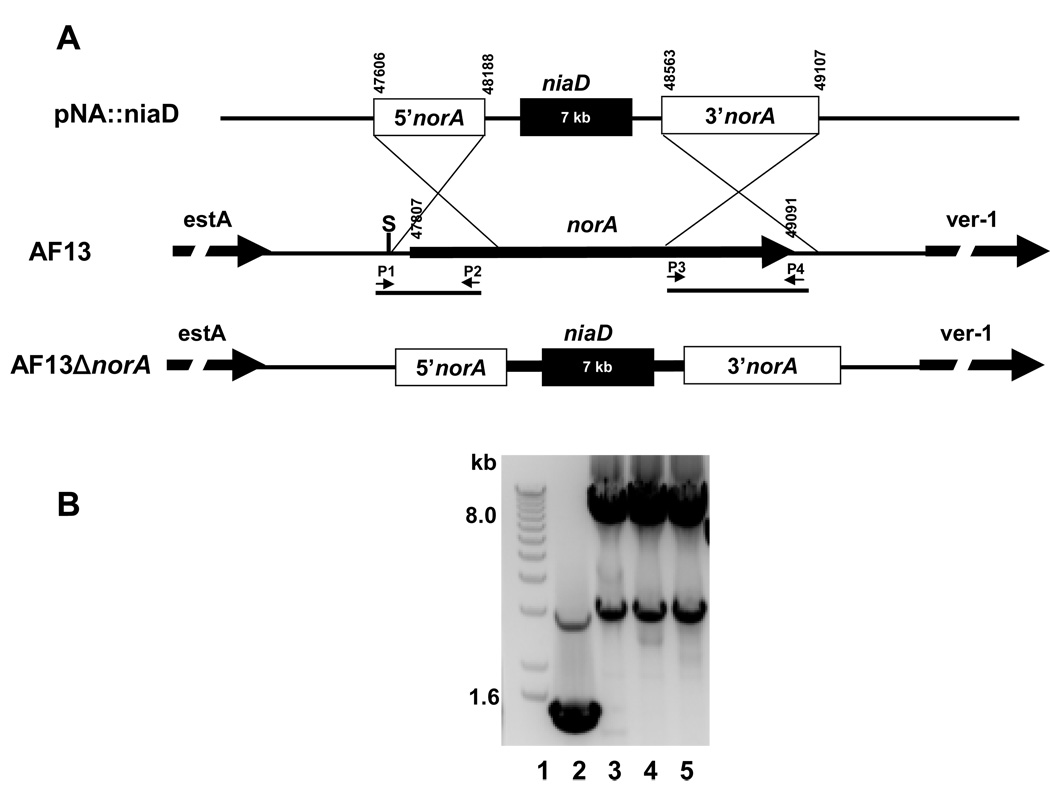
A vector for insertional inactivation of norA in A. flavus was constructed by PCR with the oligonucleotide primers P1, 5’-acgactacaagaatagcggtgacat and P2, 5’-tattctagagacgcagactcttggtatgg (Genbank Accession #AY510451; 47574 to 48188) and P3, 5’-tattctagagtactgggccgcggtcagtt and P4, 5’-aatggtacctcgagtccgcgacaactaggctcattttg (48516–49107) to amplify 5’- and 3’-portions of AF13 norA, respectively (Fig. 1A). The resulting 614 and 591 bp PCR fragments were cloned into the SphI/XbaI and XbaI/KpnI sites of pUC18, respectively. An XbaI fragment from the niaD-containing plasmid, pSL82 (Chang, et al., 1996), was then inserted into the internal XbaI site of the norA fragments in pUC18 to create the knockout vector.
Fig. 1.
Disruption of norA in A. flavus AF13. (A) Schematic showing design of the knockout vector. Numbers denote the position of the region in AF13 DNA (Genbank accession number AY51051). P1 to P4 are the oligonucleotides primers used for amplification of the 5’- and 3’-ends of the gene. The neighboring genes estA and ver-1 are shown. niaD was obtained from an XbaI digest of pSL82. The construct was prepared in pUC18. (B) PCR with oligonucleotide primers P1 and P4 of AF13 DNA from different transformant clones. Lane 1, 1 kb+ marker (Invitrogen); Lane 2, AF13 (pSL82) control; Lanes 3–5, AF13ΔnorA-clones 3, 15, 21, respectively.
Transformation of A. flavus AF13ΔniaD protoplasts was done as previously described using the PEG procedure (Ehrlich, et al., 2004) with 10 µg of XhoI/SphI-linearized plasmid. Confirmation that norA was insertionally inactivated (double crossover event) in the resulting transformants was done by PCR using the outer oligonucleotide primers (P1 and P4, Fig 1B) with DNA from the putative transformants or from pSL82-transformed AF13 as the control.
Thin layer chromatography (TLC) and liquid chromatography/mass spectrometry (LC/MS)
Fungal cultures grown from spores at 30°C for 3 days on potato dextrose agar (PDA, Difco, Voigt Global Distribution, Lawrence, KS) were extracted with acetone and chloroform as previously described (Ehrlich, et al., 2004). Aliquots of the extract were analyzed by TLC on 250 µm silica gel plates (J.T Baker, Philipsburg NJ) developed with toluene:ethyl acetate:acetic acid (TEA) (8:1:1). A prominent blue-fluorescent compound from ΔnorA cultures was partially purified by preparative TLC. The unpurified extract, the TLC-purified metabolite, and authentic standards (AFB1, synthetic aflatoxicol, synthetic deoxyAFB1, OMST, and synthetic HOMST) were analyzed in the positive ion mode by LC/MS. The materials were dissolved in methanol, injected on a Luna C18 100×4.6 mm column (5 µm, 100Å, Phenomenex) equilibrated in 10% acetonitrile/0.1% formic acid and 90% aqueous formic acid (0.1%), and eluted with a gradient to 100% acetonitrile/0.1% formic acid over 30 min. Metabolites were monitored by both diode array UV-visible spectrophotometry and quadrupole MS (Agilent 6130).
Feeding studies
Aflatoxicol (AFOH) and deoxyAFB1 were prepared by zinc borohydride reduction of AFB1 (Sigma, St Louis, MO) (Hsia & Chu, 1977). Aflatoxicol (AFOH) was partially purified from the reaction mix by preparative TLC. Synthetic AFOH was dissolved in 200 µl dimethyl sulfoxide and added to 3-day mycelial cultures of A. parasiticus SRRC2043 (accumulates OMST only) in low sugar replacement medium (Bhatnagar, et al., 1987) or to S. cerevisiae expressing norA or ordA after induction with galactose (Yu, et al., 1998). Following 4-hr incubation, metabolites were extracted into methylene chloride and aliquots examined by TLC.
BLAST searches
BLAST searches (tBLASTX and BLASTP) were done against the sequenced fungal genome datasets in Pubmed (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), the Broad Institute fungal database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html) and the A. flavus genomic sequence (http://www.aspergillusflavus.org/genomics). The cut-off for matches was E -30.
Results
Transformation of A. flavus AF13ΔniaD with the linearized norA knockout vector (Fig. 1A) yielded approximately 60 colonies, three of which had slightly darker orange mycelia when re-grown on PDA plates. The three darker orange transformants were confirmed to be double crossover norA disruptants by PCR (Fig 1B). A 1.5 kb PCR band was obtained for intact norA in the AF13 control strain and an 8 kb product for the positive ΔnorA transformants (Fig 1B). The latter product is consistent with the size expected with the 7 kb niaD selection marker inserted into the norA gene.
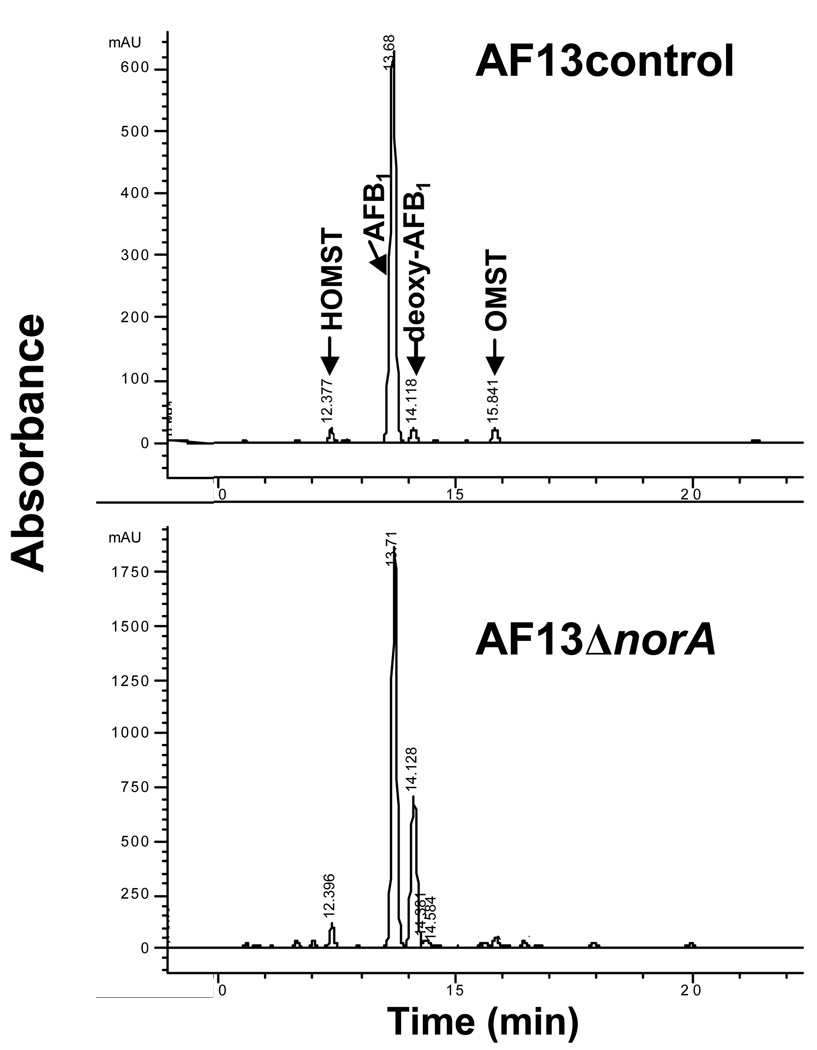
Acetone extracts of the norA knockout cultures and cultures transformed with the selection marker only were examined by liquid chromatography combined with mass spectrometry (LC/MS; Fig. 2 and Table 1). A metabolite eluted after AFB1 (14.1 min compared to 13.7 min) and exhibited a blue-shifted (λmax = 332 nm) chromophore compared to that of AFB1 (λmax = 362 nm). This less polar compound was identified as deoxyAFB1 by its positive ion mass spectrum (M+H = 297; deoxyAFB1 M = 296 Da) and its having a retention time and UV-visible chromophore identical to that of deoxyAFB1 prepared by established synthetic methods (Hsia & Chu, 1977). The LC data showed that deoxyAFB1 accumulated in at least 20-fold greater amounts in the norA knockout strain than in the selection marker-only transformed strain (Fig. 2).
Fig. 2.
Liquid chromatography profile of the A. flavus AF13ΔnorA extract and the AF13 control extract obtained during the LC-MS analysis.
Table 1.
LC/MS data from extracts of AF13ΔnorA and control cultures
| AF13ΔnorA (RTa) |
control (RT) |
UV λmax | m/zb | Identityc |
|---|---|---|---|---|
| 10.31, 10.48 | NDd | 330 | 315 | AFOH |
| 10.6 | 10.6 | 318, 360she | 371 | |
| 10.9 | 10.9 | 360 | 329 | |
| 12.4 | 12.4 | 318, 360sh | 355 | HOMST |
| 13.0 | ND | * | 329 | |
| 13.8 | 13.8 | 362 | 313 | AFB1 |
| 14.1 | 14.1f | 332 | 297 | deoxy-AFB1 |
| 15.6 | ND | 448 | 371 | |
| 15.9 | 15.9 | 312, 340sh | 339 | OMST |
RT-retention time (min)
M+H ions
Comparisons were to authenticated standards.
ND-not detected.
sh-shoulder
Control had 20-fold lower amount of AFB1 by LC
The amount was too low for determination.
Comparison of other metabolites in the acetone extracts of an AF13ΔnorA clone (#15) and the AF13 control with natural or synthetic standards by UV-visible spectrophotometry and positive ion LC/MS confirmed the presence of OMST (15.9 min), HOMST (12.4 min, M+H = 355, M = 354), and AFB1 (13.8 min) (Table 1). The metabolites shared identical LC retention times, UV-visible chromophores, and mass spectra with their respective standard. Several unknown compounds were also observed in extracts of fungi with both mutant and intact norA. One exhibited a chromophore (λmax = 318 nm, shoulder at 360 nm; M+H = 371, M= 370) similar to those of OMST and HOMST, suggesting that it could be a related intermediate in the pathway. Two unknown compounds eluting at 10.9 and 13.0 min with the same mass (M+H = 329, M = 328) were found in extracts from control and norA mutant fungi. One of them eluted at 10.9 and 13.0 min, and exhibited a chromophore similar to that of AFB1 (λmax = 360 nm). This metabolite was found mainly in the AF13ΔnorA extract. Another compound with M+H = 371, identified only in the AF13ΔnorA extract, eluted at 15.6 min. Taken together, the observed alteration in the metabolic flux between the control and knockout transformants suggests the presence of other minor natural products and intermediates in the biosynthetic pathway to AFB1.
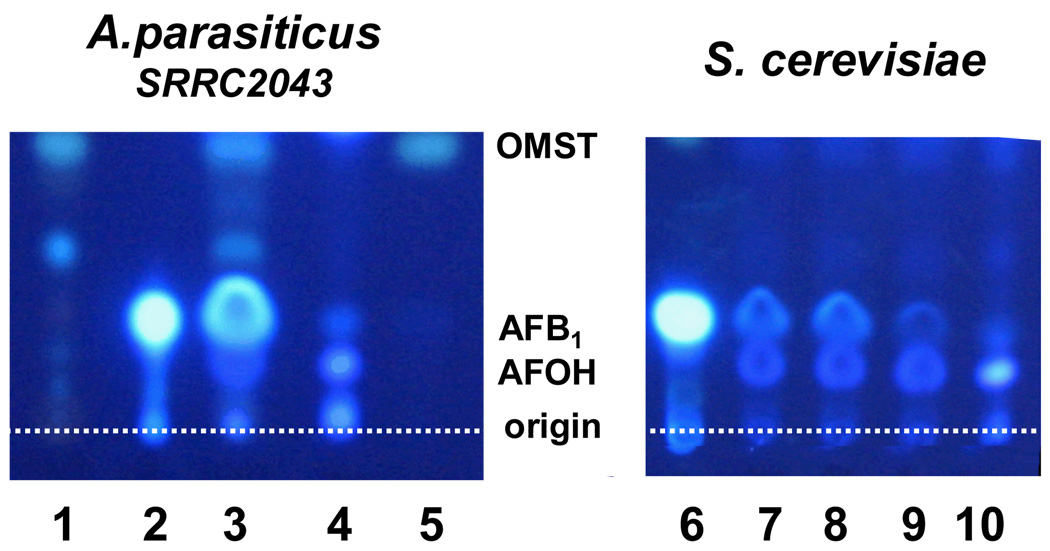
An ion with the expected mass, elution time, and chromophore for AFOH (314 Da, 10.3 min) was detected in extracts of a 2-day A. flavus norA knockout culture but not in the control culture extract. AFOH, after feeding to a strain of A. parasiticus with defective ordA but intact norA, was readily oxidized to AFB1 (Fig. 3, lane 3); deoxyAFB1 was not detected. Similarly, AFOH was oxidized to AFB1 by yeast cells whether or not they expressed norA or ordA (Fig. 3, lanes 7–9).
Fig. 3.
Feeding of AFOH to A. parasiticus SRRC2043 or S. cerevisiae: Lane 1, A. parasiticus SRRC2043 (accumulates OMST only) extract; Lanes 2 and 6, AFB1 standard; Lane 3, A. parasiticus SRRC2043 incubated with partially purified AFOH (AFOH is converted to AFB1); Lanes 4 and 10, partially purified synthetic AFOH; Lane 5, OMST standard; Lanes 7–9, AFOH fed to S. cerevisiae expressing norA, ordA, and non-transformed control, respectively. The dotted line indicates the TLC origin. Fluorescent materials at the top of the TLC plates are not shown.
Discussion
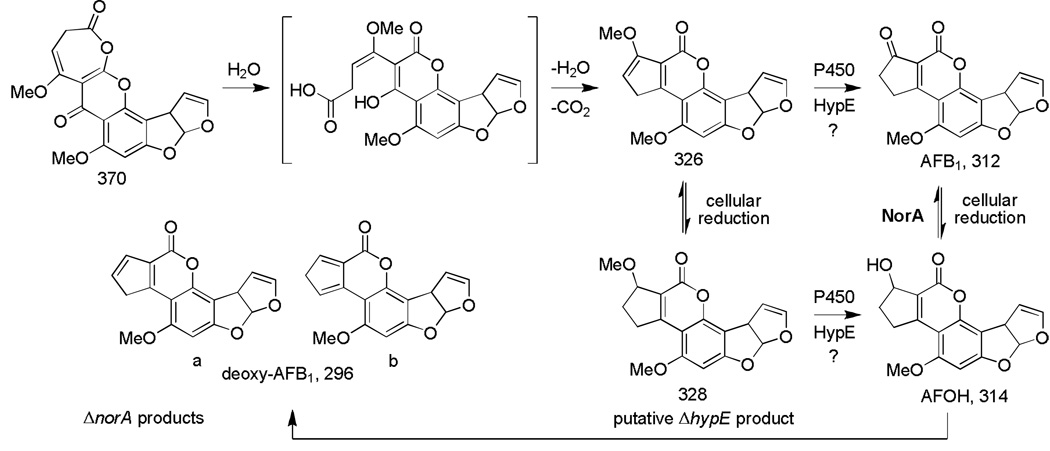
Orthologs of the aryl alcohol dehydrogenase-encoding gene norA are found in the gene clusters of all AF- and ST-producing Aspergillus species (Ehrlich, et al., 2005). The role of NorA in AF biosynthesis has not yet been defined. In previous studies, mutants of norA in A. parasiticus failed to show a detectable phenotype (Cary and Ehrlich, Chang and Ehrlich, unpublished data). Our results show that A. flavus lacking norA accumulate deoxyAFB1. This is the first time deoxyAFB1 has been shown to be a natural metabolite of AF-producing Aspergillus cultures. DeoxyAFB1 most likely results from dehydration of aflatoxicol (AFOH) as had been previously demonstrated in synthetic studies and confirmed here (Lau & Chu, 1983). AFOH is a natural enzymatic reduction product of AFB1. Therefore we suggest that A. flavus norA mutants lacking the aryl alcohol dehydrogenase accumulate an increased amount of the presumed NorA substrate AFOH, compared to cultures with intact norA, and that AFOH undergoes acid-catalyzed dehydration in the acidic growth medium to give deoxyAFB1 (Scheme 2).
Scheme 2.
Scheme showing possible intermediates in the conversion of the putative 370 Da lactone intermediate to AFB1 and deoxyAFB1. Molecular weights are shown below the compound.
The presence of AFB1 in AF13ΔnorA mutant extracts indicates that only a portion of AFB1 is reduced to AFOH in the absence of NorA, suggesting an oxidative role for NorA that minimizes accumulation of AFOH. This provides insight into the previously reported phenomenon that aflatoxin producers and non-producers are capable of interconverting AFB1 and AFOH (Nakazato, et al., 1990). The counterpart reductive enzymes involved in this oxidation-state balance as well as the underlying ecological rationale for the activity remain undefined. A BlastP search of the translated A. flavus genomic DNA database with the A. flavus NorA sequence revealed the presence of six genes predicted to encode proteins (AFLA_134080, E = 0; AFLA_077060, E = 0; AFLA_124600, E = -175; AFLA_096620, E = −107; AFLA_027250, E = −42; AFLA_093600, NorB, E = −44) with a high degree of homology (E value < −40). It is possible that these homologs could complement the function of NorA to some extent, even in the absence of NorB.
The LC/MS results show that the presumptive first OrdA oxidation product HOMST is present in the extracts of both AF13ΔnorA mutants and the AF13 control strain. HOMST was implicated as a potential intermediate in synthetic feeding studies with either A. parasiticus cultures or with yeast expressing ordA (Udwary, et al., 2002), and this intermediate was confirmed here in our product analysis. Our results indicate that NorA is involved in a catalytic step after OrdA oxidation and are consistent with the route proposed in Scheme 2 where OrdA is predicted to catalyze oxidation of HOMST to a putative 370 Da lactone.
The subsequent rearrangement steps of the presumptive 370 Da lactone are less clear. Ultimately, these are likely to result in formation of the 326 Da methyl enolether shown in Scheme 2, which is likely to be the immediate AFB1 precursor. Recent results suggest that the AF biosynthesis gene, hypE, encodes a protein with an EthD domain that may be involved in the oxidative demethylation of this methyl enolether (Holmes, 2008). Proteins with an EthD domain, previously only reported in bacteria, are required for oxidative ethyl-t-butyl ether degradation in the presence of a cytochrome P450 monooxygenase (Chauvaux, et al., 2001). Disruption of hypE in A. flavus led to accumulation of a compound with the intense blue fluorescence characteristic of deoxyAFB1 and aflatoxins, but which migrated faster than AFB1 on TLC. This new metabolite exhibited a mass of 328 Da, which is consistent with the methyl ether shown in Scheme 2. Oxidation of the methyl ether in either the 326 Da or 328 Da intermediates may occur with HypE and an unknown cytochrome P450 enzyme (possibly OrdA or CypX (AflV)) to cause loss of the methyl as formaldehyde and directly give AFB1 or AFOH, respectively. AFOH resulting from demethylation of the 328 Da ether would require NorA-catalyzed oxidation to AFB1.
In the absence of NorA, the 326 Da methyl enolether or AFB1 may be partially reduced to the 328 Da methyl ether in the reductive metabolic environment of the cell as shown in Scheme 2. As suggested previously, formation of increased quantities of deoxyAFB1 rather than AFOH in the absence of NorA could be a consequence of the precursor metabolites being produced and isolated under acidic culture conditions. In our studies, synthetic AFOH was found to dehydrate readily under mild acidic conditions. In the fungal cell, the pH is likely to be significantly higher and therefore, if AFOH is formed, it is unlikely that it would be subjected to acid-catalyzed dehydration. The balance in the cellular environment between oxidation and reduction as well as the availability of active transport out of the cell of AFB1 would be expected to play critical roles in determining the levels of the individual precursors and in maintaining the oxidation state of AFB1.
Because synthetic HOMST fed to yeast expressing ordA was converted to AFB1 [5–6], NorA is not required for formation of AFB1 by yeast or by the norA knockout mutants (Fig. 3). Conversion of AFB1 to AFOH was found by Nakazato, et al. when AFB1 was fed to strains of fungi incapable of toxin production (Nakazato, et al., 1990). NorA, therefore, may serve as a maintenance alcohol dehydrogenase to prevent derailment of AFB1 production. Our study suggests that, while conversion of OMST to AFB1 may only require a single cytochrome P450 monooxygenase, other enzymes are important to minimize derailment of AFB1 production.
Acknowledgements
We wish to thank Beverly Montalbano for early contributions to this work. The work at Southern Regional Research Center was supported by CRIS 6435-41420-004-P and at Johns Hopkins by U.S. National Institutes of Health grant ES001670 awarded to C.A.T. J.M.C. is currently a Damon Runyon Cancer Research Foundation Fellow (DRG-2002-09) in the Department of Biological Chemistry & Molecular Pharmacology, Harvard Medical School.
References
- Bhatnagar D, McCormick SP, Lee LS. Averufanin is an aflatoxin B1 precursor between averantin and averufin in the biosynthetic pathway. Appl. Environ Microbiol. 1987;53:14–16. doi: 10.1128/aem.53.1.14-16.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar D, McCormick SP, Lee LS, Hill RA. Identification of O-methylsterigmatocystin as an aflatoxin B1 and G1 precursor in Aspergillus parasiticus. Appl. Environ. Microbiol. 1987;53:1028–1033. doi: 10.1128/aem.53.5.1028-1033.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary JW, Wright M, Bhatnagar D, Lee R, Chu FS. Molecular characterization of an Aspergillus parasiticus gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl. Environ. Microbiol. 1996;62:360–366. doi: 10.1128/aem.62.2.360-366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P-K, Ehrlich KC, Linz JE, Bhatnagar D, Cleveland TE, Bennett JW. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet. 1996;30:68–75. doi: 10.1007/s002940050102. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Townsend CA. Evidence for the probable final steps in aflatoxin biosynthesis. J. Org.Chem. 1994;59:4424–4429. [Google Scholar]
- Chauvaux S, Chevalier F, Le Dantec C, Fayolle F, Miras I, Kunst F, Beguin P. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J Bacteriol. 2001;183:6551–6557. doi: 10.1128/JB.183.22.6551-6557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. J. Applied Microbiol. 2005;99:518–527. doi: 10.1111/j.1365-2672.2005.02637.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich KC, Chang P-K, Yu J, Cotty PJ. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 2004;70:6518–6524. doi: 10.1128/AEM.70.11.6518-6524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich KC, Scharfenstein LL, Montalbano BG, Chang P-K. Are the genes nadA and norB involved in formation of aflatoxin G1. Int. J. Mol. Sci. 2008;9:1717–1729. doi: 10.3390/ijms9091717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Larson TO, de Vries R, et al. Secondary metabolite profiling, growth profiles and other tools for species recognition and important Aspergillus mycotoxins. Stud Mycol. 2007;59:31–37. doi: 10.3114/sim.2007.59.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RA. PhD Thesis. Raleigh, NC: North Carolina State University; 2008. Characterization of an Aflatoxin Biosynthetic Gene and Resistance in Maize Seeds to Aspergillus flavus. [Google Scholar]
- Hsia MT, Chu FS. Reduction of aflatoxin B1 with zinc borohydride: an efficient preparation of aflatoxicol. Experientia. 1977;33:1132–1133. doi: 10.1007/BF01922282. [DOI] [PubMed] [Google Scholar]
- Lau HP, Chu FS. Preparation and characterization of acid dehydration products of aflatoxicol. J Assoc Off Anal Chem. 1983;66:98–101. [PubMed] [Google Scholar]
- Nakazato M, Morozumi S, Saito K, Fujinuma K, Nishima T, Kasai N. Interconversion of aflatoxin B1 and aflatoxicol by several fungi. Appl Environ Microbiol. 1990;56:1465–1470. doi: 10.1128/aem.56.5.1465-1470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto R, Yousibova GL, Woloshuk CP. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl. Environ. Microbiol. 1996;62:3567–3571. doi: 10.1128/aem.62.10.3567-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TJ, Dejesus AE, Steyn PS, Vleggaar R. Biosynthesis of Aflatoxins - incorporation of [2-H-2(3)] acetate into aflatoxin-B1 by Aspergillus flavus. J Chem Soc, Chem Commun. 1983:338–340. DOI: 10.1039/C39830000338. [Google Scholar]
- Singh R, Hsieh DP. Enzymatic conversion of sterigmatocystin into aflatoxin B1 by cell-free extracts of Aspergillus parasiticus. Appl. Environ. Microbiol. 1976;31:743–745. doi: 10.1128/aem.31.5.743-745.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Casillas LK, Townsend CA. Synthesis of 11-hydroxy-O-methylsterigmatocystin and the role of a cytochrome P-450 in the final step of aflatoxin biosynthesis. J. Am. Chem. Soc. 2002;124:5294–5303. doi: 10.1021/ja012185v. [DOI] [PubMed] [Google Scholar]
- Watanabe CM, Townsend CA. Incorporation of molecular oxygen in aflatoxin B1 biosynthesis. J. Org. Chem. 1996;61:1990–1993. [Google Scholar]
- Yu J, Chang P-K, Ehrlich KC, et al. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 1998;64:4834–4841. doi: 10.1128/aem.64.12.4834-4841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chang P-K, Ehrlich KC, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]